Sus_circPAPPA2 Regulates Fat Deposition in Castrated Pigs through the miR-2366/GK Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. RNA Isolation
2.3. CircRNA Sequencing Library Construction
2.4. CircRNA Identification and Differential Expression Analysis
2.5. Functional Enrichment Analysis
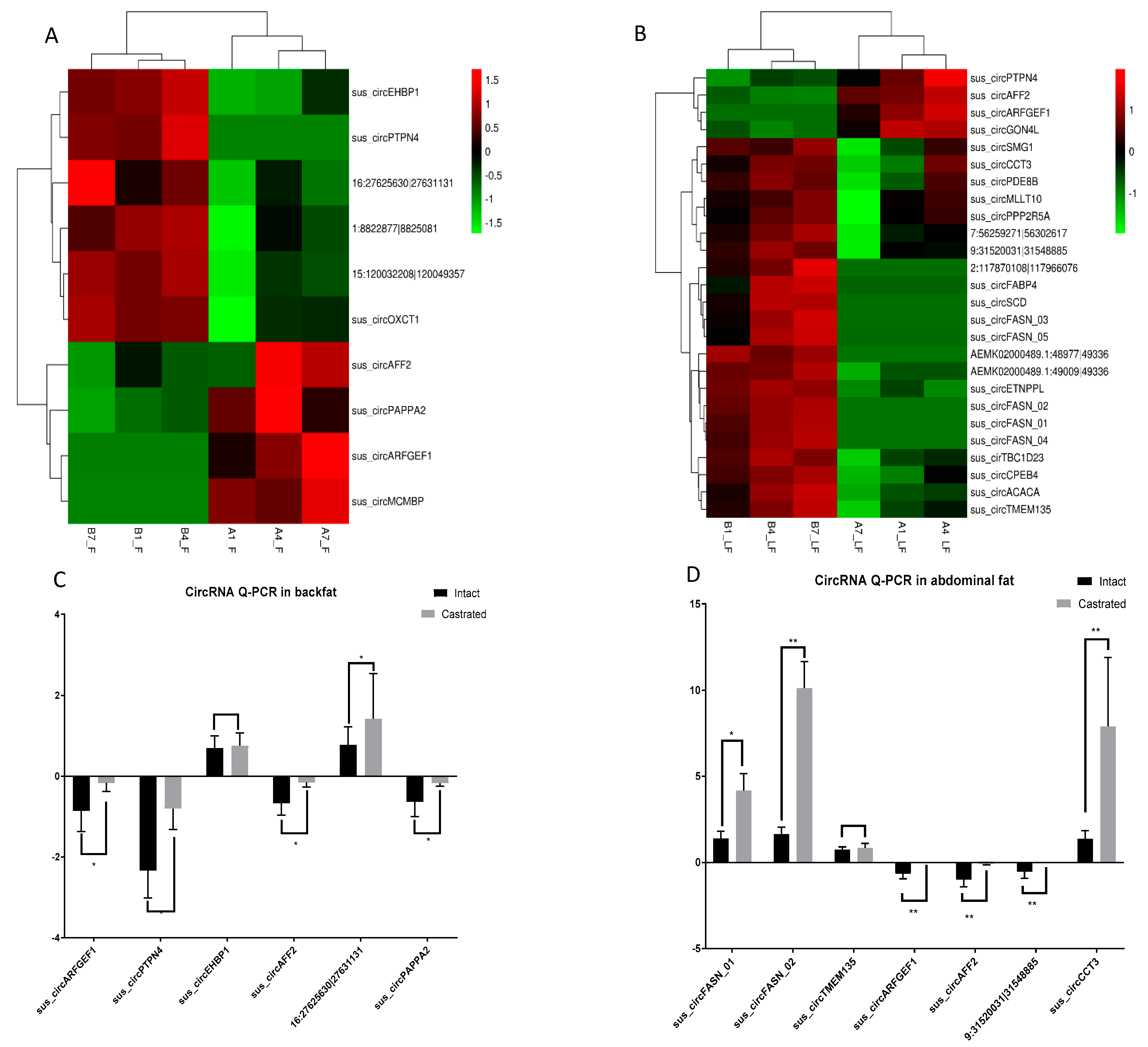
2.6. Validation of circRNAs by RT–qPCR
2.7. Vector Construction
2.8. Dual Luciferase Validation of miRNA Binding Sites in Target Genes
2.9. 293T Cell Culture and Transfection
2.10. Overexpression and Differentiation of Preadipocytes
2.11. Statistical Analysis
3. Results
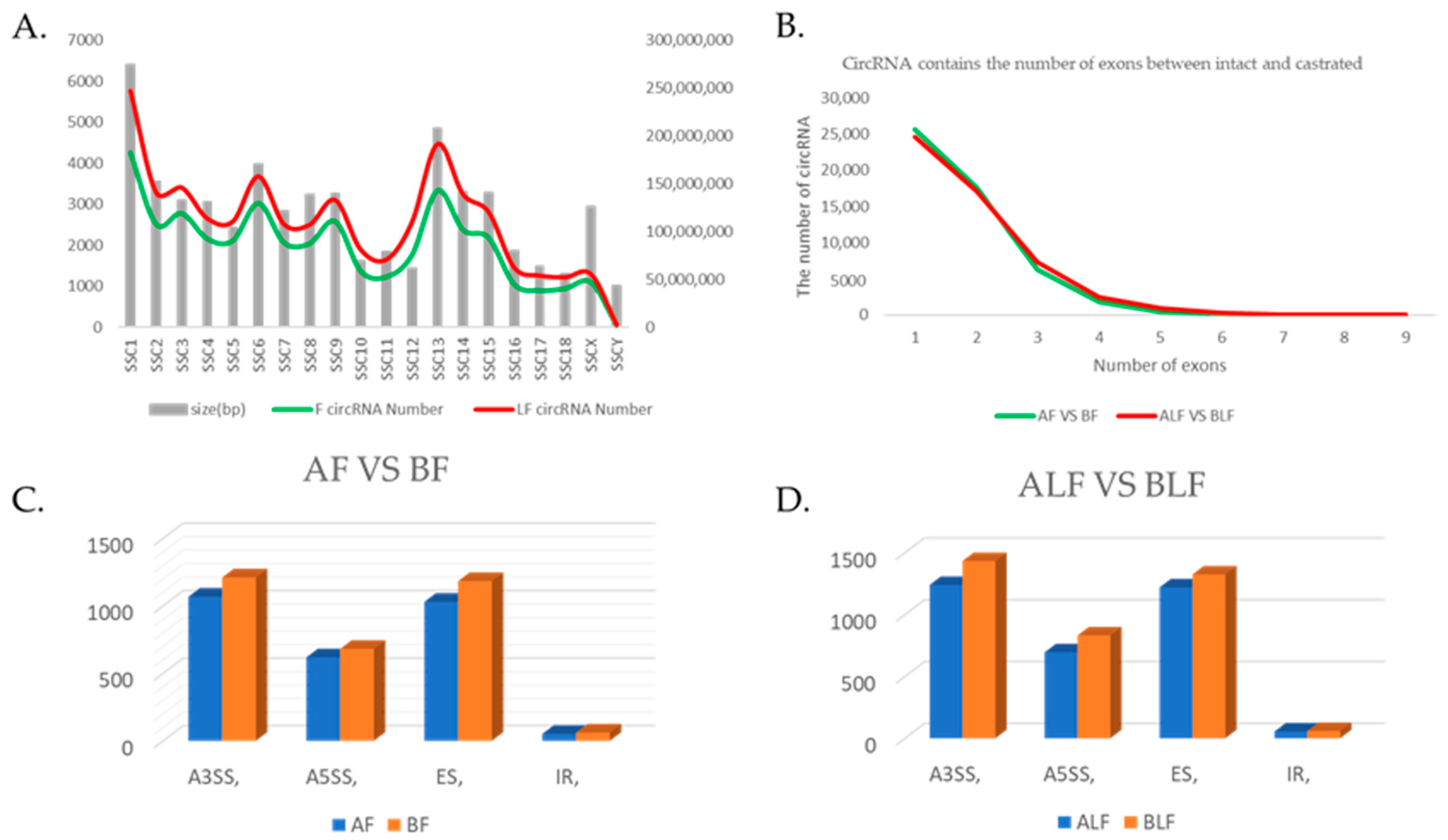
3.1. Overview of circRNA Expression Profiles
3.2. Differential circRNA Expression Analysis between Intact and Castrated Male Pigs
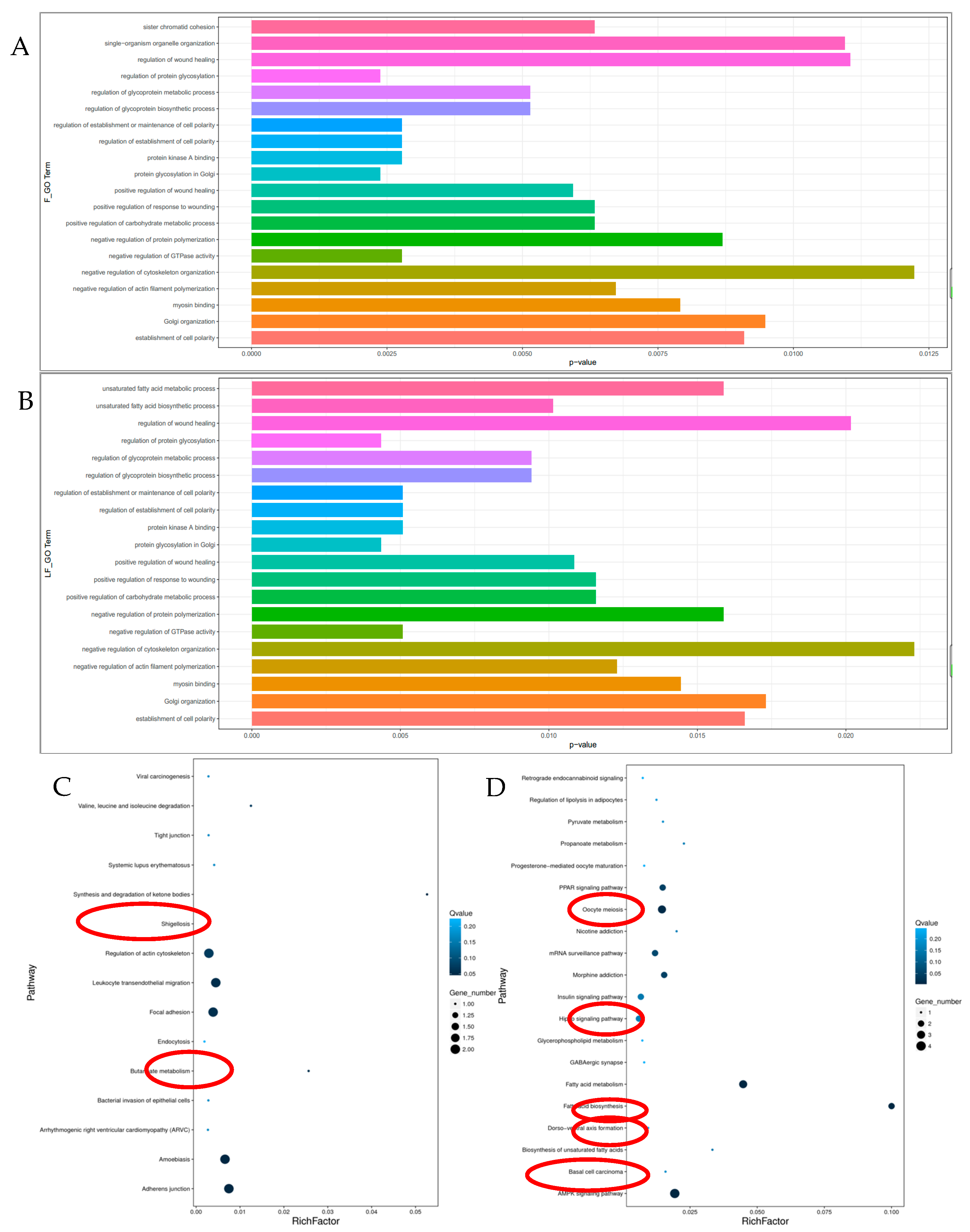
3.3. Functional Analyses of the Host Genes of Differentially Expressed circRNAs
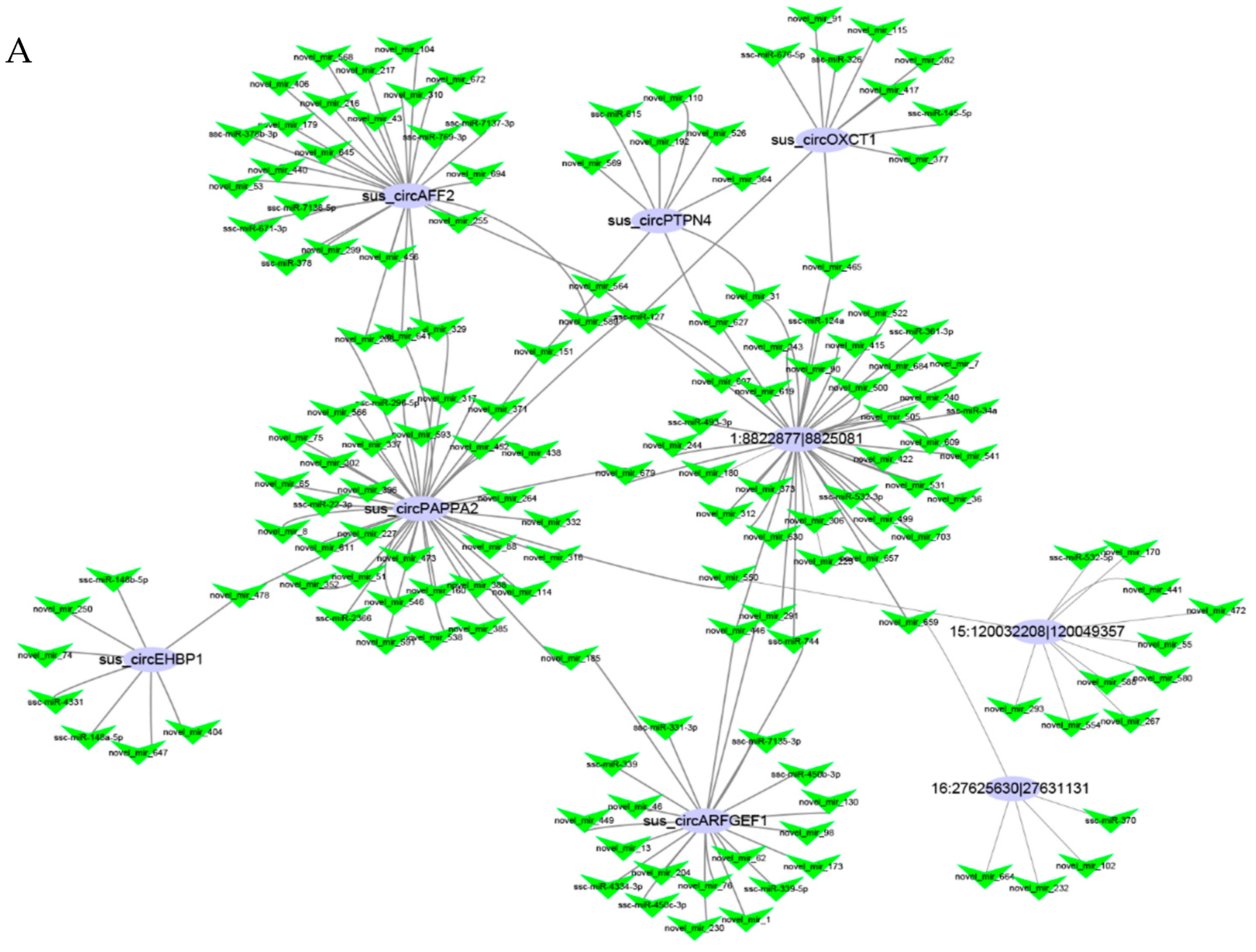
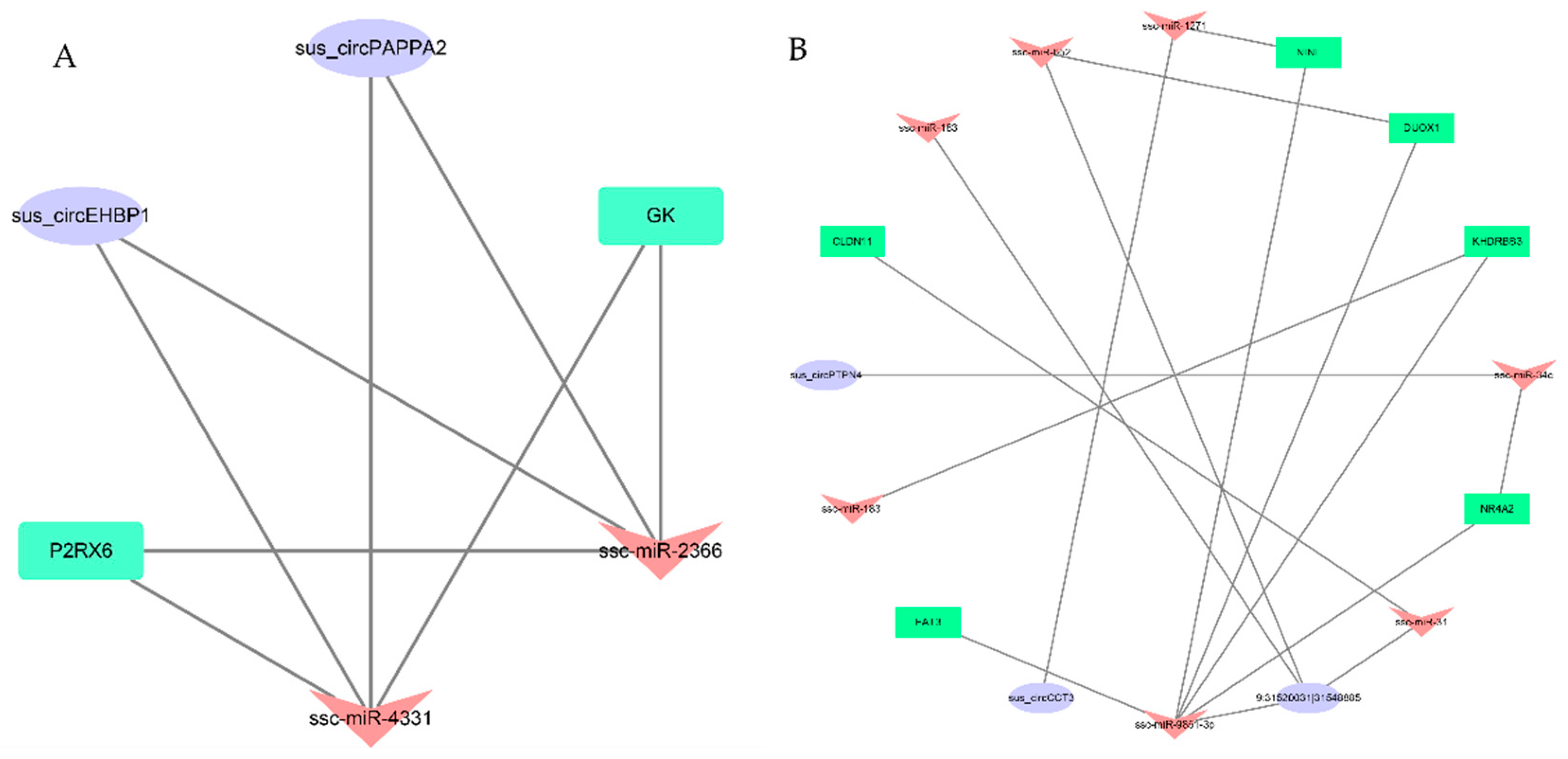
3.4. Networks and Pathway Prediction of Target miRNAs or Genes
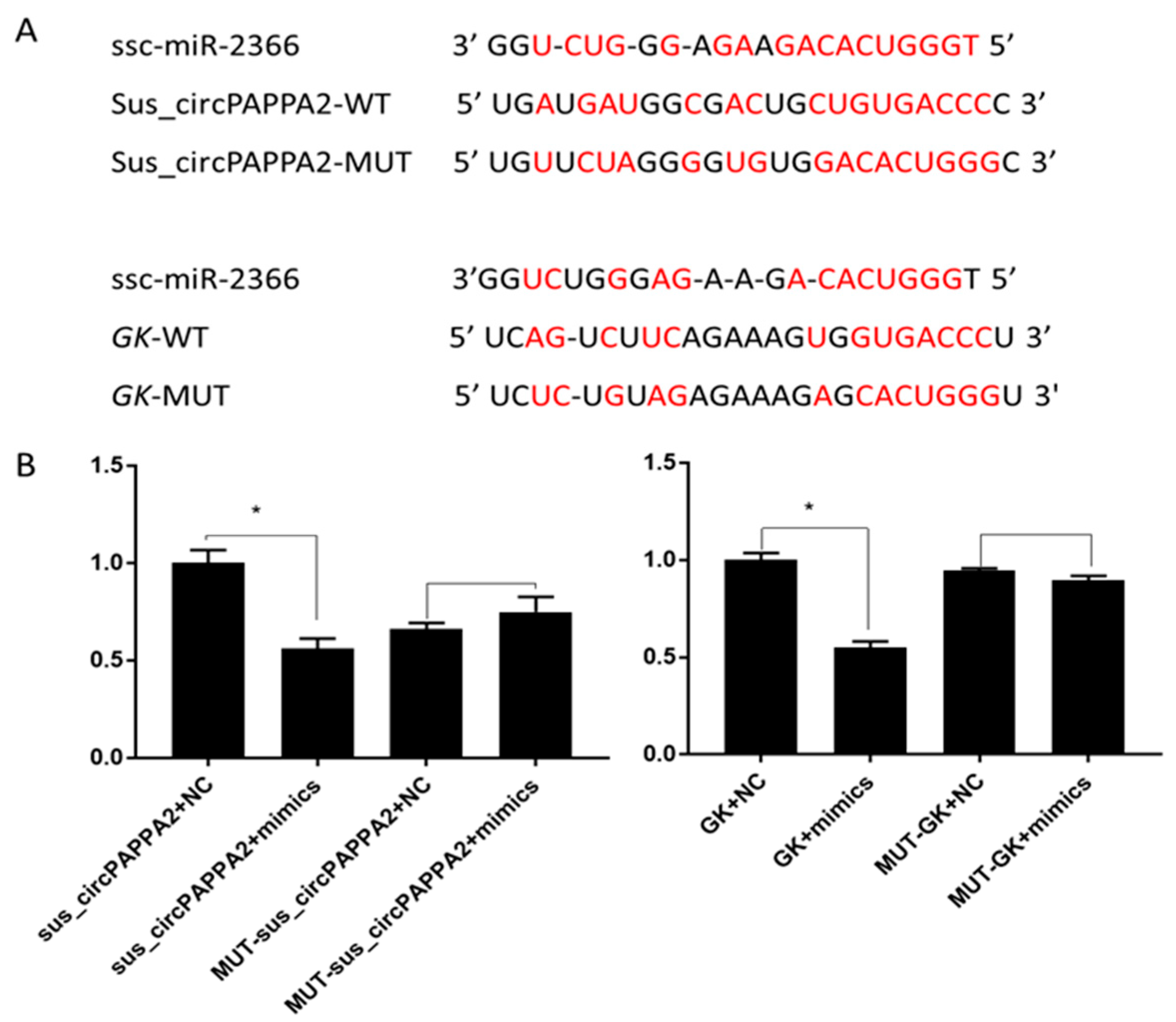
3.5. The Relationship among Sus_circPAPPA2, miR-2366 and GK Was Verified by Double Fluorescence
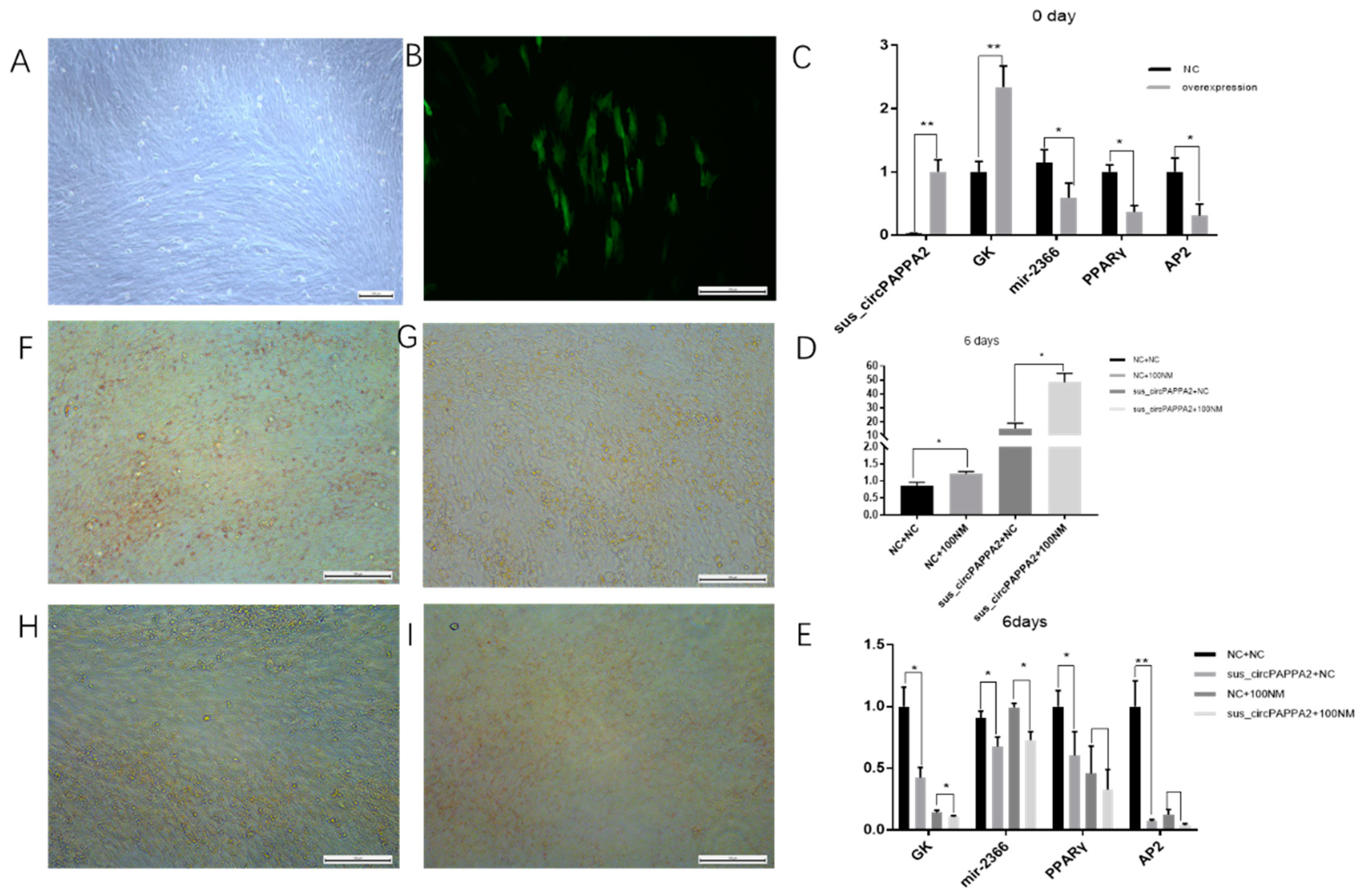
3.6. Sus_circPAPPA2 Inhibits Preadipocyte Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DE | Differential expression |
| FDR | false discovery rate |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PCR | polymerase chain reaction |
| RT-qPCR | real-time fluorescence quantification |
| Sus_circPAPPA2 | 9:118402663|118403734 |
| Sus_circEHBP1 | 3:78952890|78971884 |
| Sus_circAFF2 | X:120944619|120945464 |
| C-9-885 | 9:31520031|31548885 |
| sus_circOXCT1 | 16:26576176|26646919 |
| sus_circARFGEF1 | 4:67582515|67589305 |
| Sus_circPTPN4 | 15:31320424|31387416 |
| sus_circFASN_05 | 12:928972|929774 |
| sus_circFASN_01 | 12:929379|929774 |
| sus_circFASN_02 | 12:929837|932581 |
| sus_circFASN_03 | 12:932535|933265 |
| sus_circFASN_04 | 12:936096|936848 |
| sus_circTMEM135 | 9:20809498|20812474 |
References
- Christoffersen, B.O.; Gade, L.P.; Golozoubova, V.; Svendsen, O.; Raun, K. Influence of castration-induced testosterone and estradiol deficiency on obesity and glucose metabolism in male Gttingen minipigs. Steroids 2010, 75, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.M.; Jones, T.H. Testosterone and obesity. Obes. Rev. 2015, 16, 581–606. [Google Scholar] [CrossRef] [PubMed]
- Bekaert, M.; Van Nieuwenhove, Y.; Calders, P.; Cuvelier, C.A.; Batens, A.H.; Kaufman, J.M.; Ouwens, D.M.; Ruige, J.B. Determinants of testosterone levels in human male obesity. Endocrine 2015, 50, 202–211. [Google Scholar] [CrossRef]
- Xue, J.; Dial, G.D.; Pettigrew, J.E. Production: Performance, carcass, and meat quality advantages of boars over barrows: A literature review. J. Swine Health Prod. 1997, 5, 21–28. [Google Scholar]
- Bender, J.M.; See, M.T.; Hanson, D.J.; Lawrence, T.E.; Cassady, J.P. Correlated responses in growth, carcass, and meat quality traits to divergent selection for testosterone production in pigs. J. Anim. Sci. 2006, 84, 1331–1337. [Google Scholar] [CrossRef] [PubMed]
- Umapathy, E.; Makinde, M.O.; Mandisodza, K.T. Effect of castration on serum concentrations of gonadal hormones, insulin-like growth factor-I and its binding proteins in male pigs. Reprod. Nutr. Dev. 1997, 37, 541–549. [Google Scholar] [CrossRef][Green Version]
- Anderson, L.A.; Mcternan, P.G.; Barnett, A.H.; Sudhesh, K. The effects of androgens and estrogens on preadipocyte proliferation in human adipose tissue influence of gender and site. J. Clin. Endocrinol. Metab. 2001, 86, 5045–5051. [Google Scholar] [CrossRef]
- Escobar-Morreale, H.F.; Alvarez-Blasco, F.; Botella-Carretero, J.I.; Luque-Ramirez, M. The striking similarities in the metabolic associations of female androgen excess and male androgen deficiency. Hum. Reprod. 2014, 29, 2083–2091. [Google Scholar] [CrossRef]
- Blouin, K.; Boivin, A.; Tchernof, A. Androgens and body fat distribution. J. Steroid Biochem. Mol. Biol. 2008, 108, 272–280. [Google Scholar] [CrossRef]
- Kershaw, E.E.; Flier, J.S. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef]
- Jensen, M.D. Role of body fat distribution and the metabolic complications of obesity. J. Clin. Endocrinol. Metab. 2008, 93 (Suppl. S1), S57–S63. [Google Scholar] [CrossRef] [PubMed]
- Zerradi, M.; Dereumetz, J.; Boulet, M.M.; Tchernof, A. Androgens, body fat Distribution and Adipogenesis. Curr. Obes. Rep. 2014, 3, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, H.; Jin, L.; Li, G.; Hu, S.; Ning, C.; Guo, J.; Shuai, S.; Li, X.; Li, M. Long Noncoding RNA GAS5 Suppresses 3T3-L1 Cells Adipogenesis through miR-21a-5p/PTEN Signal Pathway. DNA Cell Biol. 2018, 37, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Santulli, G. microRNAs Distinctively Regulate Vascular Smooth Muscle and Endothelial Cells: Functional Implications in Angiogenesis, Atherosclerosis, and In-Stent Restenosis. Adv. Exp. Med. Biol. 2015, 887, 53–77. [Google Scholar]
- Xu, T.; Wu, J.; Han, P.; Zhao, Z.; Song, X. Circular RNA expression profiles and features in human tissues: A study using RNA-seq data. BMC Genom. 2017, 18 (Suppl. S6), 680. [Google Scholar] [CrossRef]
- Kaur, S.; Mirza, A.H.; Pociot, F. Cell Type-Selective Expression of Circular RNAs in Human Pancreatic Islets. Noncoding RNA 2018, 4, 38. [Google Scholar] [CrossRef]
- Li, A.; Huang, W.; Zhang, X.; Xie, L.; Miao, X. Identification and Characterization of CircRNAs of Two Pig Breeds as a New Biomarker in Metabolism-Related Diseases. Cell. Physiol. Biochem. 2018, 47, 2458–2470. [Google Scholar] [CrossRef]
- Liu, X.; Liu, K.; Shan, B.; Wei, S.; Li, D.; Han, H.; Wei, W.; Chen, J.; Liu, H.; Zhang, L. A genome-wide landscape of mRNAs, lncRNAs, and circRNAs during subcutaneous adipogenesis in pigs. J. Anim. Sci. Biotechnol. 2018, 9, 76. [Google Scholar] [CrossRef]
- Zhang, M.; Han, Y.; Zhai, Y.; Ma, X.; An, X.; Zhang, S.; Li, Z. Integrative analysis of circRNAs, miRNAs, and mRNAs profiles to reveal ceRNAs networks in chicken intramuscular and abdominal adipogenesis. BMC Genom. 2020, 21, 594. [Google Scholar] [CrossRef]
- Wang, L.; Liang, W.; Wang, S.; Wang, Z.; Bai, H.; Jiang, Y.; Bi, Y.; Chen, G.; Chang, G. Circular RNA expression profiling reveals that circ-PLXNA1 functions in duck adipocyte differentiation. PloS ONE 2020, 15, e0236069. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, J.; Zheng, Q.; Wang, S.; Wei, X.; Li, F.; Shang, J.; Lei, C.; Ma, Y. Characterization of Circular RNAs in Chinese Buffalo (Bubalus bubalis) Adipose Tissue A Focus on Circular RNAs Involved in Fat Deposition. Animals 2019, 9, 403. [Google Scholar] [CrossRef] [PubMed]
- Ebbesen, K.K.; Kjems, J.; Hansen, T.B. Circular RNAs: Identification, biogenesis and function. Biochim. Biophys. Acta 2016, 1859, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, J.; Wei, X.; Song, C.; Dong, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Ma, Y. CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J. Cell. Physiol. 2017, 233, 4643–4651. [Google Scholar] [CrossRef]
- Wang, L.; Yi, J.; Lu, L.-Y.; Zhang, Y.-Y.; Wang, L.; Hu, G.-S.; Liu, Y.-C.; Ding, J.-C.; Shen, H.-F.; Zhao, F.-Q.; et al. Estrogen-induced circRNA, circPGR, functions as a ceRNA to promote estrogen receptor-positive breast cancer cell growth by regulating cell cycle-related genes. Theranostics 2021, 11, 1732–1752. [Google Scholar] [CrossRef]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.-M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. J. Bioinform. 2009, 25, 1966–1967. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zheng, Y.; Zhang, J.; Chen, S.; Zhao, F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun. 2016, 7, 12060. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Traish, A.M.; Guay, A.; Feeley, R.; Saad, F. The dark side of testosterone deficiency: I. Metabolic syndrome and erectile dysfunction. J. Androl. 2009, 30, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Vishnu, P.G.; Punyakumari, B.; Ekambaram, B.; Prakash, M.G.; Subramanyam, B.V. Chromosomal profile of indigenous pig (Sus scrofa). Vet. World 2015, 8, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Rink, A.; Santschi, E.M.; Eyer, K.M.; Roelofs, B.; Hess, M.; Godfrey, M.; Karajusuf, E.K.; Yerle, M.; Milan, D.; Beattie, C.W. A first-generation EST RH comparative map of the porcine and human genome. Mamm. Genome 2002, 13, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.P.; He, Y.J.; Hou, J.C.; Chen, X.; Zhou, S.Y.; Yang, S.J.; Li, J.; Zhang, H.D.; Hu, J.H.; Zhong, S.L.; et al. The role of circRNAs in cancers. Biosci. Rep. 2017, 37, BSR20170750. [Google Scholar] [CrossRef]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, T.; Xiao, J. Circular RNAs Promising Biomarkers for Human Diseases. EBioMedicine 2018, 34, 267–274. [Google Scholar] [CrossRef]
- Svartberg, J.; Agledahl, I.; Figenschau, Y.; Sildnes, T.; Waterloo, K.; Jorde, R. Testosterone treatment in elderly men with subnormal testosterone levels improves body composition and BMD in the hip. Int. J. Impot. Res. 2008, 20, 378–387. [Google Scholar] [CrossRef]
- Allan, C.A.; Strauss, B.J.; Burger, H.G.; Forbes, E.A.; McLachlan, R.I. Testosterone therapy prevents gain in visceral adipose tissue and loss of skeletal muscle in nonobese aging men. J. Clin. Endocrinol. Metab. 2008, 93, 139–146. [Google Scholar] [CrossRef]
- Ibrahim, M.M. Subcutaneous and visceral adipose tissue structural and functional differences. Obes. Rev. 2010, 11, 11–18. [Google Scholar] [CrossRef]
- Zeng, J.; Zhou, S.W.; Zhao, J.; Jin, M.H.; Kang, D.J.; Yang, Y.X.; Wang, X.L.; Chen, Y.L. Role of OXCT1 in ovine adipose and preadipocyte differentiation. Biochem. Biophys. Res. Commun. 2019, 512, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Al Batran, R.; Gopal, K.; Capozzi, M.E.; Chahade, J.J.; Saleme, B.; Tabatabaei-Dakhili, S.A.; Greenwell, A.A.; Niu, J.; Almutairi, M.; Byrne, N.J.; et al. Pimozide Alleviates Hyperglycemia in Diet-Induced Obesity by Inhibiting Skeletal Muscle Ketone Oxidation. Cell Metab. 2020, 31, 909–919.e908. [Google Scholar] [CrossRef]
- Hesse, D.; Jaschke, A.; Chung, B.; Schurmann, A. Trans-Golgi proteins participate in the control of lipid droplet and chylomicron formation. Biosci. Rep. 2013, 33, e00001. [Google Scholar] [CrossRef] [PubMed]
- Takanabe, R.; Ono, K.; Abe, Y.; Takaya, T.; Horie, T.; Wada, H.; Kita, T.; Satoh, N.; Shimatsu, A.; Hasegawa, K. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem. Biophys. Res. Commun. 2008, 376, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Fang, S.; Chen, Y.; Yang, Z.; Yuan, Y.; Zhang, J.; Ye, L.; Gu, W. Inhibition of FASN suppresses the malignant biological behavior of non-small cell lung cancer cells via deregulating glucose metabolism and AKT/ERK pathway. Lipids Health Dis. 2019, 18, 118. [Google Scholar] [CrossRef]
- Hale, C.; Lloyd, D.J.; Pellacani, A.; Veniant, M.M. Molecular targeting of the GK-GKRP pathway in diabetes. Expert Opin. Ther. Targets 2015, 19, 129–139. [Google Scholar] [CrossRef]
- Meng, Y.; Cui, Y.; Zhang, W.; Fu, S.; Huang, L.; Dong, H.; Du, H. Integrative Analysis of Genome and Expression Profile Data Reveals the Genetic Mechanism of the Diabetic Pathogenesis in Goto Kakizaki (GK) Rats. Front. Genet. 2018, 9, 724. [Google Scholar] [CrossRef]
- Zhao, X.; Bai, X.; Guan, L.; Li, J.; Song, X.; Ma, X.; Guo, J.; Zhang, Z.; Du, Q.; Huang, Y.; et al. microRNA-4331 Promotes Transmissible Gastroenteritis Virus (TGEV)-induced Mitochondrial Damage via Targeting RB1, Upregulating Interleukin-1 Receptor Accessory Protein (IL1RAP), and Activating p38 MAPK Pathway In Vitro. Mol. Cell Proteom. 2018, 17, 190–204. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- McInnes, K.J.; Smith, L.B.; Hunger, N.I.; Saunders, P.T.; Andrew, R.; Walker, B.R. Deletion of the androgen receptor in adipose tissue in male mice elevates retinol binding protein 4 and reveals independent effects on visceral fat mass and on glucose homeostasis. Diabetes 2012, 61, 1072–1081. [Google Scholar] [CrossRef]
- De Vera, I.M.; Giri, P.K.; Munoz-Tello, P.; Brust, R.; Fuhrmann, J.; Matta-Camacho, E.; Shang, J.; Campbell, S.; Wilson, H.D.; Granados, J.; et al. Identification of a Binding Site for Unsaturated Fatty Acids in the Orphan Nuclear Receptor Nurr1. ACS Chem. Biol. 2016, 11, 1795–1799. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Benoit, G.; Liu, J.; Prasad, S.; Aarnisalo, P.J. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 2003, 423, 555–560. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Bai, Y.; Cui, R.; He, S.; Zhao, X.; Wu, K.; Fang, M. Sus_circPAPPA2 Regulates Fat Deposition in Castrated Pigs through the miR-2366/GK Pathway. Biomolecules 2022, 12, 753. https://doi.org/10.3390/biom12060753
Liu X, Bai Y, Cui R, He S, Zhao X, Wu K, Fang M. Sus_circPAPPA2 Regulates Fat Deposition in Castrated Pigs through the miR-2366/GK Pathway. Biomolecules. 2022; 12(6):753. https://doi.org/10.3390/biom12060753
Chicago/Turabian StyleLiu, Ximing, Ying Bai, Ran Cui, Shuaihan He, Xingbo Zhao, Keliang Wu, and Meiying Fang. 2022. "Sus_circPAPPA2 Regulates Fat Deposition in Castrated Pigs through the miR-2366/GK Pathway" Biomolecules 12, no. 6: 753. https://doi.org/10.3390/biom12060753
APA StyleLiu, X., Bai, Y., Cui, R., He, S., Zhao, X., Wu, K., & Fang, M. (2022). Sus_circPAPPA2 Regulates Fat Deposition in Castrated Pigs through the miR-2366/GK Pathway. Biomolecules, 12(6), 753. https://doi.org/10.3390/biom12060753





