Abstract
Over two decades of studies on small noncoding RNA molecules illustrate the significance of microRNAs (miRNAs/miRs) in controlling multiple physiological and pathological functions through post-transcriptional and spatiotemporal gene expression. Among the plethora of miRs that are essential during animal embryonic development, in this review, we elaborate the indispensable role of the miR-200 family (comprising miR-200a, -200b, 200c, -141, and -429) in governing the cellular functions associated with epithelial homeostasis, such as epithelial differentiation and neurogenesis. Additionally, in pathological contexts, miR-200 family members are primarily involved in tumor-suppressive roles, including the reversal of the cancer-associated epithelial–mesenchymal transition dedifferentiation process, and are dysregulated during organ fibrosis. Moreover, recent eminent studies have elucidated the crucial roles of miR-200s in the pathophysiology of multiple neurodegenerative diseases and tissue fibrosis. Lastly, we summarize the key studies that have recognized the potential use of miR-200 members as biomarkers for the diagnosis and prognosis of cancers, elaborating the application of these small biomolecules in aiding early cancer detection and intervention.
1. Introduction
In classical molecular biology, the flow of genetic information from DNA to messenger RNA (mRNA) and thereafter to protein is regarded as the “central dogma” of life. However, the fraction of protein-coding genes accounts for merely ~3% of the human genome, whereas almost 97% of the genome is non-protein coding, which remained puzzling for several years. Until the early 1970s, this bulk of genetic material was largely considered to be functionally inactive and even termed as “junk DNA” [1]. However, with the advent of systemic whole-genomic sequencing analyses and extensive genome-wide association studies, thousands of “silent” genetic elements such as cis/trans-gene-regulatory elements, introns, repetitive sequences, transposable elements, telomeres, and pseudogenes were identified and functionally characterized within the so-called “junk DNA”. Such non-protein-coding, gene regulatory elements were attributed as one of the paramount discoveries in genomics ever since the decoding of the DNA structure in 1953 [2,3,4]. Further exploration of these non-protein-coding segments of the genome showed that a significant portion of these genes are transcribed as non-coding RNA (ncRNA), which are capable of influencing multiple and highly specific biochemical and molecular processes such as chromatin accessibility, RNA splicing, and transcription and/or protein translation-rate determination. Besides the fundamental ncRNAs (transfer RNA (tRNA) and ribosomal RNA (rRNA)) that are integral to the central dogma, eukaryotic genomes encode for equally important classes of ncRNAs such as microRNAs (miRNA/miR), small interfering RNAs (siRNA), small nuclear RNAs (snRNA), Piwi-interacting RNAs (piRNA), and long non-coding RNAs (lncRNA).
MiRNAs/miRs are a prominent subclass of single-stranded ncRNAs, whose transcripts are about 19–22 nucleotides in length. MiRNAs bind to the 3′ untranslated region (3′UTR) of target mRNAs to elicit mRNA degradation and/or protein translational repression [5]. The most salient feature of miRNA–mRNA targeting is the presence of a 7 nt “seed” region (nucleotides 2–8) in every miRNA that exhibit sequence complementarity to its target mRNA, among which the complementarity of the seed sequence is absolutely crucial for target specificity [6]. According to the recent release, the miRBase database (http://mirbase.org/, accessed on 1 June 2022, exclusive public repository for miRNA sequences and annotation) contains over 38,000 precursors and 48,000 mature miRNA entries from 271 organisms [7]. RNA polymerase II transcribes miRs in the nucleus to form pri-miR transcripts. Next, the RNase III enzyme Drosha processes pre-miRNA transcripts into ~70-nucleotide pre-miR with imperfect stem-loop structures, which is exported into the cytoplasm. Subsequently, Dicer (another RNase III enzyme) processes the pre-miRNA and generates a transient ~22-nucleotide miRNA: miRNA* duplex molecule. The miRNA: miRNA* duplex is incorporated into the miRISC complex, which includes the Argonaute proteins and the RNA-binding protein TRBP. The mature miR strand is preferentially retained in the functional miRISC complex. The miRISC complex finally delivers the mature miR to its target mRNA and mediates the site-specific cleavage and degradation of the mRNA or inhibits its translation [8]. In addition to the canonical miRNA targeting mechanism, recent small RNA deep-sequencing data reveal the presence of differential and/or the distinct subcellular localization of miRNAs to execute unconventional functions in cellular homeostasis [9,10,11,12]. Based on the large volume of studies investigating miRs, it is fair to presume that miRs have equally indispensable roles during development and in a variety of human diseases (Figure 1).
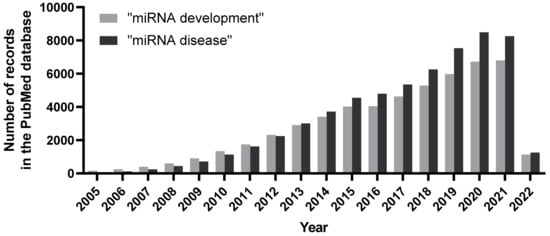
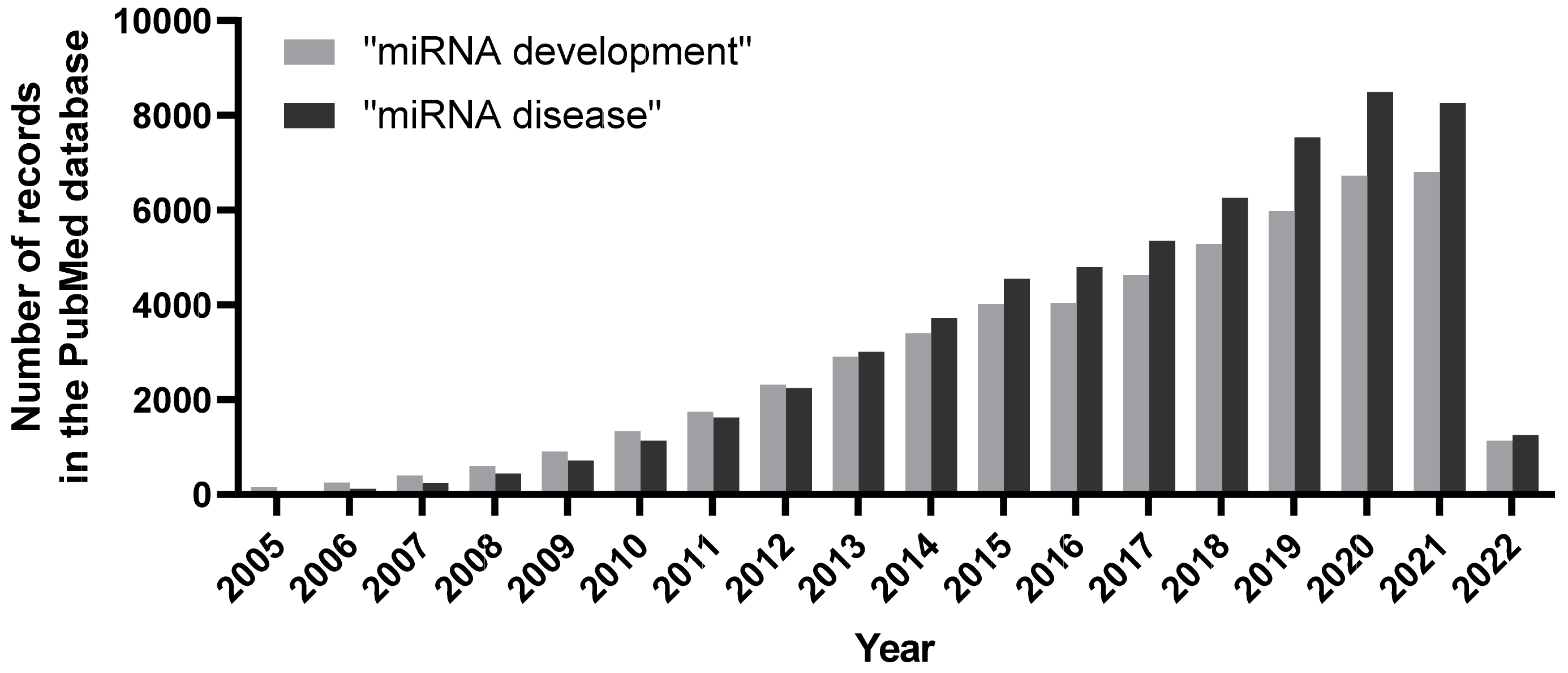
Figure 1.
Number of publications per year associated with search terms “miRNA development” and “miRNA disease” in the PubMed database. An increasing trend of publications unearthing miRNAs and their roles during organismal development, as well as during pathological scenarios, clearly denote their vital biological implications.
The human genome encodes for more than 2500 mature miRNAs that exert their functions in during several physiological processes such as embryogenesis, muscle differentiation, and stem cell division, as well as in numerous pathological conditions such as cardiovascular disease, autoimmune disease, and cancer [7,13]. As far as the role of miRNAs in cancer is concerned, these molecules can variably function either as tumor suppressors or as oncogenes. As tumor suppressors, miRNAs regulate the expression of several oncogenic factors. For example, miR-15a and miR-16-1 post-transcriptionally downregulate B cell lymphoma 2 (BCL2), an anti-apoptotic gene that is often overexpressed in different types of leukemia and lymphomas [14]. Significant reductions of miR-143 and miR-145 are observed in colorectal cancers, denoting that the expression of certain miRNAs is abrogated during carcinogenesis [15]. As oncogenes, the aberrant activation of certain miRNAs could directly or indirectly lead to cancer progression. The mitogen-activated protein kinase (MAPK) signaling-mediated overexpression of miR-21 induces tumorigenesis- and metastasis-associated phenotypes, including cell migration and invasion in lung cancer [16], EGF-induced pancreatic cancer [17], and Her2/neu-overexpressing breast cancer [18]. Mir-21 expression is associated with cancer stem cell properties and is involved in stemness maintenance in multiple cancers, including pancreatic ductal adenocarcinoma cells [19,20,21,22]. In addition, miR-21 contributes to drug resistance in head and neck squamous cell carcinoma [23], breast cancer [24], acute myeloid leukemia [25], ovarian cancer [26], and colon adenocarcinoma [27]. The abundant expression of circulating miRNAs such as miR-629*, miR-660, and miR-141 were detected in the plasma of prostate cancer xenograft model system, leading to the potential use of circulating miRNAs as blood-based biomarkers of human cancer detection [28]. Furthermore, miRNA-based clinical therapy has made significant progress over the past decade. MiR-16-based nanoparticle-encapsulated microRNA mimics have been employed in phase I trials in recurrent malignant pleural mesothelioma patients [29]. Another prospective phase II clinical trial encompassing the stratification of chemo-refractory metastatic colorectal cancer patients with low miR-31-3p positively correlated with clinical benefits from anti-EGFR monoclonal antibodies [30]. Hence, in this special issue focusing on “MicroRNAs—Small Molecules with Great Potential in Tumorigenesis”, we intend to focus on a specific family of miRNAs that is indispensable for animal development and its deregulation among the pathogenesis of various diseases: the miR-200 family.
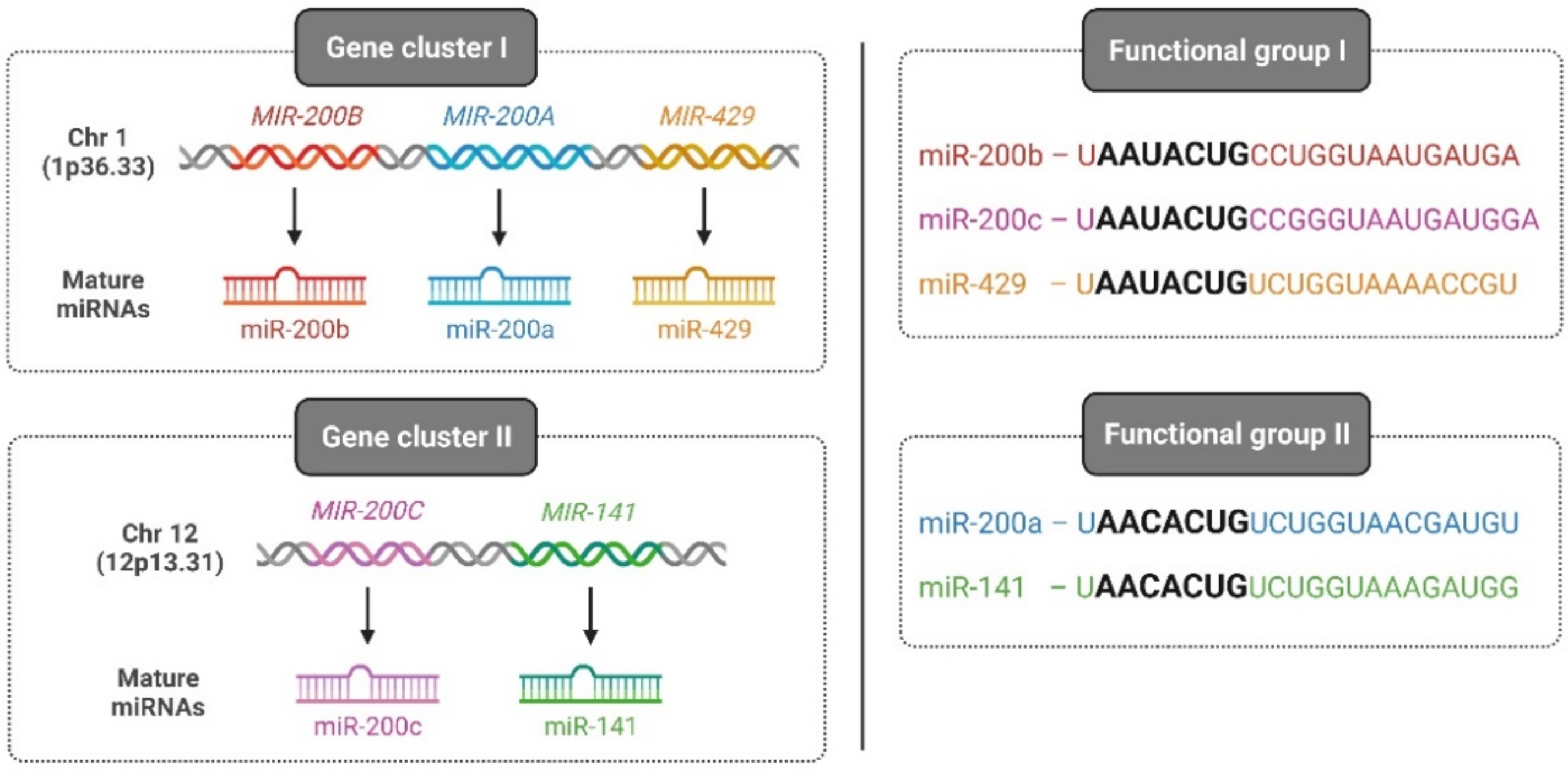
Among vertebrates, the miR-200 family is one of the most-conserved miRNAs and consists of five members: miR-200a, miR-200b, miR-200c, miR-141, and miR-429. On one hand, based on its chromosomal location, the miR-200 family is divided into two clusters: cluster I, which is located on human chromosome 1 (1p36.33) encoding for miR-200a, -200b, and -429, and cluster II, which is located on chromosome 12 (12p13.31) encoding for miR-141 and -200c. On the other hand, based on the seed sequence that binds to target genes, the miR-200 family is classified into two groups: group I, comprising miR-200b, -200c, and -429 with a seed sequence: 5′-AAUACUG-3′, and group II, consisting of miR-200a and -141 with a seed sequence: 5′-AACACUG-3′ (Figure 2).
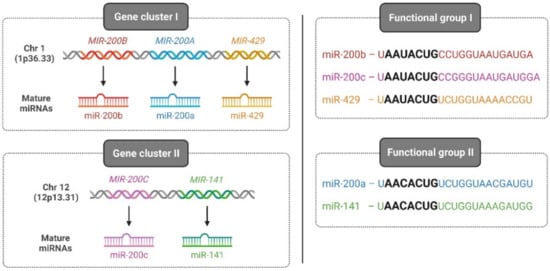
Figure 2.
Classification of the human miR-200 family. Two categories of the human miR-200 family classification exist based on the chromosomal location of the gene (left—gene cluster I and II) and based on seed sequences that bind to 3′ UTR of cognate genes (right—functional group I and II). Illustration created with Biorender.com (https://biorender.com/, accessed on 1 June 2022).
It is also speculated that members of the same miRNA family are segregated in two different locations on the genome, rendering miR-200s with more flexibility in imposing spatial, temporal, and tissue-specific control of target gene expression, since the expression of miR-200s is also subjected to direct regulation through histone modification [31,32]. Nevertheless, several exogenous stimuli and signaling cascades activate miR-200 members to mediate key cellular functions, which we will elaborate upon in the coming sections. Similarly, the deregulation of miR-200s is frequently observed in multiple pathological conditions such as tissue fibrosis, neurodegenerative diseases, and cancer, which are discussed in the later part of the article.
2. miR-200 Family in Development
The formation and establishment of two distinct layers of tissue, epithelium and mesenchyme, demarcate the basis of organ development and coordinated multicellularity in metazoans. The epithelium is characterized by a layer of cells adjacently linked through interconnecting cellular structures such as adherens junctions, tight junctions, desmosomes, and gap junctions. The obligate roles of each of these structures in epithelial cellular organization, adhesion, and selective permeability are extensively discussed in recent reviews [33,34,35,36]. In addition, epithelial cells display characteristic apical–basolateral cellular polarity (the apical pole facing the outer lumen and the basal pole facing the basement extracellular matrix), which is crucial for the asymmetric localization of proteins at distinct membrane domains and the orientation of microtubule networks during intra-cellular trafficking [37,38]. During early embryonic events such as embryonic implantation, gastrulation, and neural crest formation, a subset of epithelial cells from the epithelial layer of the embryo systematically loses its epithelial features through a process called epithelial-to-mesenchymal transition (EMT), thereby acquiring a mesenchymal phenotype. Unlike epithelial cells, mesenchymal cells show weakened cell–cell adhesion properties and lack apical–basal cellular polarity, resulting in cells with enhanced migratory behavior as well as the potential to degrade the underlying extracellular matrix. The salient bifurcation of these two contrasting cellular types during early embryogenesis is vital for successive vertebral column development and organogenesis [39]. Recently, studies focusing on the role of miRNAs during early embryogenesis have identified that the miR-200 family plays essential roles in the establishment and functioning of the epithelial phenotype during early embryogenesis and organogenesis that are detailed below, and a summary of specific miR-200 family members, their target genes, and the regulated function is listed in Table 1.

Table 1.
miR-200s, target genes, and their roles in development.
In order to establish and safeguard epithelial cell identity during development, members of the miR-200 family deploy a multi-pronged approach, depending on the cellular context. In certain tissues, miR-200s function as potent repressors of the EMT process. For example, during neural crest cell migration, miR-200c and miR-145 target Sox-1 and Sox-9, respectively, to upregulate E-cadherin expression and suppress EMT [42]. miR-200s target ZEB1 during the differentiation of human embryonic stem cells into hepatocytes and target ZEB2 in order to promote the late steps of postnatal forebrain neurogenesis [46,47]. Similarly, miR-200s’ mediated repression of ZEB1, ZEB2, and components of the Indian hedgehog signaling such as PTCH1, GLI2, and GLI3 is crucial during the endometrial development of embryo implantation [48]. In addition to directly curbing EMT, miR-200s suppress EMT-associated stem cell and self-renewal properties, which are crucial for the development and maintenance of unspecialized cellular niches. By directly targeting the transcription of stem cell self-renewal factor BMI1, miR-200c overexpressing murine mammary cells, when injected into the mammary fat pad of female mice, displayed aberrant, disorganized clusters of cells with non-functional mammary duct formation, potentially initiating myoepithelial-cell rather than luminal-cell differentiation [43]. Similarly, in the mesenchymal derivative of the breast epithelial cell line D492, the expression of miR-200c-141 reversed the EMT phenotype, and the co-expression of miR-200c-141 and ΔNp63 (a transcription factor crucial for stem-cell maintenance) restored epithelial differentiation and branching morphogenesis [50].
Recently, a few studies have demonstrated the unique role of miR-200s during the development of mammalian skin, which encompasses skin and hair follicle development and hair morphogenesis. The quantitative miRNA sequencing analysis of mouse neonatal skin has identified that the miR-200 family is among the most abundantly expressed miRNAs during embryonic skin development [51]. Mouse embryonic stem cells with activated sonic hedgehog signaling decreased miR-200s and activated the nuclear expression of ZEB1/ZEB2, leading to enhanced migration and skin wound healing [52]. Additionally, using double-end sequencing of cashmere goats during fetal periods, the expression of miR-200s was upregulated in pregnancy samples from day 55 and 66 when compared to pregnancy samples from day 45, underlining the significance of miR-200s in hair follicle development [53].
The murine dental epithelial stem cell niche is solely responsible for maintaining the stem cell population through self-renewal, while the spatiotemporal elevated expression of transcription factors such as Sox2, Bmi1, and Pitx2 stimulate the differentiation of dental epithelial cells that make up the lower incisor [54,55]. Therefore, the maintenance and differentiation of the lower incisor epithelial population offers an ideal model system to investigate the underlying role of miR-200s in epithelial stem cell renewal and differentiation. Along this line, recent histological and RNA sequencing analyses of murine dental epithelial stem cells show that miR-200 expression is required for the differentiation of terminally located progenitor cells and ameloblasts, as well as during the maintenance of the epithelial dental stem cell niche [56,57,58]. Furthermore, embryos depleted of miR-200c show defects in oral epithelial tissue invagination, leading to a reduction in incisor tissue composition and length. Using a conditional knockout system of dental stem cells and ameloblasts (cells that generate enamel), Cao et al. have shown that Pitx2 directly activates the expression of the miR-200c/141 cluster and miR-203, which subsequently inhibits noggin expression and leads to increased BMP signaling activity, leading to epithelial cell differentiation [45]. In addition, another independent investigation has shown that elevated BMP signaling during the early stages of mouse somatic cell reprogramming induces the expression of miR-205 and miR-200 family members, to reinforce an MET phenotype. This reveals the presence of miR-200-mediated epithelial differentiation as being vital during the initiation phase of reprogramming [59]. Therefore, these findings illustrate that BMP signaling-mediated miR-200 expression is crucial during the maintenance of dental epithelial stem cells, as well as for the differentiation of progenitor cells.
While the significance of the miR-200 family during EMT suppression was initially recognized in cancers, investigations along the same period have shown that the miR-200 family indeed plays distinct roles during embryogenesis and development, which is not confined just to the establishment and maintenance of the epithelial phenotype. In this section, we elaborate upon the role of the miR-200 family in the neurosensory epithelium such as olfactory neurogenesis and taste sensory organs. In mammals, the vomeronasal organ (VNO) and the main olfactory epithelium (MOE) are made of pseudostratified epithelial cells and bipolar sensory neurons, which are essential for the detection of pheromones and volatile odorants, respectively [60]. In situ hybridization experiments in mouse embryonic MOE showed strong expressions of miR-200a, -b, and -429, detected as early as E9.5 (first distinguishable stage of olfactory development) and maintained stable expression until E13.5 [61]. Moreover, the expression of all miR-200 members was observed in immature and mature neuronal cell layers of MOE and VNO, highlighting the persistent role of miR-200s in adult MOE and VNO neurogenesis. Intriguingly, the intranasal delivery of CRISPR-Cas9-mediated miR-200b/a knockdown in the MOE caused a significant loss of differentiated olfactory sensory neurons, accompanied by a dramatic reduction in olfactory-mediated male–male aggressive behavior and male–female mating behavior, which is mechanistically mediated through miR-200/TET3/REST signaling [40]. Mutant mice null for Dlx5, a homeogene that controls olfactory receptor neuron differentiation, showed a reduced expression of miR-9 and miR-200s, and the subsequent knockdown of miR-9 and miR-200s in zebrafish embryos led to defective olfactory placode organization as well as the altered differentiation and migration of olfactory receptor neurons [49]. In addition to regulating olfactory epithelial differentiation, miR-200s are implicated in the development of other essential neurosensory organs. Zebrafish embryos carrying morpholinos (anti-sense oligonucleotide analogs that are used for generating knockdown embryos) targeted against miR-200 expression resulted in a reduction in taste bud cells, and the upstream activation of FGF and Notch signaling is essential for miR-200 activity [41]. Similarly, the characterization of miRNAs in the developing submandibular gland revealed that miR-200c specifically regulates FGFR-dependent epithelial end bud proliferation and branching morphogenesis [44]. Based on the abovementioned investigations, the function of miR-200s is evidently essential for the development and functioning of selected sensory organs.
Furthermore, miR-8, the insect homolog of miR-200, is regarded as a pleiotropic regulator of Drosophila development, including neuroepithelial expansion [62], steroid signaling-mediated body size regulation [63,64], and pigmentation patterning [65]. miR-8 activity is also essential during mosquito oogenesis, such as the proper secretion of yolk protein precursors like vitellogenin and lipophorin in developing oocytes, through the Wingless signaling pathway [66].
3. miR-200 Family in Pathophysiology
The aberrant expression of the miR-200 family is involved in a group of pathophysiologic conditions such as cancer, diabetes, asthma, autoimmune diseases, kidney diseases, and neurodegenerative diseases. In this section, we would like to elaborate upon the role of miR-200s on the pathophysiology of neurogenerative diseases and fibrosis.
3.1. miR-200 Family in Neurodegenerative Diseases
In general, disorders characterized by the progressive degeneration of the structure or function of the central nervous system (CNS) are collectively referred as neurodegenerative diseases. Representative neurodegenerative diseases include Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), Huntington’s disease (HD), multiple sclerosis, and prion diseases. Consequently, they cause significant defects in motor and cognitive ability [67]. An eminent study of the miR-8 mutants has shown that these mutants displayed high levels of apoptosis in the brain, behavioral defects, and an impaired neuromuscular coordination phenotype in Drosophila legs, which prompted the potential role of miR-200s in neurodegenerative disorders in mammals [68]. Recently, a number of studies have revealed that members of the miR-200 family are differentially expressed in the human brain and, more importantly, modulate genes associated with specific neurodegenerative disorders (Table 2).

Table 2.
miR-200s, target genes and their roles in neurodegenerative diseases.
3.1.1. miR-200 Family in Alzheimer’s Disease
Alzheimer’s disease (AD) is a progressive neurological disorder, characterized by an increasing level of memory loss and deterioration of cognitive functions that eventually lead to dementia [78]. In particular, the pathological landscapes of AD brains are well recognized by the deposition of beta amyloid peptide (Aβ) and the formation of intracellular neurofibrillary tangles (NFT) [79], and ER stress is believed to be the initial driver for neuronal cell loss in AD [80]. Several studies have demonstrated that members of the miR-200 family are found to be linked with AD progression. The overexpression of miR-200a in peripheral blood mononuclear cells from AD patients targets genes related to cell cycle and DNA repair [81]. Another report showed that the upregulation of miR-200a-3p targets SIRT1, an anti-apoptotic protein, in order to stimulate Aβ-induced neuronal apoptosis [70]. Similarly, miR-200c is overexpressed in the plasma of patients with moderate to severe forms of AD, indicating its association with the progression of the disease [78].
In turn, the decreased expression of miR-200b and -429 by Aβ42 leads to the expression of the amyloid precursor protein (APP). Increased APP levels, in turn, lead to an increase in Aβ42, which causes a further reduction of miR-200b. This generates a kind of vicious cycle that contributes to the progression of AD [71]. Furthermore, folic acid deficiency can also decrease the level of miR-200b. Thus, a folic acid-deficient diet stimulates APP overexpression and promotes Aβ generation [72,82]. Higaki et al. report that miR-200b/c reduces insulin resistance by targeting the ribosomal protein kinase S6 B1 [73]. This contributes to a reduction in the production of Aβ peptide and thus alleviates the Aβ-induced toxicity. On the contrary, the upregulation of miR-200c levels was observed in the serum of AD patients [78], denoting that the precise role of miR-200s in Aβ-induced ER stress remains to be clearly investigated. These findings highlight that the miR-200 family is an important player in the pathogenesis of AD and could be explored for the possibility of diagnostic markers of the disease.
3.1.2. miR-200 Family in Parkinson’s Disease
Parkinson’s disease (PD) is the most common neurological movement disorder [83]. PD involves a progressive loss of neurons in the brain, especially dopamine-producing (“dopaminergic”) neurons in a specific area of the brain called the substantia nigra. The loss of more than 50% of DA neurons causes a corresponding reduction in the synthesis of dopamine neurotransmitters. This, in turn, is manifested by motor dysfunction and clinical symptoms such as resting tremor, slowness of movement, speech changes, and impaired posture and balance [84]. Moreover, the neurons of PD patients are enriched in the aggregated protein α-synuclein (α-syn). This causes impairment in pathways such as vesicle trafficking or activating neuroinflammation disorders [85]. Although the exact molecular mechanisms of PD are still unknown, many studies suggest a key role of the miR-200 family in the pathogenesis of Parkinson’s disease.
The point mutations in human α-syn (A53T) and transgenic murine model of α-synucleinopathy (M83 SCNA∗A53T) have been directly implicated in the onset of familial early PD [86,87]. The miRNA profiling of transgenic mice and the cerebrospinal fluid of PD patients showed a significant enrichment of miR-200a-3p expressions, denoting their potential role in PD pathogenesis [88]. Moreover, miR-200 expression was correlated with the severity of PD, as confirmed by high Hoehn and Yahr (H&Y) scores [89]. These results may indicate that miR-200a expression may be correlated with the severity of PD, and miR-200a may be an effective marker of PD disease progression. In addition, sirtuin (SIRT1), a histone deacetylases family of proteins, have a protective role in PD through the amelioration of oxidative stress-induced neural cell death and the suppression of α-syn-induced aggregate formation [90]. SIRT1 has also been shown to render anti-apoptotic functions by suppressing p53 activity through deacetylation and promote cell survival [91,92,93]. In the context of the miR-200 family, miR-200a also has the ability to induce SIRT1 downregulation and trigger the apoptosis of dopaminergic neurons, consequently contributing to the development of PD [74,94]. Interestingly, Delavar et al. [75] showed that miR-141 also targets SIRT1 expression and correlates with PD-related pathogenic processes. Mechanistically, in a 1-methyl-4-phenylpyridinium- (MPP+-) induced in vitro PD model, the upregulation of miR-141-3p induced increased apoptosis, oxidative stress, and mitochondrial membrane potential through the direct targeting of SIRT1 expression [69]. The same study also shows that resveratrol (a SIRT1 activator) blocked and sirtinol (a SIRT1 inhibitor) reversed the abovementioned biological effects of miR-141-3p, respectively. Therefore, these studies highlight that the miR-200 family has potential roles in the onset and progression of PD.
3.1.3. miR-200 Family in Amyotrophic Lateral Sclerosis
Amyotrophic lateral sclerosis (ALS) is another progressive neurodegenerative disease that primarily affects motor neurons controlling voluntary muscles, and the resulting loss of these motor neurons leads to the deterioration of the coordinated muscle movements involved in walking, talking, eating, and, eventually, breathing [95]. Although the origins of ALS are sporadic in nature, a small number of cases are associated with genetic changes [96]. Over the years, about 20 genes have been associated with familial ALS, and many of these genes encode RNA-binding proteins, including Fused in sarcoma (FUS), a DNA/RNA-binding protein [97]. miR-141 is shown to regulate the expression of FUS, EWS, and TAF15 in differentiating neuronal cells, denoting that miR-141-mediated FUS regulation is observed during neurogenesis [76]. Interestingly, miR-141/200a and FUS are linked by a feed-forward regulatory loop where the prevalent FUS mutation in ALS masks miR-141/200a binding sites and contributes to the excessive accumulation of the FUS protein, eventually augmenting ALS pathogenesis [95].
In addition, miR-200c is directly regulated by FUS, which contributes to gene silencing. In turn, mutations in FUS reduce the silencing of genes targeted by miR-200c, which may be one of mechanisms involved in the development of ALS [77]. Additionally, Zhou et al. [98] found that the expression level of miR-200a was increased in the early stage and decreased in the later stage in ALS transgenic mice, indicating this microRNA as a potential marker for detecting the progression of ALS.
3.1.4. miR-200 Family in Multiple Sclerosis and Prion Disease
Multiple sclerosis (MS) is the most common demyelinating disease of the CNS. It is considered an autoimmune disease in which the body’s immune system attacks its own tissues [99]. The epidemiology of multiple sclerosis in developing countries shows that there has been a sharp increase in the incidence of patients, with an overall incidence of 85.8 per 100,000 [100]. The cause of MS is unknown, but it appears that a combination of environmental factors, epigenetics, and genetics lead to ongoing immune attacks on the CNS [101]. Two preliminary assessments on MS patient samples showed that miR-141 and -200a levels were increased in the relapsing phase of MS patients compared to the remission and control groups [102,103]. In addition, elevated miR levels induce the differentiation of Th17 cells that are involved in the development of MS [102]. Prion disease is a rare, fatal, neurodegenerative disease caused by abnormally folded prion proteins in brain (PrP) [104]. Most cases of prion disease in humans arise spontaneously and their signs and symptoms typically begin in adulthood and worsen with time [105]. A pilot study has potentially revealed a correlation between prion disease and miR-200 family expression, where decreased levels of all members of the miR-200 family correlated with morphological changes of dendritic spines and synaptic dysfunction [106]. Although the differential expressions of the miR-200 family members are beginning to be documented in such neurodegenerative conditions, specific roles of these miRNAs in their etiology remain largely unknown.
3.2. miR-200 Family in Fibrosis
Fibrosis is one of the major pathological processes that affect vital organs such as the kidneys, liver, lungs, and intestines. It is characterized by impaired epithelial architecture and excessive deposition of extracellular matrix and fibrous connective tissue, which generates multiple inflamed scar tissues within the organ, eventually leading to organ dysfunction and failure. Furthermore, patients diagnosed with cystic fibrosis pose increased risk towards cancer progression, denoting that fibrosis might serve as a gateway to life-threatening illnesses [107]. At the molecular level, several studies have validated the activation of EMT during the early stages of fibrosis and is referred to as type 2 EMT [108,109]. Accordingly, with EMT-inhibiting roles, miR-200s have been also implicated during tissue fibrosis.
Using a unilateral ureter obstruction model, Oba et al. have shown that the injection of 0.5 nM of pre-miR-200b (precursor) efficiently inhibited the rise of collagen and fibronectin levels in obstructed kidneys, and could ameliorate renal tubulointerstitial fibrosis [110]. Similarly, the collection duct-specific inhibition of miR-200 activity in a transgenic mouse model evoked the expression of profibrotic target genes and inflammatory cytokines such as Mcp1, Il6, and Cxcl2 [111]. Furthermore, the downregulation of miR-200 members in renal fibrosis is primarily mediated through TGF-β1, and accordingly, the overexpression of miR-141 or -200b hindered Smad-dependent TGF-β signaling and downstream EMT phenotypes [112,113,114].
Levels of miR-200a and miR-200c were also significantly downregulated in murine lungs in a bleomycin-mediated fibrosis model as well as in patients with idiopathic pulmonary fibrosis (IPF), and the restoration of miR-200 expression in senescent IPF cells resumed normal regenerative functions [115,116,117]. In the case of liver fibrosis, miR-200s seem to play a pro-fibrotic role. In a CCL4-induced liver fibrosis model, miR-199 and miR-200 levels were significantly upregulated in comparison to the control groups [118]. Accordingly, the expression of miR-200s were elevated in serums of patients diagnosed with non-alcoholic fatty liver disease (NAFLD), and NAFLD mice treated with miR-200 inhibitor ameliorated liver fibrosis. Mechanistically, GRHL2 was overexpressed in the serum of NAFLD patients, which in turn negatively targeted SIRT1 and also activated miR-200 and the MAPK signaling pathway, aggravating liver fibrosis and intestinal mucosal barrier dysfunction [119]. GRHL2 is regarded as a gatekeeper of epithelial phenotype and differentiation, also involved in the direct activation of the miR-200 family [120].
3.3. miR-200 Family in Cancer
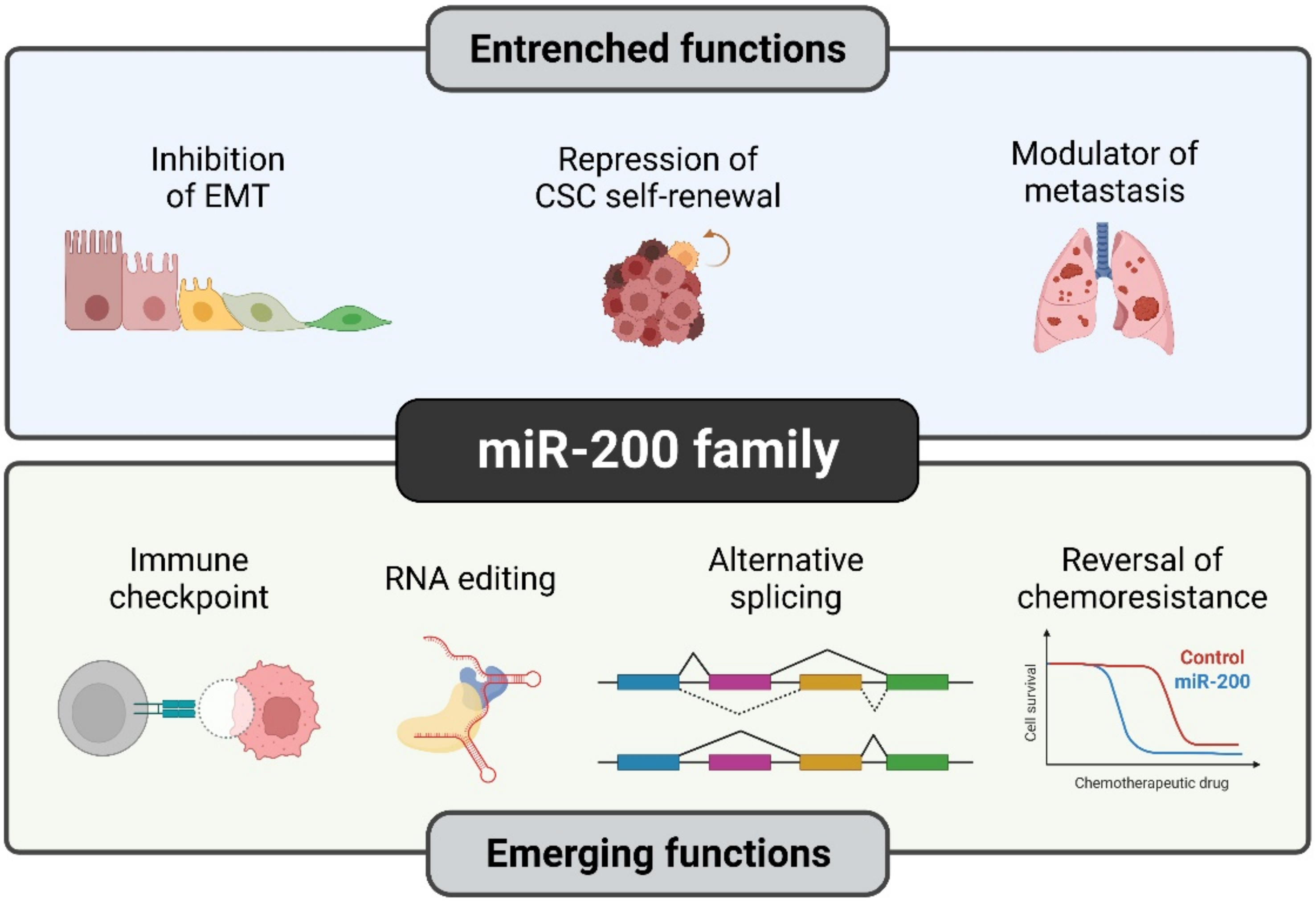
Over the past two decades, the role of miR-200s in cancer has been extensively studied, which has led us to distinguish the established (well-explored) and emerging functions that are exclusively observed during cancer progression (Figure 3).
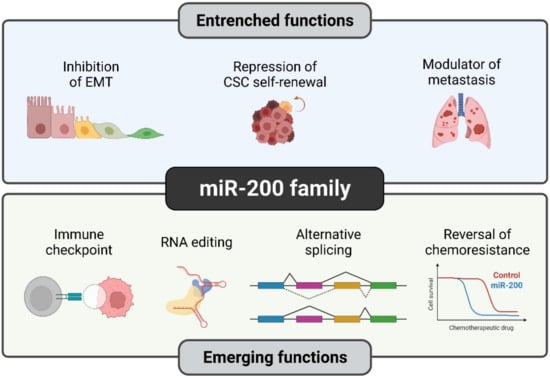
Figure 3.
Established and novel functions of the miR-200 family in human cancers. Although certain functions of miR-200s such as EMT suppression are quite well understood, members of the miR-200 family have recently been studied in detail for their novel roles such as RNA editing and reversal of chemoresistance. Illustration created with Biorender.com (https://biorender.com/, accessed on 1 June 2022).
The dysregulation of miR-200 expression in various entities of human cancer (reviewed in [121]) assigns it with tumor-promoting as well as tumor-suppressive effects. Through the direct targeting of genes that are involved in tumor-promoting mechanisms, members of the miR-200 family, individually or collectively, have been reported to be involved in the regulation of EMT/MET and metastasis [122,123], in the regulation of the cell cycle and apoptosis [74,124], as well as its deregulation during chemoresistance [125], cancer stemness [43,126], and, more recently, also in modulating intra-tumoral immune cell function [127,128,129]
3.3.1. miR-200 Family in Cancer-Associated EMT and MET
In the metastatic cascade, miR-200 exerts a context-dependent role. In general, the expression of miR-200 family members in the primary tumor strengthens an epithelial phenotype, thereby preventing EMT. Members of the miR-200 family primarily block EMT through direct repression of the EMT inducers ZEB1 and ZEB2, and impede tumor cell dissociation, migration, and invasion, which potentially combats the initial stages of the metastatic cascade, thus implicating the miRNAs’ tumor-suppressive effect. During the late 2000s, five research groups independently validated that miR-200s strongly repress EMT in multiple cancer types by directly targeting the major EMT transcription factors ZEB1 and ZEB2 [130,131,132,133,134]. The 3′UTR of ZEB1 and ZEB2 contain eight and nine miR-200 binding sites, respectively, which results in a strong and effective repression of the ZEB1/2 transcripts [132,134]. ZEB1 and ZEB2, on the other hand, suppress the transcription of all members of the miR-200 family [131,135]. These data illustrate that ZEB1/2 and miR-200s are not just functional rivals during cellular differentiation (EMT) and dedifferentiation (MET) processes in cancer, but mutually control the expression of each other, generating a double-negative feedback loop [136].
miR-200s directly target members of the Notch signaling pathway such as Jag1, Jag2, Maml2, and Maml3 to suppress pancreatic and lung adenocarcinoma proliferation and metastasis [137,138,139]. Several studies have reported that miR-200 family members influence cancer cell invasion by regulating the proteins involved in remodeling of the cytoskeleton. This interaction may be dependent on [140] or independent of the ZEB1/miR-200 axis [141,142].
Although cancer-associated EMT is regarded as a binary process driving cellular epithelial identity towards a mesenchymal phenotype, meticulous studies in the recent decade have acknowledged that this process is executed in a rather gradual mode, with the presence of one or more intermediary cellular states, each exhibiting distinct phenotypical, transcriptional, and epigenetic signatures [143,144,145]. Cells harboring such intermediate or hybrid EMT characteristics are prevalent across multiple cancer types such as breast [146,147], prostate [148], ovarian [143], and non-small lung cancer [149]. Moreover, cells with hybrid EMT state display increased tumorigenicity [150] and metastatic potential [145,151]. Subsequent mathematical and computational studies have highlighted that the core EMT decision-making circuit consists of two dynamically interconnected inhibitory feedback loops: one between miR-200 and ZEB1/2 [137], and the other between miR-34 and SNAIL1/2 [152]. Such regulatory feedback mechanisms are crucial during the generation and maintenance of hybrid EMT cellular states [153,154,155,156]. Recently, Garinet et al. profiled 176 resected non-small cell lung carcinoma specimens and identified that most tumors were associated with an EMT-hybrid state and miR-200s expression profiles derived from those samples were utilized for the identification of good prognostic groups that are unrelated to conventional EMT scores [157]. Furthermore, a recent TGF-β1-induced single-cell RNA sequencing study re-established that the gradual deregulation of miR-200 family expression is observed during intermediate EMT cellular clusters [158]. All the above studies denote that cancer-associated EMT is a dynamic, continual process instead of a terminal differentiation program and that sequential deregulation of the miR-200 family is observed during the generation of intermediate EMT cellular states.
3.3.2. miR-200 Family’s Tumor-Promoting Roles during Tumorigenesis and Metastasis
Although numerous studies have elucidated the indispensable role of the miR-200 family in cancer metastasis-associated functions such as tumor cell dissemination, invasion and angiogenesis, miR-200s are active players during the early stages of tumorigenesis such as neoplastic transformation. MiR-200s seem to render a pro-tumorigenic factor in many cancers, which is in contrast to their tumor-suppressive roles in later stages of the metastatic process. Xenografts carrying miR-141 or miR-200a overexpressing Kras-transformed ovarian fibroblasts formed faster and larger tumors by inhibiting p38α and stimulating the oxidative stress response [159]. In the case of ovarian carcinoma, a conditional mouse model showed that the loss of Dicer, a miRNA biogenesis factor, promoted the epithelialization of fallopian tube stromal cells and initiated tumorigenesis [160]. Accordingly, elevated expressions of miR-200 and E-cadherin were identified in human and white Leghorn hen ovarian cancer tissues, highlighting their roles in the initial development of ovarian carcinoma [161,162]. Ovarian cancer cells with miR-200 knockdown when grown on a three-dimensional (3D) organotypic setting showed an increased number of lumina, which is due to mitotic spindle disorientation, loss of cellular polarity, and a collective migration phenotype, facilitating ovarian carcinogenesis [163]. At the molecular level, the loss of collective migration in these 3D structures is due to the disruption of ROCK-mediated myosin-II phosphorylation and SRC signaling. The increased expression of miR-200 in early-stage lung adenocarcinomas in vitro and in vivo activated PI3K/AKT signaling through FOG2 targeting and promoted tumor-initiating spheroid growth formation [164]. Early ductal carcinomas in situ (DCIS) lesions of the breast tissue are non-obligatory precursors of invasive breast cancer, yet their severe microenvironment with hypoxia, nutrient deprivation, and acidosis facilitate tumorigenesis [165,166]. Subtype-specific miRNA profiling of normal, DCIS, and invasive breast carcinoma cohorts showed significant enrichment of miR-21-5p and all members of the miR-200 family in the DCIS samples, denoting their potential tumorigenic role in breast carcinogenesis [167].
In the context of cancer progression and metastasis, it is important to note that miR-200s support an MET of mesenchymal tumor cells at distant organ sites, thereby promoting metastatic colonization. In breast cancer, miR-200s promote the metastatic colonization of secondary organs by targeting the cancer cell secretome, subsequently influencing the tumor microenvironment [168,169]. In addition, miR-200c, miR-192, and miR-17 were identified as targeting genes involved in extracellular matrix remodeling. The expression of the three miRNAs in tumor-associated fibroblasts may therefore suppress the invasion of colorectal cancer cells [170]. Further studies on miR-200 in the tumor microenvironment recently revealed that apoptotic MCF7 breast cancer cells release miR-200c, which is taken up by tumor-associated macrophages. This leads to the overexpression of miR-200c in the tumor-associated macrophages, followed by the reduced expression of a set of pro-migratory miR-200c targets and a reduced capacity of the macrophages to infiltrate into tumor spheroids [129]. Taken together, these findings show that miR-200 is a crucial player in the crosstalk between tumor cells and tumor-associated stromal and immune cells. This crosstalk may modulate the cells’ invasive behavior with tumor-promoting or suppressing effects, depending on the releasing and receiving cell. This points to the fact that the role of miR-200s in tumor progression extends beyond the regulation of transition between EMT and MET and the ZEB/miR-200 feedback loop.
3.3.3. miR200 Family in Radiotherapy
Radiotherapy still remains one of the main front-line treatment options in several cancer types including breast, lung, prostate, cervical, and head and neck cancers. The response of cancer cells towards radiotherapy is mostly affected by the tumor size. However, recent preclinical studies have highlighted that miRNAs, especially miR-200s, contribute towards radiotherapy response in multiple cancer types [171]. In breast cancer cells, it has been shown that high miR-200c expression is important for radiosensitivity, while decreased expression is associated with radioresistance [172]. This has been demonstrated in studies with cell lines showing different basal levels of miR-200c expression. The MCF-7 cell line, which has a high level of miR-200c expression, showed a higher sensitivity to radiotherapy compared to the MDA-MB-231 cell line. Additionally, higher expression of miR-200c increases the sensitivity to radiation by inhibiting cell proliferation and by increasing apoptosis and DNA double-strand breaks [173]. Further studies showed that the sensitization of cancer cells to radiation therapy with high levels of miR-200c is mediated by targeting TBK1 and VEGF-VEGFR2, resulting in cell apoptosis [174]. Moreover, Sun et al. identified ubiquilin 1 (UBQLN1) as a functional target of miR-200c, where miR-200c sensitizes breast cancer cells to radiation in a manner associated with the inhibition of radiation-induced autophagy [174]. In turn, Wang et al. speculated that radiation resistance is promoted by the interaction of the long coding RNA, LINC02582, with USP7 to deubiquitinate and stabilize CHK1. Therefore, they proposed that the miR-200c/LINC02582/USP7/CHK1 signaling axis could be a therapeutic target to improve the breast cancer response to radiotherapy [175].
In addition, miR-200c plays a role in the effectiveness of radiotherapy in lung cancer. The high expression of miR-200c increases radiation sensitivity by regulating the oxidative stress response by targeting PRDX2, GAPB/Nrf2, and SESN1 and by inhibiting the repair of radiation-induced double-strand breaks [176]. Next, miR-200a affects the radiosensitivity of non-small cell lung cancer (NSCLC) cells. The analysis of clinical data in NSCLC patients showed that miR-200a negatively regulates HGF expression, resulting in reduced cancer cell invasion and metastasis. Moreover, the miR-200a/HGF pathway also influences the radiosensitivity of NSCLC cells. This was confirmed in experiments on cell lines A549 and H1299, where the overexpression of miR-200a after irradiation resulted in the increased apoptosis and DNA double-stranded breaks of these cancer cells [177].
Interestingly, miR-200a is detected in the saliva of patients with head and neck squamous cell carcinoma after radiation treatment. The analysis of the distribution of miR-200a expression during and after radiotherapy showed that the miR level is significantly higher 12 months after treatment compared with the baseline state [178]. It may constitute a potential tool in monitoring the response to radiotherapy in patients with HNSCC. Along this line, radiation-induced oral mucositis (RIOM) is one of the more prevalent side effects of radiotherapy in HNSCC patients. A preliminary study identified that all members of the miR-200 family were significantly upregulated in RIOM formation, and the knockdown of miR-200c led to a reduction of proinflammatory cytokine synthesis and reactive oxygen species generation [179].
In addition to all of the abovementioned studies, two new reports this year implicate the association of miR-200s with radiosensitivity in other cancer types. The suppression of miR-200b/c in esophageal squamous cell carcinoma impaired with cell sensitivity to concurrent chemoradiotherapy treatment with or without surgery [180]. Similarly, tumor xenografts carrying miR-200a/b/-429 overexpressing cervical carcinoma cell lines were significantly sensitized to radiotherapy, which is also independent of tumor hypoxia [181]. Therefore, the prospective role of the miR-200 family as a predictor of response to cancer radiotherapy has been revealed in work over the last decade and certainly demands more research.
3.3.4. miR-200 Family in Chemotherapy
Dealing with the emergence of therapy resistance poses a major challenge in the treatment of cancer patients. Recent literature has reported that miR-200s are involved in modulating the response of cancer cells to anti-cancer therapy by influencing the status of certain adenocarcinomas towards chemo- and radiosensitivity. Importantly, therapy resistance occurs through numerous resistance mechanisms such as increased DNA repair capacity, elevated drug efflux through ATP-binding cassette (ABC) transporters, varying expression of β-tubulin isotypes, changes in cell cycle, inhibition of apoptosis, and response to oxidative stress [182]. The participation of miR-200 in these processes has been well described, thereby miR-200s may enhance the therapeutic possibilities of EMT-associated cancer metastasis and may even predict therapeutic response [140,183,184]. Crudele et al. reviewed the miR-200 family network’s connection with resistance to six different anti-cancer treatments [185]. In this regard, the restoration of miR-200c expression in ovarian and endometrial cancer cells leads to increased sensitivity to microtubule-binding chemotherapeutic agents through the reduction of class III β-tubulin (TUBB3) [125]. Similarly, in chemoresistant pancreatic cells, the induction of miR-200s and miR-203 expression through the class I HDAC inhibitor mocetinostat restored drug sensitivity to standard chemotherapeutics such as gemcitabine and docetaxel [186]. The oxaliplatin-resistant colorectal cancer cell line showed increased expression of SUZ12, a Polycomb-repressive complex 2 subunit, and decreased expression of the miR-200 family [187]. In a similar fashion, the expression of miR-200c alone enhanced the chemosensitivity and reduced the metastatic potential of p53null claudin-low breast cancer mouse models [188]. The depletion of miR-200c significantly reduced 5-flurouracil-induced apoptosis and caspase 3 activity in colorectal HCT-116 cells [189], and the inhibition of miR-141/200c in the ovarian cancer cell line OVCAR-3 led to resistance to paclitaxel and carboplatin [190]. The overexpression of miR-200c in melanoma cells decreased resistance to cisplatin as well as a BRAF- and a MEK-inhibitor through the downregulation of the ABC transporters ABCG2, ABCG5, and MDR1 directly or indirectly through the downregulation of BMI-1 [183,191]. Intriguingly, a recent pilot study has shown that the anticancer drug cisplatin interacts with pre-miR-200b and decreases the production of mature miR-200b expression in ovarian cancer cells [184]. Therefore, a direct correlation between miR-200s and a drug sensitivity phenotype has been established primarily using adenocarcinomas; however, the underlying mechanistic insights are still being delineated.
It is important to mention that there are also reports that observe the opposite effects, and describe miR-200 as conferring chemoresistance. In a study by Yu et al., the expression of miR-200a correlated with poor response to preoperative chemotherapy and poor prognosis in breast cancer patients. Additionally, miR-200a was described to confer chemoresistance to gemcitabine by targeting TP53INP1 and YAP1 [192]. TP53INP1 is a p73 target gene that inhibits cell growth and promotes cell death, and is induced by stress in response to DNA damage, i.e., by cisplatin [193]. In hepatocellular carcinoma, the overexpression of miR-200a-3p increased resistance to 5-FU by targeting DUSP6. In this setting, the inhibition of miR-200a-3p decreased cell growth and viability after 5-FU treatment, thereby sensitizing Hep3B cells to different anti-cancer drugs, including 5-FU, cisplatin, and doxorubicin [194].
The administration of anticancer drugs or radiotherapy is proposed to lead to an accumulation of reactive oxygen species (ROS). These ROS interfere with different cellular processes such as cell survival and motility, and the oxidative stress influences miRNA expression patterns. The miR-200 family was described to be involved in the oxidative stress response by regulating KEAP1 expression in breast and ovarian cancer [149,150,179]. The introduction of miR-141/-200c into a paclitaxel-resistant ovarian cell line confers resistance to carboplatin, while altering the expression of genes involved in balancing oxidative stress [190].
In summary, miR-200 family members target a plethora of genes involved in fate determining processes within cancer cells as well as in cells of the tumor microenvironment. A selection of these miR-200 target genes is compiled in Table 3. Seemingly contradictory effects may occur, which require a precise analysis of cellular context and spatiotemporal expression pattern of the miR-200s in order to predict the outcome of a potential therapeutic interference. The fact that cells may release miRNAs assigns miR-200s a role in inter cellular communication and makes them detectable in body fluids for a potential use as biomarkers which is discussed in the following section.

Table 3.
Summary of miR-200s, target genes, and intervening mechanism in multiple cancer types.
4. miR-200 Family as Predictive Markers
The miR-200 family has substantial influence on cancer cell motility and invasive behavior through the direct suppression of EMT and metastasis formation in different tumor types. Consequently, many studies have suggested that the miR-200 family has great potential to be key biomarkers in a variety of diseases, including cancer (recently reviewed in [199], Table 4) and neurodegenerative diseases [200,201]. For instance, a recent meta-analysis study encompassing 24 eligible breast cancer articles and 16,565 subjects showed a significant correlation between high miR-200s expression and the poor overall survival of breast cancer patients, while the downregulation of miR-200s was associated with the poor survival of triple-negative and luminal breast cancer patients [202]. Another meta-analysis study involving eight studies with data from 1150 bladder cancer patients showed that the high expression of the miR-200 family was associated with better overall survival and relapse-free survival, which is regarded as a reliable prognostic biomarker in bladder cancer patients [203]. Likewise, the low expression of miR-200c in breast cancer was correlated with a poor response to neoadjuvant chemotherapy [200]. Furthermore, the profiling of candidate miRNAs in plasma samples from metastatic breast cancer patients showed that circulating tumor cells enrich miR-200s, miR-203, and miR-375 expressions and are associated with the onset of metastatic disease, further highlighting their prognostic significance [204,205]. A similar trend of higher levels of circulating miR-200s expression in plasma and exosomes of colorectal cancer patients were correlated with poor prognosis [206,207]. The overexpression of miR-200b and -200c was observed in the serum exosomes of pancreatic ductal adenocarcinoma patients, in addition to a correlation between their expression and shorter overall survival [208]. Furthermore, elevated expressions of miR-141 and -200c in non-small cell lung cancer and colorectal cancer were associated with poor prognosis and shorter overall survival [209,210].

Table 4.
A summary of differential changes in miR-200s expression as predictive biomarkers of multiple cancer types. The type of sample used and method of detection are also included.
In addition to the potential utility of miR-200s in solid carcinoma diagnosis/prognosis, certain studies have highlighted the prospect of miR-200s as diagnostic markers in liquid biopsies. Zuberi et al. identified a direct correlation of miR-200a/c expression in the serum of epithelial ovarian cancer patients in advanced stages of disease (stages III and IV) when compared with early stages (I and II) [216]. In this study, the authors demonstrate a correlation between increased miR200a/c serum levels, which are associated with an aggressive tumor progression, combined with poor prognosis in ovarian cancer. Similarly, miR-21, miR-200s, and miR-205 were among the abundant RNA biomarkers detected in the biofluids of ovarian cancer patients when compared to controls [223]. The RNA profiling of exosomes isolated from serum, plasma, or pleural effusions of patients with lung cancer [224], cholangiocarcinoma [222], colon cancer [207], and pancreatic ductal adenocarcinomas [208] showed the miR-200 family as being one of the top differentially expressed microRNAs in distinguishing those diagnosed with benign disease. Taken together, the differential expression of miR-200s during varying stages of cancer progression and their distinctive expression in different cancer types render a prognostic and/or predictive value in multiple tumor types.
5. Conclusions and Future Perspectives
In spite of being single-stranded, non-coding oligonucleotides and encoded by less than 1% of the human genome, miRNAs are known to regulate over 60% of human protein-coding genes [225]. Therefore, it is imperative to claim that miRNAs are one of the major gene-regulatory elements, which exert control on almost every aspect of cellular events such as growth, differentiation, homeostasis, and death. It is also apparent that the deregulation of miRNAs is directly associated with the onset and progression of multiple diseases and pathological conditions. Furthermore, recent reviews have determined that miRNAs have roles in newer avenues, including drug addiction [226], neonatal sepsis [227], and the alteration of synaptic plasticity in depression [228,229], illustrating that this class of ncRNAs displays versatile functions in several aspects of cellular development as well as in pathological conditions.
Among the most versatile miRNAs, one or multiple members of the miR-200 family are directly involved in regulating almost all of the abovementioned aspects of cellular events, which we have comprehensively elaborated in the earlier sections. The initial function of miR-200s begin as early as the migration of neural crest cells and are crucial players in epithelial differentiation and branching morphogenesis. In particular, miR-200-mediated gene expression regulation is essential during mammalian skin, hair follicle, dental, and sensory organ development. Interestingly, recent studies are beginning to understand the miR-200s’ role in stem cell/progenitor cell populations [58,230], denoting that the role of miR-200s in embryonic and organ development is still underexplored.
In terms of the role of miR-200s in cancer, five pioneering yet independent publications in 2008 [130,131,132,133,134] revealing the EMT- and metastasis-suppressing roles of miR-200s in multiple cancer entities not only cemented an indispensable spot for the miR-200 family in comprehending cancer progression, but also prompted the scientific community to contemplate and investigate miR-200s and other miRNAs from a pathological perspective. Since then, an incessant surge in the number of publications identifying the association between miR-200s expressions and the pathogenesis of several human ailments has been observed. In this review, we have reviewed both the established functions of miR-200s, such as EMT suppression and cancer stemness inhibition, as well as newly emerging functions, such as RNA editing, alternative splicing, and the reversal of chemoresistance. Such new emerging functions underline the fact that miR-200s are not mere deregulators of oncogenes, but possess diverse potentials in combating and intervening in cancer metastasis. In contrast to the suppressive roles of miR-200s listed here, researchers have also found that members of the miR-200 family impose pro-tumorigenic functions, such as during tumor initiation and metastasis progression. This paradoxical phenomenon is not unique to the miR-200 family, serving as a reminder that these molecules execute functions based on the cellular context and upstream signaling events. Furthermore, the expression and, thereby, the functionality of miR-200s are often directly regulated or deregulated by transcription factors that are on the other side of the functional spectrum. Recent studies on three-dimensional (3D) chromatin interaction studies at the genomic level have speculated that local chromatin conformational changes at the 3D genome levels influence the expression of ncRNAs, including miRNAs [231,232]. Therefore, genome-wide chromatin conformation studies on the miR-200 locus across multiple pathological situations (chromatin conformational changes of cancer cells on miR-200 loci before and after EMT, drug sensitivity, etc.) could expose novel mechanisms through which miR-200-mediated disease progression or repression could be achieved.
Lastly, several studies and systemic reviews have already highlighted the prospective functions of miR-200s as a prognostic biomarker. However, efforts to utilize miR-200s and other related ncRNAs as commercial biomarker test kits are still lacking. On the other hand, simple and inexpensive commercial test kits are now available for quantifying other genetic abnormalities such as TERT promoter mutations [233], and the number of CGG repeats in the fragile X mental retardation 1 (FMR1) gene in Fragile X syndrome (FXS) [234]. Such initiatives in the coming years are essential for the successful utilization of miRNA-based biomarkers in the preclinical and clinical settings.
Author Contributions
Conceptualization, V.S.; writing—original draft preparation, review and editing, V.S., U.C.B. and K.B.-R.; funding acquisition, K.B.-R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ohno, S. So Much “Junk” DNA in Our Genome. Brookhaven Symp. Biol. 1972, 23, 366–370. [Google Scholar] [PubMed]
- Lander, E.S. Initial Impact of the Sequencing of the Human Genome. Nature 2011, 470, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, E. Genomics. ENCODE Project Writes Eulogy for Junk DNA. Science 2012, 337, 1159–1161. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of Mammalian MicroRNA Targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a Role in Cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Dragomir, M.P.; Knutsen, E.; Calin, G.A. SnapShot: Unconventional MiRNA Functions. Cell 2018, 174, 1038–1038.e1. [Google Scholar] [CrossRef]
- Jie, M.; Feng, T.; Huang, W.; Zhang, M.; Feng, Y.; Jiang, H.; Wen, Z. Subcellular Localization of MiRNAs and Implications in Cellular Homeostasis. Genes 2021, 12, 856. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef]
- Xu, M.; Chen, Y.; Xu, Z.; Zhang, L.; Jiang, H.; Pian, C. MiRLoc: Predicting MiRNA Subcellular Localization by Incorporating MiRNA-MRNA Interactions and MRNA Subcellular Localization. Brief. Bioinform. 2022, 23, bbac044. [Google Scholar] [CrossRef]
- Chang, T.-C.; Mendell, J.T. MicroRNAs in Vertebrate Physiology and Human Disease. Annu. Rev. Genom. Hum. Genet. 2007, 8, 215–239. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. MiR-15 and MiR-16 Induce Apoptosis by Targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Michael, M.Z.; O’ Connor, S.M.; van Holst Pellekaan, N.G.; Young, G.P.; James, R.J. Reduced Accumulation of Specific MicroRNAs in Colorectal Neoplasia. Mol. Cancer Res. 2003, 1, 882–891. [Google Scholar]
- Shi, L.; Middleton, J.; Jeon, Y.-J.; Magee, P.; Veneziano, D.; Laganà, A.; Leong, H.-S.; Sahoo, S.; Fassan, M.; Booton, R.; et al. KRAS Induces Lung Tumorigenesis through MicroRNAs Modulation. Cell Death Dis. 2018, 9, 219. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, S.; Zhu, Z.; Yu, L.; Ren, Y.; Jiang, M.; Weng, J.; Li, B. MiR-21 Promotes EGF-Induced Pancreatic Cancer Cell Proliferation by Targeting Spry2. Cell Death Dis. 2018, 9, 1157. [Google Scholar] [CrossRef]
- Huang, T.-H.; Wu, F.; Loeb, G.B.; Hsu, R.; Heidersbach, A.; Brincat, A.; Horiuchi, D.; Lebbink, R.J.; Mo, Y.-Y.; Goga, A.; et al. Up-Regulation of MiR-21 by HER2/Neu Signaling Promotes Cell Invasion. J. Biol. Chem. 2009, 284, 18515–18524. [Google Scholar] [CrossRef]
- Xia, H.; Zhang, W.; Zhang, B.; Zhao, Y.; Zhao, Y.; Li, S.; Liu, Y. MiR-21 Modulates the Effect of EZH2 on the Biological Behavior of Human Lung Cancer Stem Cells in Vitro. Oncotarget 2017, 8, 85442–85451. [Google Scholar] [CrossRef]
- Gao, Z.; Liu, H.; Shi, Y.; Yin, L.; Zhu, Y.; Liu, R. Identification of Cancer Stem Cell Molecular Markers and Effects of has-MiR-21-3p on Stemness in Esophageal Squamous Cell Carcinoma. Cancers 2019, 11, 518. [Google Scholar] [CrossRef]
- Li, A.; Yang, P.-M. Overexpression of MiR-21-5p in Colorectal Cancer Cells Promotes Self-Assembly of E-Cadherin-Dependent Multicellular Tumor Spheroids. Tissue Cell 2020, 65, 101365. [Google Scholar] [CrossRef]
- Mortoglou, M.; Miralles, F.; Arisan, E.D.; Dart, A.; Jurcevic, S.; Lange, S.; Uysal-Onganer, P. MicroRNA-21 Regulates Stemness in Pancreatic Ductal Adenocarcinoma Cells. Int. J. Mol. Sci. 2022, 23, 1275. [Google Scholar] [CrossRef]
- Sheng, S.; Su, W.; Mao, D.; Li, C.; Hu, X.; Deng, W.; Yao, Y.; Ji, Y. MicroRNA-21 Induces Cisplatin Resistance in Head and Neck Squamous Cell Carcinoma. PLoS ONE 2022, 17, e0267017. [Google Scholar] [CrossRef]
- Najjary, S.; Mohammadzadeh, R.; Mokhtarzadeh, A.; Mohammadi, A.; Kojabad, A.B.; Baradaran, B. Role of MiR-21 as an Authentic Oncogene in Mediating Drug Resistance in Breast Cancer. Gene 2020, 738, 144453. [Google Scholar] [CrossRef]
- Vandewalle, V.; Essaghir, A.; Bollaert, E.; Lenglez, S.; Graux, C.; Schoemans, H.; Saussoy, P.; Michaux, L.; Valk, P.J.M.; Demoulin, J.-B.; et al. MiR-15a-5p and MiR-21-5p Contribute to Chemoresistance in Cytogenetically Normal Acute Myeloid Leukaemia by Targeting PDCD4, ARL2 and BTG2. J. Cell Mol. Med. 2021, 25, 575–585. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Li, Y.-F.; Ma, H.; Gao, Y.-H. Regulation of MYB Mediated Cisplatin Resistance of Ovarian Cancer Cells Involves MiR-21-Wnt Signaling Axis. Sci. Rep. 2020, 10, 6893. [Google Scholar] [CrossRef]
- Sun, L.-H.; Tian, D.; Yang, Z.-C.; Li, J.-L. Exosomal MiR-21 Promotes Proliferation, Invasion and Therapy Resistance of Colon Adenocarcinoma Cells through Its Target PDCD4. Sci. Rep. 2020, 10, 8271. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating MicroRNAs as Stable Blood-Based Markers for Cancer Detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and Activity of MicroRNA-Loaded Minicells in Patients with Recurrent Malignant Pleural Mesothelioma: A First-in-Man, Phase 1, Open-Label, Dose-Escalation Study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Anandappa, G.; Lampis, A.; Cunningham, D.; Khan, K.H.; Kouvelakis, K.; Vlachogiannis, G.; Hedayat, S.; Tunariu, N.; Rao, S.; Watkins, D.; et al. MiR-31-3p Expression and Benefit from Anti-EGFR Inhibitors in Metastatic Colorectal Cancer Patients Enrolled in the Prospective Phase II PROSPECT-C Trial. Clin. Cancer Res. 2019, 25, 3830–3838. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.-Y.; Wright, J.A.; Attema, J.L.; Gregory, P.A.; Bert, A.G.; Smith, E.; Thomas, D.; Lopez, A.F.; Drew, P.A.; Khew-Goodall, Y.; et al. Epigenetic Modulation of the MiR-200 Family Is Associated with Transition to a Breast Cancer Stem-Cell-like State. J. Cell Sci. 2013, 126, 2256–2266. [Google Scholar] [CrossRef]
- Humphries, B.; Wang, Z.; Oom, A.L.; Fisher, T.; Tan, D.; Cui, Y.; Jiang, Y.; Yang, C. MicroRNA-200b Targets Protein Kinase Cα and Suppresses Triple-Negative Breast Cancer Metastasis. Carcinogenesis 2014, 35, 2254–2263. [Google Scholar] [CrossRef]
- Green, K.J.; Jaiganesh, A.; Broussard, J.A. Desmosomes: Essential Contributors to an Integrated Intercellular Junction Network. F1000Research 2019, 8, F1000. [Google Scholar] [CrossRef]
- Landschaft, D. Gaps and Barriers: Gap Junctions as a Channel of Communication between the Soma and the Germline. Semin. Cell Dev. Biol. 2020, 97, 167–171. [Google Scholar] [CrossRef]
- Otani, T.; Furuse, M. Tight Junction Structure and Function Revisited. Trends Cell Biol. 2020, 30, 805–817. [Google Scholar] [CrossRef]
- Veeraval, L.; O’Leary, C.J.; Cooper, H.M. Adherens Junctions: Guardians of Cortical Development. Front. Cell Dev. Biol. 2020, 8, 6. [Google Scholar] [CrossRef]
- Akhtar, N.; Streuli, C.H. An Integrin-ILK-Microtubule Network Orients Cell Polarity and Lumen Formation in Glandular Epithelium. Nat. Cell Biol. 2013, 15, 17–27. [Google Scholar] [CrossRef]
- Rodriguez-Boulan, E.; Macara, I.G. Organization and Execution of the Epithelial Polarity Programme. Nat. Rev. Mol. Cell Biol. 2014, 15, 225–242. [Google Scholar] [CrossRef]
- Hay, E.D. The Mesenchymal Cell, Its Role in the Embryo, and the Remarkable Signaling Mechanisms That Create It. Dev. Dyn. 2005, 233, 706–720. [Google Scholar] [CrossRef]
- Yang, D.; Wu, X.; Zhou, Y.; Wang, W.; Wang, Z. The MicroRNA/TET3/REST Axis Is Required for Olfactory Globose Basal Cell Proliferation and Male Behavior. EMBO Rep. 2020, 21, e49431. [Google Scholar] [CrossRef]
- Kapsimali, M.; Kaushik, A.-L.; Gibon, G.; Dirian, L.; Ernest, S.; Rosa, F.M. Fgf Signaling Controls Pharyngeal Taste Bud Formation through MiR-200 and Delta-Notch Activity. Development 2011, 138, 3473–3484. [Google Scholar] [CrossRef]
- Banerjee, P.; Dutta, S.; Pal, R. Dysregulation of Wnt-Signaling and a Candidate Set of MiRNAs Underlie the Effect of Metformin on Neural Crest Cell Development. Stem Cells 2016, 34, 334–345. [Google Scholar] [CrossRef][Green Version]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of MiRNA-200c Links Breast Cancer Stem Cells with Normal Stem Cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef]
- Rebustini, I.T.; Hayashi, T.; Reynolds, A.D.; Dillard, M.L.; Carpenter, E.M.; Hoffman, M.P. MiR-200c Regulates FGFR-Dependent Epithelial Proliferation via Vldlr during Submandibular Gland Branching Morphogenesis. Development 2012, 139, 191–202. [Google Scholar] [CrossRef]
- Cao, H.; Jheon, A.; Li, X.; Sun, Z.; Wang, J.; Florez, S.; Zhang, Z.; McManus, M.T.; Klein, O.D.; Amendt, B.A. The Pitx2:MiR-200c/141:Noggin Pathway Regulates Bmp Signaling and Ameloblast Differentiation. Development 2013, 140, 3348–3359. [Google Scholar] [CrossRef]
- Beclin, C.; Follert, P.; Stappers, E.; Barral, S.; Coré, N.; de Chevigny, A.; Magnone, V.; Lebrigand, K.; Bissels, U.; Huylebroeck, D.; et al. MiR-200 Family Controls Late Steps of Postnatal Forebrain Neurogenesis via Zeb2 Inhibition. Sci. Rep. 2016, 6, 35729. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, N.; Park, S.-W.; Kim, H.; Park, H.-J.; Han, Y.-M. Lineage-Specific Expression of MiR-200 Family in Human Embryonic Stem Cells during In Vitro Differentiation. Int. J. Stem Cells 2017, 10, 28–37. [Google Scholar] [CrossRef]
- Jimenez, P.T.; Mainigi, M.A.; Word, R.A.; Kraus, W.L.; Mendelson, C.R. MiR-200 Regulates Endometrial Development During Early Pregnancy. Mol. Endocrinol. 2016, 30, 977–987. [Google Scholar] [CrossRef] [PubMed]
- Garaffo, G.; Conte, D.; Provero, P.; Tomaiuolo, D.; Luo, Z.; Pinciroli, P.; Peano, C.; D’Atri, I.; Gitton, Y.; Etzion, T.; et al. The Dlx5 and Foxg1 Transcription Factors, Linked via MiRNA-9 and -200, Are Required for the Development of the Olfactory and GnRH System. Mol. Cell Neurosci. 2015, 68, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Hilmarsdóttir, B.; Briem, E.; Sigurdsson, V.; Franzdóttir, S.R.; Ringnér, M.; Arason, A.J.; Bergthorsson, J.T.; Magnusson, M.K.; Gudjonsson, T. MicroRNA-200c-141 and ∆Np63 Are Required for Breast Epithelial Differentiation and Branching Morphogenesis. Dev. Biol. 2015, 403, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Hoefert, J.E.; Bjerke, G.A.; Wang, D.; Yi, R. The MicroRNA-200 Family Coordinately Regulates Cell Adhesion and Proliferation in Hair Morphogenesis. J. Cell Biol. 2018, 217, 2185–2204. [Google Scholar] [CrossRef]
- Suh, H.N.; Han, H.J. Sonic Hedgehog Increases the Skin Wound-Healing Ability of Mouse Embryonic Stem Cells through the MicroRNA 200 Family. Br. J. Pharmacol. 2015, 172, 815–828. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Li, X.; Han, W.; Yang, K.; Wang, H.; Zhang, Y.; Su, R.; Liu, Z.; Wang, R.; et al. High-Throughput Sequencing of Hair Follicle Development-Related Micrornas in Cashmere Goat at Various Fetal Periods. Saudi J. Biol. Sci. 2018, 25, 1494–1508. [Google Scholar] [CrossRef]
- Sun, Z.; Yu, W.; Sanz Navarro, M.; Sweat, M.; Eliason, S.; Sharp, T.; Liu, H.; Seidel, K.; Zhang, L.; Moreno, M.; et al. Sox2 and Lef-1 Interact with Pitx2 to Regulate Incisor Development and Stem Cell Renewal. Development 2016, 143, 4115–4126. [Google Scholar] [CrossRef]
- Sanz-Navarro, M.; Seidel, K.; Sun, Z.; Bertonnier-Brouty, L.; Amendt, B.A.; Klein, O.D.; Michon, F. Plasticity within the Niche Ensures the Maintenance of a Sox2+ Stem Cell Population in the Mouse Incisor. Development 2018, 145, dev155929. [Google Scholar] [CrossRef]
- Michon, F.; Tummers, M.; Kyyrönen, M.; Frilander, M.J.; Thesleff, I. Tooth Morphogenesis and Ameloblast Differentiation Are Regulated by Micro-RNAs. Dev. Biol. 2010, 340, 355–368. [Google Scholar] [CrossRef]
- Jheon, A.H.; Li, C.-Y.; Wen, T.; Michon, F.; Klein, O.D. Expression of MicroRNAs in the Stem Cell Niche of the Adult Mouse Incisor. PLoS ONE 2011, 6, e24536. [Google Scholar] [CrossRef]
- Sweat, M.; Sweat, Y.; Yu, W.; Su, D.; Leonard, R.J.; Eliason, S.L.; Amendt, B.A. The MiR-200 Family Is Required for Ectodermal Organ Development through the Regulation of the Epithelial Stem Cell Niche. Stem Cells 2021, 39, 761–775. [Google Scholar] [CrossRef]
- Samavarchi-Tehrani, P.; Golipour, A.; David, L.; Sung, H.; Beyer, T.A.; Datti, A.; Woltjen, K.; Nagy, A.; Wrana, J.L. Functional Genomics Reveals a BMP-Driven Mesenchymal-to-Epithelial Transition in the Initiation of Somatic Cell Reprogramming. Cell Stem Cell 2010, 7, 64–77. [Google Scholar] [CrossRef]
- Trinh, K.; Storm, D.R. Detection of Odorants through the Main Olfactory Epithelium and Vomeronasal Organ of Mice. Nutr. Rev. 2004, 62, S189–S192. [Google Scholar] [CrossRef]
- Choi, P.S.; Zakhary, L.; Choi, W.-Y.; Caron, S.; Alvarez-Saavedra, E.; Miska, E.A.; McManus, M.; Harfe, B.; Giraldez, A.J.; Horvitz, H.R.; et al. Members of the MiRNA-200 Family Regulate Olfactory Neurogenesis. Neuron 2008, 57, 41–55. [Google Scholar] [CrossRef]
- Morante, J.; Vallejo, D.M.; Desplan, C.; Dominguez, M. Conserved MiR-8/MiR-200 Defines a Glial Niche That Controls Neuroepithelial Expansion and Neuroblast Transition. Dev. Cell 2013, 27, 174–187. [Google Scholar] [CrossRef]
- Hyun, S.; Lee, J.H.; Jin, H.; Nam, J.; Namkoong, B.; Lee, G.; Chung, J.; Kim, V.N. Conserved MicroRNA MiR-8/MiR-200 and Its Target USH/FOG2 Control Growth by Regulating PI3K. Cell 2009, 139, 1096–1108. [Google Scholar] [CrossRef]
- Jin, H.; Kim, V.N.; Hyun, S. Conserved MicroRNA MiR-8 Controls Body Size in Response to Steroid Signaling in Drosophila. Genes Dev. 2012, 26, 1427–1432. [Google Scholar] [CrossRef]
- Kennell, J.A.; Cadigan, K.M.; Shakhmantsir, I.; Waldron, E.J. The MicroRNA MiR-8 Is a Positive Regulator of Pigmentation and Eclosion in Drosophila. Dev. Dyn. 2012, 241, 161–168. [Google Scholar] [CrossRef]
- Lucas, K.J.; Roy, S.; Ha, J.; Gervaise, A.L.; Kokoza, V.A.; Raikhel, A.S. MicroRNA-8 Targets the Wingless Signaling Pathway in the Female Mosquito Fat Body to Regulate Reproductive Processes. Proc. Natl. Acad. Sci. USA 2015, 112, 1440–1445. [Google Scholar] [CrossRef]
- Pentón-Rol, G.; Cervantes-Llanos, M. Report on the Symposium “Molecular Mechanisms Involved in Neurodegeneration”. Behav. Sci. 2018, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Karres, J.S.; Hilgers, V.; Carrera, I.; Treisman, J.; Cohen, S.M. The Conserved MicroRNA MiR-8 Tunes Atrophin Levels to Prevent Neurodegeneration in Drosophila. Cell 2007, 131, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Dong, L.; Liu, N.; Luo, X.; He, Z. Mir-141-3p Regulates Apoptosis and Mitochondrial Membrane Potential via Targeting Sirtuin1 in a 1-Methyl-4-Phenylpyridinium in Vitro Model of Parkinson’s Disease. BioMed Res. Int. 2020, 2020, e7239895. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-S.; Liu, W.; Lu, G.-X. MiR-200a-3p Promotes b-Amyloid-Induced Neuronal Apoptosis through down-Regulation of SIRT1 in Alzheimer’s Disease. J. Biosci. 2017, 42, 397–404. [Google Scholar] [CrossRef]
- Liu, C.-G.; Wang, J.-L.; Li, L.; Xue, L.-X.; Zhang, Y.-Q.; Wang, P.-C. MicroRNA-135a and -200b, Potential Biomarkers for Alzheimer׳s Disease, Regulate β Secretase and Amyloid Precursor Protein. Brain Res. 2014, 1583, 55–64. [Google Scholar] [CrossRef]
- Liu, R.; Wang, M.; Su, L.; Li, X.; Zhao, S.; Yu, M. The Expression Pattern of MicroRNAs and the Associated Pathways Involved in the Development of Porcine Placental Folds That Contribute to the Expansion of the Exchange Surface Area. Biol. Reprod. 2015, 93, 62. [Google Scholar] [CrossRef]
- Higaki, S.; Muramatsu, M.; Matsuda, A.; Matsumoto, K.; Satoh, J.-I.; Michikawa, M.; Niida, S. Defensive Effect of MicroRNA-200b/c against Amyloid-Beta Peptide-Induced Toxicity in Alzheimer’s Disease Models. PLoS ONE 2018, 13, e0196929. [Google Scholar] [CrossRef]
- Salimian, N.; Peymani, M.; Ghaedi, K.; Nasr Esfahani, M.H. Modulation in MiR-200a/SIRT1axis Is Associated with Apoptosis in MPP+-Induced SH-SY5Y Cells. Gene 2018, 674, 25–30. [Google Scholar] [CrossRef]
- Rostamian Delavar, M.; Baghi, M.; Safaeinejad, Z.; Kiani-Esfahani, A.; Ghaedi, K.; Nasr-Esfahani, M.H. Differential Expression of MiR-34a, MiR-141, and MiR-9 in MPP+-Treated Differentiated PC12 Cells as a Model of Parkinson’s Disease. Gene 2018, 662, 54–65. [Google Scholar] [CrossRef]
- Svetoni, F.; De Paola, E.; La Rosa, P.; Mercatelli, N.; Caporossi, D.; Sette, C.; Paronetto, M.P. Post-Transcriptional Regulation of FUS and EWS Protein Expression by MiR-141 during Neural Differentiation. Hum. Mol. Genet. 2017, 26, 2732–2746. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, Y.-C.; Mullane, P.; Ji, Y.J.; Liu, H.; He, L.; Arora, A.; Hwang, H.-Y.; Alessi, A.F.; Niaki, A.G.; et al. FUS Regulates Activity of MicroRNA-Mediated Gene Silencing. Mol. Cell 2018, 69, 787–801.e8. [Google Scholar] [CrossRef]
- Wu, Q.; Ye, X.; Xiong, Y.; Zhu, H.; Miao, J.; Zhang, W.; Wan, J. The Protective Role of MicroRNA-200c in Alzheimer’s Disease Pathologies Is Induced by Beta Amyloid-Triggered Endoplasmic Reticulum Stress. Front. Mol. Neurosci. 2016, 9, 140. [Google Scholar] [CrossRef]
- Vinters, H.V. Emerging Concepts in Alzheimer’s Disease. Annu. Rev. Pathol. 2015, 10, 291–319. [Google Scholar] [CrossRef]
- Cornejo, V.H.; Hetz, C. The Unfolded Protein Response in Alzheimer’s Disease. Semin. Immunopathol. 2013, 35, 277–292. [Google Scholar] [CrossRef]
- Schipper, H.M.; Maes, O.C.; Chertkow, H.M.; Wang, E. MicroRNA Expression in Alzheimer Blood Mononuclear Cells. Gene Regul. Syst. Biol. 2007, 1, 263–274. [Google Scholar] [CrossRef]
- Shen, L.; Ji, H.-F. Associations between Homocysteine, Folic Acid, Vitamin B12 and Alzheimer’s Disease: Insights from Meta-Analyses. J. Alzheimer’s Dis 2015, 46, 777–790. [Google Scholar] [CrossRef]
- Goh, S.Y.; Chao, Y.X.; Dheen, S.T.; Tan, E.-K.; Tay, S.S.-W. Role of MicroRNAs in Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 5649. [Google Scholar] [CrossRef]
- Jankovic, J. Parkinson’s Disease: Clinical Features and Diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Hallett, P.J.; Engelender, S.; Isacson, O. Lipid and Immune Abnormalities Causing Age-Dependent Neurodegeneration and Parkinson’s Disease. J. Neuroinflamm. 2019, 16, 153. [Google Scholar] [CrossRef]
- Li, J.; Uversky, V.N.; Fink, A.L. Conformational Behavior of Human Alpha-Synuclein Is Modulated by Familial Parkinson’s Disease Point Mutations A30P and A53T. Neurotoxicology 2002, 23, 553–567. [Google Scholar] [CrossRef]
- Puentes, L.N.; Lengyel-Zhand, Z.; Lee, J.Y.; Hsieh, C.-J.; Schneider, M.E.; Edwards, K.J.; Luk, K.C.; Lee, V.M.-Y.; Trojanowski, J.Q.; Mach, R.H. Poly (ADP-Ribose) Interacts With Phosphorylated α-Synuclein in Post Mortem PD Samples. Front. Aging Neurosci. 2021, 13, 704041. [Google Scholar] [CrossRef]
- Mo, M.; Xiao, Y.; Huang, S.; Cen, L.; Chen, X.; Zhang, L.; Luo, Q.; Li, S.; Yang, X.; Lin, X.; et al. MicroRNA Expressing Profiles in A53T Mutant Alpha-Synuclein Transgenic Mice and Parkinsonian. Oncotarget 2017, 8, 15–28. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Wee, H.L.; Chan, Y.-H.; Seah, S.H.; Au, W.L.; Lau, P.N.; Pica, E.C.; Li, S.C.; Luo, N.; Tan, L.C.S. Progression of Parkinson’s Disease as Evaluated by Hoehn and Yahr Stage Transition Times. Mov. Disord. 2010, 25, 710–716. [Google Scholar] [CrossRef]
- Singh, P.; Hanson, P.S.; Morris, C.M. SIRT1 Ameliorates Oxidative Stress Induced Neural Cell Death and Is Down-Regulated in Parkinson’s Disease. BMC Neurosci. 2017, 18, 46. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-L.; Mostoslavsky, R.; Saito, S.; Manis, J.P.; Gu, Y.; Patel, P.; Bronson, R.; Appella, E.; Alt, F.W.; Chua, K.F. Developmental Defects and P53 Hyperacetylation in Sir2 Homolog (SIRT1)-Deficient Mice. Proc. Natl. Acad. Sci. USA 2003, 100, 10794–10799. [Google Scholar] [CrossRef]
- Vaziri, H.; Dessain, S.K.; Eaton, E.N.; Imai, S.-I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. HSIR2SIRT1 Functions as an NAD-Dependent P53 Deacetylase. Cell 2001, 107, 149–159. [Google Scholar] [CrossRef]
- Kim, E.-J.; Kho, J.-H.; Kang, M.-R.; Um, S.-J. Active Regulator of SIRT1 Cooperates with SIRT1 and Facilitates Suppression of P53 Activity. Mol. Cell 2007, 28, 277–290. [Google Scholar] [CrossRef]
- Talepoor Ardakani, M.; Rostamian Delavar, M.; Baghi, M.; Nasr-Esfahani, M.H.; Kiani-Esfahani, A.; Ghaedi, K. Upregulation of MiR-200a and MiR-204 in MPP+ -Treated Differentiated PC12 Cells as a Model of Parkinson’s Disease. Mol. Genet. Genom. Med. 2019, 7, e548. [Google Scholar] [CrossRef]
- Dini Modigliani, S.; Morlando, M.; Errichelli, L.; Sabatelli, M.; Bozzoni, I. An ALS-Associated Mutation in the FUS 3’-UTR Disrupts a MicroRNA-FUS Regulatory Circuitry. Nat. Commun. 2014, 5, 4335. [Google Scholar] [CrossRef]
- Figueroa-Romero, C.; Hur, J.; Lunn, J.S.; Paez-Colasante, X.; Bender, D.E.; Yung, R.; Sakowski, S.A.; Feldman, E.L. Expression of MicroRNAs in Human Post-Mortem Amyotrophic Lateral Sclerosis Spinal Cords Provides Insight into Disease Mechanisms. Mol. Cell Neurosci. 2016, 71, 34–45. [Google Scholar] [CrossRef]
- Taylor, J.P.; Brown, R.H.; Cleveland, D.W. Decoding ALS: From Genes to Mechanism. Nature 2016, 539, 197–206. [Google Scholar] [CrossRef]
- Zhou, F.; Zhang, C.; Guan, Y.; Chen, Y.; Lu, Q.; Jie, L.; Gao, H.; Du, H.; Zhang, H.; Liu, Y.; et al. Screening the Expression Characteristics of Several MiRNAs in G93A-SOD1 Transgenic Mouse: Altered Expression of MiRNA-124 Is Associated with Astrocyte Differentiation by Targeting Sox2 and Sox9. J. Neurochem. 2018, 145, 51–67. [Google Scholar] [CrossRef]
- Brownlee, W.J.; Hardy, T.A.; Fazekas, F.; Miller, D.H. Diagnosis of Multiple Sclerosis: Progress and Challenges. Lancet 2017, 389, 1336–1346. [Google Scholar] [CrossRef]
- Milo, R.; Kahana, E. Multiple Sclerosis: Geoepidemiology, Genetics and the Environment. Autoimmun. Rev. 2010, 9, A387–A394. [Google Scholar] [CrossRef]
- Waubant, E.; Lucas, R.; Mowry, E.; Graves, J.; Olsson, T.; Alfredsson, L.; Langer-Gould, A. Environmental and Genetic Risk Factors for MS: An Integrated Review. Ann. Clin. Transl. Neurol. 2019, 6, 1905–1922. [Google Scholar] [CrossRef] [PubMed]
- Naghavian, R.; Ghaedi, K.; Kiani-Esfahani, A.; Ganjalikhani-Hakemi, M.; Etemadifar, M.; Nasr-Esfahani, M.H. MiR-141 and MiR-200a, Revelation of New Possible Players in Modulation of Th17/Treg Differentiation and Pathogenesis of Multiple Sclerosis. PLoS ONE 2015, 10, e0124555. [Google Scholar] [CrossRef] [PubMed]
- Izadi, M.; Tahmasebi, S.; Pustokhina, I.; Yumashev, A.V.; Lakzaei, T.; Alvanegh, A.G.; Roshangar, L.; Dadashpour, M.; Yousefi, M.; Ahmadi, M. Changes in Th17 Cells Frequency and Function after Ozone Therapy Used to Treat Multiple Sclerosis Patients. Mult. Scler. Relat. Disord. 2020, 46, 102466. [Google Scholar] [CrossRef] [PubMed]
- Vallabh, S.M.; Minikel, E.V.; Schreiber, S.L.; Lander, E.S. Towards a Treatment for Genetic Prion Disease: Trials and Biomarkers. Lancet Neurol. 2020, 19, 361–368. [Google Scholar] [CrossRef]
- Thompson, A.G.B.; Lowe, J.; Fox, Z.; Lukic, A.; Porter, M.-C.; Ford, L.; Gorham, M.; Gopalakrishnan, G.S.; Rudge, P.; Walker, A.S.; et al. The Medical Research Council Prion Disease Rating Scale: A New Outcome Measure for Prion Disease Therapeutic Trials Developed and Validated Using Systematic Observational Studies. Brain 2013, 136, 1116–1127. [Google Scholar] [CrossRef]
- Boese, A.S.; Saba, R.; Campbell, K.; Majer, A.; Medina, S.; Burton, L.; Booth, T.F.; Chong, P.; Westmacott, G.; Dutta, S.M.; et al. MicroRNA Abundance Is Altered in Synaptoneurosomes during Prion Disease. Mol. Cell Neurosci. 2016, 71, 13–24. [Google Scholar] [CrossRef]
- Maisonneuve, P.; Marshall, B.C.; Knapp, E.A.; Lowenfels, A.B. Cancer Risk in Cystic Fibrosis: A 20-Year Nationwide Study From the United States. J. Natl. Cancer Inst. 2013, 105, 122–129. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The Basics of Epithelial-Mesenchymal Transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Li, M.; Luan, F.; Zhao, Y.; Hao, H.; Zhou, Y.; Han, W.; Fu, X. Epithelial-Mesenchymal Transition: An Emerging Target in Tissue Fibrosis. Exp. Biol. Med. 2016, 241, 1–13. [Google Scholar] [CrossRef]
- Oba, S.; Kumano, S.; Suzuki, E.; Nishimatsu, H.; Takahashi, M.; Takamori, H.; Kasuya, M.; Ogawa, Y.; Sato, K.; Kimura, K.; et al. MiR-200b Precursor Can Ameliorate Renal Tubulointerstitial Fibrosis. PLoS ONE 2010, 5, e13614. [Google Scholar] [CrossRef]
- Hajarnis, S.; Yheskel, M.; Williams, D.; Brefort, T.; Glaudemans, B.; Debaix, H.; Baum, M.; Devuyst, O.; Patel, V. Suppression of MicroRNA Activity in Kidney Collecting Ducts Induces Partial Loss of Epithelial Phenotype and Renal Fibrosis. JASN 2018, 29, 518–531. [Google Scholar] [CrossRef]
- Kato, M.; Arce, L.; Wang, M.; Putta, S.; Lanting, L.; Natarajan, R. A MicroRNA Circuit Mediates Transforming Growth Factor-Β1 Autoregulation in Renal Glomerular Mesangial Cells. Kidney Int. 2011, 80, 358–368. [Google Scholar] [CrossRef]
- Xiong, M.; Jiang, L.; Zhou, Y.; Qiu, W.; Fang, L.; Tan, R.; Wen, P.; Yang, J. The MiR-200 Family Regulates TGF-Β1-Induced Renal Tubular Epithelial to Mesenchymal Transition through Smad Pathway by Targeting ZEB1 and ZEB2 Expression. Am. J. Physiol. Renal. Physiol. 2012, 302, F369–F379. [Google Scholar] [CrossRef]
- Huang, Y.; Tong, J.; He, F.; Yu, X.; Fan, L.; Hu, J.; Tan, J.; Chen, Z. MiR-141 Regulates TGF-Β1-Induced Epithelial-Mesenchymal Transition through Repression of HIPK2 Expression in Renal Tubular Epithelial Cells. Int. J. Mol. Med. 2015, 35, 311–318. [Google Scholar] [CrossRef]
- Yang, S.; Banerjee, S.; de Freitas, A.; Sanders, Y.Y.; Ding, Q.; Matalon, S.; Thannickal, V.J.; Abraham, E.; Liu, G. Participation of MiR-200 in Pulmonary Fibrosis. Am. J. Pathol. 2012, 180, 484–493. [Google Scholar] [CrossRef]
- Chilosi, M.; Caliò, A.; Rossi, A.; Gilioli, E.; Pedica, F.; Montagna, L.; Pedron, S.; Confalonieri, M.; Doglioni, C.; Ziesche, R.; et al. Epithelial to Mesenchymal Transition-Related Proteins ZEB1, β-Catenin, and β-Tubulin-III in Idiopathic Pulmonary Fibrosis. Mod. Pathol. 2017, 30, 26–38. [Google Scholar] [CrossRef]
- Moimas, S.; Salton, F.; Kosmider, B.; Ring, N.; Volpe, M.C.; Bahmed, K.; Braga, L.; Rehman, M.; Vodret, S.; Graziani, M.L.; et al. MiR-200 Family Members Reduce Senescence and Restore Idiopathic Pulmonary Fibrosis Type II Alveolar Epithelial Cell Transdifferentiation. ERJ Open Res. 2019, 5, 00138–02019. [Google Scholar] [CrossRef]
- Hassan, Z.K.; Al-Olayan, E.M. Curcumin Reorganizes MiRNA Expression in a Mouse Model of Liver Fibrosis. Asian Pac. J. Cancer Prev. 2012, 13, 5405–5408. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, Z.; Guan, L.; Ao, R. GRHL2 Induces Liver Fibrosis and Intestinal Mucosal Barrier Dysfunction in Non-Alcoholic Fatty Liver Disease via MicroRNA-200 and the MAPK Pathway. J. Cell Mol. Med. 2020, 24, 6107–6119. [Google Scholar] [CrossRef]
- Sundararajan, V.; Pang, Q.Y.; Choolani, M.; Huang, R.Y.-J. Spotlight on the Granules (Grainyhead-Like Proteins)—From an Evolutionary Conserved Controller of Epithelial Trait to Pioneering the Chromatin Landscape. Front. Mol. Biosci. 2020, 7, 213. [Google Scholar] [CrossRef]
- Feng, X.; Wang, Z.; Fillmore, R.; Xi, Y. MiR-200, a New Star MiRNA in Human Cancer. Cancer Lett. 2014, 344, 166–173. [Google Scholar] [CrossRef]
- Lamouille, S.; Subramanyam, D.; Blelloch, R.; Derynck, R. Regulation of Epithelial-Mesenchymal and Mesenchymal-Epithelial Transitions by Micrornas. Curr. Opin. Cell Biol. 2013, 25, 200–207. [Google Scholar] [CrossRef]
- Gollavilli, P.N.; Parma, B.; Siddiqui, A.; Yang, H.; Ramesh, V.; Napoli, F.; Schwab, A.; Natesan, R.; Mielenz, D.; Asangani, I.A.; et al. The Role of MiR-200b/c in Balancing EMT and Proliferation Revealed by an Activity Reporter. Oncogene 2021, 40, 2309. [Google Scholar] [CrossRef]
- Uhlmann, S.; Zhang, J.D.; Schwäger, A.; Mannsperger, H.; Riazalhosseini, Y.; Burmester, S.; Ward, A.; Korf, U.; Wiemann, S.; Sahin, Ö. MiR-200bc/429 Cluster Targets PLCγ1 and Differentially Regulates Proliferation and EGF-Driven Invasion than MiR-200a/141 in Breast Cancer. Oncogene 2010, 29, 4297–4306. [Google Scholar] [CrossRef]
- Cochrane, D.R.; Spoelstra, N.S.; Howe, E.N.; Nordeen, S.K.; Richer, J.K. MicroRNA-200c Mitigates Invasiveness and Restores Sensitivity to Microtubule-Targeting Chemotherapeutic Agents. Mol. Cancer Ther. 2009, 8, 1055–1066. [Google Scholar] [CrossRef]
- Ceppi, P.; Peter, M.E. MicroRNAs Regulate Both Epithelial-to-Mesenchymal Transition and Cancer Stem Cells. Oncogene 2014, 33, 269–278. [Google Scholar] [CrossRef]
- Noman, M.Z.; Janji, B.; Abdou, A.; Hasmim, M.; Terry, S.; Tan, T.Z.; Mami-Chouaib, F.; Thiery, J.P.; Chouaib, S. The Immune Checkpoint Ligand PD-L1 Is Upregulated in EMT-Activated Human Breast Cancer Cells by a Mechanism Involving ZEB-1 and MiR-200. OncoImmunology 2017, 6, e1263412. [Google Scholar] [CrossRef]
- Chen, L.; Gibbons, D.L.; Goswami, S.; Cortez, M.A.; Ahn, Y.-H.; Byers, L.A.; Zhang, X.; Yi, X.; Dwyer, D.; Lin, W.; et al. Metastasis Is Regulated via MicroRNA-200/ZEB1 Axis Control of Tumour Cell PD-L1 Expression and Intratumoral Immunosuppression. Nat. Commun. 2014, 5, 5241. [Google Scholar] [CrossRef]
- Raue, R.; Frank, A.-C.; Fuhrmann, D.C.; de la Cruz-Ojeda, P.; Rösser, S.; Bauer, R.; Cardamone, G.; Weigert, A.; Syed, S.N.; Schmid, T.; et al. MicroRNA-200c Attenuates the Tumor-Infiltrating Capacity of Macrophages. Biology 2022, 11, 349. [Google Scholar] [CrossRef]
- Hurteau, G.J.; Carlson, J.A.; Spivack, S.D.; Brock, G.J. Overexpression of the MicroRNA Hsa-MiR-200c Leads to Reduced Expression of Transcription Factor 8 and Increased Expression of E-Cadherin. Cancer Res. 2007, 67, 7972–7976. [Google Scholar] [CrossRef]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A Reciprocal Repression between ZEB1 and Members of the MiR-200 Family Promotes EMT and Invasion in Cancer Cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The MiR-200 Family and MiR-205 Regulate Epithelial to Mesenchymal Transition by Targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Lee, E.S.; Hu, G.; Kang, Y. The MiR-200 Family Inhibits Epithelial-Mesenchymal Transition and Cancer Cell Migration by Direct Targeting of E-Cadherin Transcriptional Repressors ZEB1 and ZEB2. J. Biol. Chem. 2008, 283, 14910–14914. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The MiR-200 Family Determines the Epithelial Phenotype of Cancer Cells by Targeting the E-Cadherin Repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A Double-Negative Feedback Loop between ZEB1-SIP1 and the MicroRNA-200 Family Regulates Epithelial-Mesenchymal Transition. Cancer Res. 2008, 68, 7846–7854. [Google Scholar] [CrossRef]
- Brabletz, S.; Brabletz, T. The ZEB/MiR-200 Feedback Loop—A Motor of Cellular Plasticity in Development and Cancer? EMBO Rep. 2010, 11, 670–677. [Google Scholar] [CrossRef]
- Brabletz, S.; Bajdak, K.; Meidhof, S.; Burk, U.; Niedermann, G.; Firat, E.; Wellner, U.; Dimmler, A.; Faller, G.; Schubert, J.; et al. The ZEB1/MiR-200 Feedback Loop Controls Notch Signalling in Cancer Cells: ZEB1 Activates Notch Signalling. EMBO J. 2011, 30, 770–782. [Google Scholar] [CrossRef]
- Yang, Y.; Ahn, Y.-H.; Gibbons, D.L.; Zang, Y.; Lin, W.; Thilaganathan, N.; Alvarez, C.A.; Moreira, D.C.; Creighton, C.J.; Gregory, P.A.; et al. The Notch Ligand Jagged2 Promotes Lung Adenocarcinoma Metastasis through a MiR-200-Dependent Pathway in Mice. J. Clin. Investig. 2011, 121, 1373–1385. [Google Scholar] [CrossRef]
- Xue, B.; Chuang, C.H.; Prosser, H.M.; Fuziwara, C.S.; Chan, C.; Sahasrabudhe, N.; Kühn, M.; Wu, Y.; Chen, J.; Biton, A.; et al. MiR-200 Deficiency Promotes Lung Cancer Metastasis by Activating Notch Signaling in Cancer-Associated Fibroblasts. Genes Dev. 2021, 35, 1109–1122. [Google Scholar] [CrossRef]
- Sundararajan, V.; Gengenbacher, N.; Stemmler, M.P.; Kleemann, J.A.; Brabletz, T.; Brabletz, S. The ZEB1/MiR-200c Feedback Loop Regulates Invasion via Actin Interacting Proteins MYLK and TKS5. Oncotarget 2015, 6, 27083–27096. [Google Scholar] [CrossRef]
- Li, X.; Roslan, S.; Johnstone, C.N.; Wright, J.A.; Bracken, C.P.; Anderson, M.; Bert, A.G.; Selth, L.A.; Anderson, R.L.; Goodall, G.J.; et al. MiR-200 Can Repress Breast Cancer Metastasis through ZEB1-Independent but Moesin-Dependent Pathways. Oncogene 2014, 33, 4077–4088. [Google Scholar] [CrossRef]
- Jurmeister, S.; Baumann, M.; Balwierz, A.; Keklikoglou, I.; Ward, A.; Uhlmann, S.; Zhang, J.D.; Wiemann, S.; Sahin, Ö. MicroRNA-200c Represses Migration and Invasion of Breast Cancer Cells by Targeting Actin-Regulatory Proteins FHOD1 and PPM1F. Mol. Cell Biol. 2012, 32, 633–651. [Google Scholar] [CrossRef]
- Huang, R.Y.-J.; Wong, M.K.; Tan, T.Z.; Kuay, K.T.; Ng, A.H.C.; Chung, V.Y.; Chu, Y.-S.; Matsumura, N.; Lai, H.-C.; Lee, Y.F.; et al. An EMT Spectrum Defines an Anoikis-Resistant and Spheroidogenic Intermediate Mesenchymal State That Is Sensitive to e-Cadherin Restoration by a Src-Kinase Inhibitor, Saracatinib (AZD0530). Cell Death Dis. 2013, 4, e915. [Google Scholar] [CrossRef]
- Hong, T.; Watanabe, K.; Ta, C.H.; Villarreal-Ponce, A.; Nie, Q.; Dai, X. An Ovol2-Zeb1 Mutual Inhibitory Circuit Governs Bidirectional and Multi-Step Transition between Epithelial and Mesenchymal States. PLoS Comput. Biol. 2015, 11, e1004569. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the Tumour Transition States Occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Grosse-Wilde, A.; Fouquier d’Hérouël, A.; McIntosh, E.; Ertaylan, G.; Skupin, A.; Kuestner, R.E.; del Sol, A.; Walters, K.-A.; Huang, S. Stemness of the Hybrid Epithelial/Mesenchymal State in Breast Cancer and Its Association with Poor Survival. PLoS ONE 2015, 10, e0126522. [Google Scholar] [CrossRef]
- Yamashita, N.; Tokunaga, E.; Iimori, M.; Inoue, Y.; Tanaka, K.; Kitao, H.; Saeki, H.; Oki, E.; Maehara, Y. Epithelial Paradox: Clinical Significance of Coexpression of E-Cadherin and Vimentin With Regard to Invasion and Metastasis of Breast Cancer. Clin. Breast Cancer 2018, 18, e1003–e1009. [Google Scholar] [CrossRef]
- Ruscetti, M.; Quach, B.; Dadashian, E.L.; Mulholland, D.J.; Wu, H. Tracking and Functional Characterization of Epithelial-Mesenchymal Transition and Mesenchymal Tumor Cells during Prostate Cancer Metastasis. Cancer Res. 2015, 75, 2749–2759. [Google Scholar] [CrossRef]
- Fustaino, V.; Presutti, D.; Colombo, T.; Cardinali, B.; Papoff, G.; Brandi, R.; Bertolazzi, P.; Felici, G.; Ruberti, G. Characterization of Epithelial-Mesenchymal Transition Intermediate/Hybrid Phenotypes Associated to Resistance to EGFR Inhibitors in Non-Small Cell Lung Cancer Cell Lines. Oncotarget 2017, 8, 103340–103363. [Google Scholar] [CrossRef]
- Kröger, C.; Afeyan, A.; Mraz, J.; Eaton, E.N.; Reinhardt, F.; Khodor, Y.L.; Thiru, P.; Bierie, B.; Ye, X.; Burge, C.B.; et al. Acquisition of a Hybrid E/M State Is Essential for Tumorigenicity of Basal Breast Cancer Cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7353–7362. [Google Scholar] [CrossRef]
- Jolly, M.K.; Somarelli, J.A.; Sheth, M.; Biddle, A.; Tripathi, S.C.; Armstrong, A.J.; Hanash, S.M.; Bapat, S.A.; Rangarajan, A.; Levine, H. Hybrid Epithelial/Mesenchymal Phenotypes Promote Metastasis and Therapy Resistance across Carcinomas. Pharmacol. Ther. 2019, 194, 161–184. [Google Scholar] [CrossRef] [PubMed]
- Siemens, H.; Jackstadt, R.; Hünten, S.; Kaller, M.; Menssen, A.; Götz, U.; Hermeking, H. MiR-34 and SNAIL Form a Double-Negative Feedback Loop to Regulate Epithelial-Mesenchymal Transitions. Cell Cycle 2011, 10, 4256–4271. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Jolly, M.K.; Levine, H.; Onuchic, J.N.; Ben-Jacob, E. MicroRNA-Based Regulation of Epithelial-Hybrid-Mesenchymal Fate Determination. Proc. Natl. Acad. Sci. USA 2013, 110, 18144–18149. [Google Scholar] [CrossRef]
- Jolly, M.K.; Tripathi, S.C.; Jia, D.; Mooney, S.M.; Celiktas, M.; Hanash, S.M.; Mani, S.A.; Pienta, K.J.; Ben-Jacob, E.; Levine, H. Stability of the Hybrid Epithelial/Mesenchymal Phenotype. Oncotarget 2016, 7, 27067–27084. [Google Scholar] [CrossRef]
- Burger, G.A.; Danen, E.H.J.; Beltman, J.B. Deciphering Epithelial–Mesenchymal Transition Regulatory Networks in Cancer through Computational Approaches. Front. Oncol. 2017, 7, 162. [Google Scholar] [CrossRef]
- Jia, W.; Tripathi, S.; Chakraborty, P.; Chedere, A.; Rangarajan, A.; Levine, H.; Jolly, M.K. Epigenetic Feedback and Stochastic Partitioning during Cell Division Can Drive Resistance to EMT. Oncotarget 2020, 11, 2611–2624. [Google Scholar] [CrossRef]
- Garinet, S.; Didelot, A.; Denize, T.; Perrier, A.; Beinse, G.; Leclere, J.-B.; Oudart, J.-B.; Gibault, L.; Badoual, C.; Le Pimpec-Barthes, F.; et al. Clinical Assessment of the MiR-34, MiR-200, ZEB1 and SNAIL EMT Regulation Hub Underlines the Differential Prognostic Value of EMT MiRs to Drive Mesenchymal Transition and Prognosis in Resected NSCLC. Br. J. Cancer 2021, 125, 1544–1551. [Google Scholar] [CrossRef]
- Deshmukh, A.P.; Vasaikar, S.V.; Tomczak, K.; Tripathi, S.; den Hollander, P.; Arslan, E.; Chakraborty, P.; Soundararajan, R.; Jolly, M.K.; Rai, K.; et al. Identification of EMT Signaling Cross-Talk and Gene Regulatory Networks by Single-Cell RNA Sequencing. Proc. Natl. Acad. Sci. USA 2021, 118, e2102050118. [Google Scholar] [CrossRef]
- Mateescu, B.; Batista, L.; Cardon, M.; Gruosso, T.; de Feraudy, Y.; Mariani, O.; Nicolas, A.; Meyniel, J.-P.; Cottu, P.; Sastre-Garau, X.; et al. MiR-141 and MiR-200a Act on Ovarian Tumorigenesis by Controlling Oxidative Stress Response. Nat. Med. 2011, 17, 1627–1635. [Google Scholar] [CrossRef]
- Hua, Y.; Choi, P.-W.; Trachtenberg, A.J.; Ng, A.C.; Kuo, W.P.; Ng, S.-K.; Dinulescu, D.M.; Matzuk, M.M.; Berkowitz, R.S.; Ng, S.-W. Epithelialization of Mouse Ovarian Tumor Cells Originating in the Fallopian Tube Stroma. Oncotarget 2016, 7, 66077–66086. [Google Scholar] [CrossRef]
- Ansenberger, K.; Zhuge, Y.; Lagman, J.A.J.; Richards, C.; Barua, A.; Bahr, J.M.; Hales, D.B. E-Cadherin Expression in Ovarian Cancer in the Laying Hen, Gallus Domesticus, Compared to Human Ovarian Cancer. Gynecol. Oncol. 2009, 113, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Hales, K.H.; Speckman, S.C.; Kurrey, N.K.; Hales, D.B. Uncovering Molecular Events Associated with the Chemosuppressive Effects of Flaxseed: A Microarray Analysis of the Laying Hen Model of Ovarian Cancer. BMC Genom. 2014, 15, 709. [Google Scholar] [CrossRef] [PubMed]
- Choi, P.-W.; So, W.W.; Yang, J.; Liu, S.; Tong, K.K.; Kwan, K.M.; Kwok, J.S.-L.; Tsui, S.K.W.; Ng, S.-K.; Hales, K.H.; et al. MicroRNA-200 Family Governs Ovarian Inclusion Cyst Formation and Mode of Ovarian Cancer Spread. Oncogene 2020, 39, 4045–4060. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Wang, J.; Yang, P.; Lu, Q.; Zhang, T.; Yang, Y. MicroRNA-200 Promotes Lung Cancer Cell Growth through FOG2-Independent AKT Activation. IUBMB Life 2015, 67, 720–725. [Google Scholar] [CrossRef]
- Damaghi, M.; West, J.; Robertson-Tessi, M.; Xu, L.; Ferrall-Fairbanks, M.C.; Stewart, P.A.; Persi, E.; Fridley, B.L.; Altrock, P.M.; Gatenby, R.A.; et al. The Harsh Microenvironment in Early Breast Cancer Selects for a Warburg Phenotype. Proc. Natl. Acad. Sci. USA 2021, 118, e2011342118. [Google Scholar] [CrossRef]
- Sinha, V.C.; Piwnica-Worms, H. Intratumoral Heterogeneity in Ductal Carcinoma In Situ: Chaos and Consequence. J. Mammary Gland Biol. Neoplasia 2018, 23, 191–205. [Google Scholar] [CrossRef]
- Haakensen, V.D.; Nygaard, V.; Greger, L.; Aure, M.R.; Fromm, B.; Bukholm, I.R.K.; Lüders, T.; Chin, S.-F.; Git, A.; Caldas, C.; et al. Subtype-Specific Micro-RNA Expression Signatures in Breast Cancer Progression. Int. J. Cancer 2016, 139, 1117–1128. [Google Scholar] [CrossRef]
- Dykxhoorn, D.M.; Wu, Y.; Xie, H.; Yu, F.; Lal, A.; Petrocca, F.; Martinvalet, D.; Song, E.; Lim, B.; Lieberman, J. MiR-200 Enhances Mouse Breast Cancer Cell Colonization to Form Distant Metastases. PLoS ONE 2009, 4, e7181. [Google Scholar] [CrossRef]
- Korpal, M.; Ell, B.J.; Buffa, F.M.; Ibrahim, T.; Blanco, M.A.; Celià-Terrassa, T.; Mercatali, L.; Khan, Z.; Goodarzi, H.; Hua, Y.; et al. Direct Targeting of Sec23a by MiR-200s Influences Cancer Cell Secretome and Promotes Metastatic Colonization. Nat. Med. 2011, 17, 1101–1108. [Google Scholar] [CrossRef]
- Ast, V.; Korda, T.; Oswald, M.; Kolte, A.; Eisel, D.; Osen, W.; Eichmüller, S.B.; Berndt, A.; König, R. MiR-192, MiR-200c and MiR-17 Are Fibroblast-Mediated Inhibitors of Colorectal Cancer Invasion. Oncotarget 2018, 9, 35559–35580. [Google Scholar] [CrossRef]
- Oghabi Bakhshaiesh, T.; Esmaeili, R. Effects of Noncoding RNAs in Radiotherapy Response in Breast Cancer: A Systematic Review. Cell Cycle 2022, 21, 883–893. [Google Scholar] [CrossRef]
- Kozak, J.; Jonak, K.; Maciejewski, R. The Function of MiR-200 Family in Oxidative Stress Response Evoked in Cancer Chemotherapy and Radiotherapy. Biomed. Pharm. 2020, 125, 110037. [Google Scholar] [CrossRef]
- Lin, J.; Liu, C.; Gao, F.; Mitchel, R.E.J.; Zhao, L.; Yang, Y.; Lei, J.; Cai, J. MiR-200c Enhances Radiosensitivity of Human Breast Cancer Cells. J. Cell Biochem. 2013, 114, 606–615. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, T.; Yuan, Y.; Guo, Z.; Xie, G.; Du, S.; Lin, X.; Xu, Z.; Liu, M.; Wang, W.; et al. MiR-200c Inhibits Autophagy and Enhances Radiosensitivity in Breast Cancer Cells by Targeting UBQLN1. Int. J. Cancer 2015, 136, 1003–1012. [Google Scholar] [CrossRef]
- Wang, B.; Zheng, J.; Li, R.; Tian, Y.; Lin, J.; Liang, Y.; Sun, Q.; Xu, A.; Zheng, R.; Liu, M.; et al. Long Noncoding RNA LINC02582 Acts Downstream of MiR-200c to Promote Radioresistance through CHK1 in Breast Cancer Cells. Cell Death Dis. 2019, 10, 764. [Google Scholar] [CrossRef]
- Cortez, M.A.; Valdecanas, D.; Zhang, X.; Zhan, Y.; Bhardwaj, V.; Calin, G.A.; Komaki, R.; Giri, D.K.; Quini, C.C.; Wolfe, T.; et al. Therapeutic Delivery of MiR-200c Enhances Radiosensitivity in Lung Cancer. Mol. Ther. 2014, 22, 1494–1503. [Google Scholar] [CrossRef]
- Du, M.; Wang, J.; Chen, H.; Wang, S.; Chen, L.; Xu, Y.; Su, F.; Lu, X. MicroRNA-200a Suppresses Migration and Invasion and Enhances the Radiosensitivity of NSCLC Cells by Inhibiting the HGF/C-Met Signaling Pathway. Oncol. Rep. 2019, 41, 1497–1508. [Google Scholar] [CrossRef]
- Greither, T.; Vorwerk, F.; Kappler, M.; Bache, M.; Taubert, H.; Kuhnt, T.; Hey, J.; Eckert, A.W. Salivary MiR-93 and MiR-200a as Post-Radiotherapy Biomarkers in Head and Neck Squamous Cell Carcinoma. Oncol. Rep. 2017, 38, 1268–1275. [Google Scholar] [CrossRef]
- Tao, J.; Fan, M.; Zhou, D.; Hong, Y.; Zhang, J.; Liu, H.; Sharma, S.; Wang, G.; Dong, Q. MiR-200c Modulates the Pathogenesis of Radiation-Induced Oral Mucositis. Oxid. Med. Cell Longev. 2019, 2019, 2352079. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Lin, C.-H.; Chang, W.-L.; Lin, W.-D.; Pan, J.-K.; Wang, W.-J.; Su, B.-C.; Chung, H.-H.; Tsai, C.-H.; Lin, F.-C.; et al. Concurrent Chemoradiotherapy-Driven Cell Plasticity by MiR-200 Family Implicates the Therapeutic Response of Esophageal Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 4367. [Google Scholar] [CrossRef]
- Nilsen, A.; Hillestad, T.; Skingen, V.E.; Aarnes, E.-K.; Fjeldbo, C.S.; Hompland, T.; Evensen, T.S.; Stokke, T.; Kristensen, G.B.; Grallert, B.; et al. MiR-200a/b/-429 Downregulation Is a Candidate Biomarker of Tumor Radioresistance and Independent of Hypoxia in Locally Advanced Cervical Cancer. Mol. Oncol. 2022, 16, 1402–1419. [Google Scholar] [CrossRef]
- Loret, N.; Denys, H.; Tummers, P.; Berx, G. The Role of Epithelial-to-Mesenchymal Plasticity in Ovarian Cancer Progression and Therapy Resistance. Cancers 2019, 11, 838. [Google Scholar] [CrossRef]
- Liu, S.; Tetzlaff, M.T.; Cui, R.; Xu, X. MiR-200c Inhibits Melanoma Progression and Drug Resistance through Down-Regulation of Bmi-1. Am. J. Pathol. 2012, 181, 1823–1835. [Google Scholar] [CrossRef]
- Mezencev, R.; Wartell, R.M. Cisplatin Binds to Pre-MiR-200b and Impairs Its Processing to Mature MicroRNA. Neoplasma 2018, 65, 222–227. [Google Scholar] [CrossRef]
- Crudele, F.; Bianchi, N.; Astolfi, A.; Grassilli, S.; Brugnoli, F.; Terrazzan, A.; Bertagnolo, V.; Negrini, M.; Frassoldati, A.; Volinia, S. The Molecular Networks of MicroRNAs and Their Targets in the Drug Resistance of Colon Carcinoma. Cancers 2021, 13, 4355. [Google Scholar] [CrossRef]
- Meidhof, S.; Brabletz, S.; Lehmann, W.; Preca, B.-T.; Mock, K.; Ruh, M.; Schüler, J.; Berthold, M.; Weber, A.; Burk, U.; et al. ZEB1-associated Drug Resistance in Cancer Cells Is Reversed by the Class I HDAC Inhibitor Mocetinostat. EMBO Mol. Med. 2015, 7, 831–847. [Google Scholar] [CrossRef]
- San, K.; Horita, M.; Ganapathy, A.; Chinnadurai, G.; Ezekiel, U.R. Deregulated Expression of MicroRNA-200b/c and SUZ12, a Polycomb Repressive Complex 2 Subunit, in Chemoresistant Colorectal Cancer Cells. Genes Cancer 2017, 8, 673–681. [Google Scholar] [CrossRef]
- Knezevic, J.; Pfefferle, A.D.; Petrovic, I.; Greene, S.B.; Perou, C.M.; Rosen, J.M. Expression of MiR-200c in Claudin-Low Breast Cancer Alters Stem Cell Functionality, Enhances Chemosensitivity and Reduces Metastatic Potential. Oncogene 2015, 34, 5997–6006. [Google Scholar] [CrossRef]
- Heydari, K.; Saidijam, M.; Reza Sharifi, M.; Dermani, F.K.; Soleimani Asl, S.; Shabab, N.; Najafi, R. The Effect of MiR-200c Inhibition on Chemosensitivity (5- FluoroUracil) in Colorectal Cancer. Pathol. Oncol. Res. 2018, 24, 145–151. [Google Scholar] [CrossRef]
- Brozovic, A.; Duran, G.E.; Wang, Y.C.; Francisco, E.B.; Sikic, B.I. The MiR-200 Family Differentially Regulates Sensitivity to Paclitaxel and Carboplatin in Human Ovarian Carcinoma OVCAR-3 and MES-OV Cells. Mol. Oncol. 2015, 9, 1678–1693. [Google Scholar] [CrossRef]
- Haenisch, S.; Werk, A.N.; Cascorbi, I. MicroRNAs and Their Relevance to ABC Transporters. Br. J. Clin. Pharmacol. 2014, 77, 587. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.J.; Yang, L.; Hong, Q.; Kuang, X.Y.; Di, G.H.; Shao, Z.M. MicroRNA-200a Confers Chemoresistance by Antagonizing TP53INP1 and YAP1 in Human Breast Cancer. BMC Cancer 2018, 18, 74. [Google Scholar] [CrossRef] [PubMed]
- Tomasini, R.; Seux, M.; Nowak, J.; Bontemps, C.; Carrier, A.; Dagorn, J.C.; Pébusque, M.J.; Iovanna, J.L.; Dusetti, N.J. TP53INP1 Is a Novel P73 Target Gene That Induces Cell Cycle Arrest and Cell Death by Modulating P73 Transcriptional Activity. Oncogene 2005, 24, 8093–8104. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kim, C.; Kang, H.; Tak, H.; Ahn, S.; Yoon, S.K.; Kuh, H.J.; Kim, W.; Lee, E.K. MicroRNA-200a-3p Increases 5-Fluorouracil Resistance by Regulating Dual Specificity Phosphatase 6 Expression. Exp. Mol. Med. 2017, 49, e327. [Google Scholar] [CrossRef] [PubMed]
- Choi, P.-W.; Ng, S.-W. The Functions of MicroRNA-200 Family in Ovarian Cancer: Beyond Epithelial-Mesenchymal Transition. Int. J. Mol. Sci. 2017, 18, 1207. [Google Scholar] [CrossRef]
- Yu, S.-J.; Hu, J.-Y.; Kuang, X.-Y.; Luo, J.-M.; Hou, Y.-F.; Di, G.-H.; Wu, J.; Shen, Z.-Z.; Song, H.-Y.; Shao, Z.-M. MicroRNA-200a Promotes Anoikis Resistance and Metastasis by Targeting YAP1 in Human Breast Cancer. Clin. Cancer Res. 2013, 19, 1389–1399. [Google Scholar] [CrossRef]
- Van Jaarsveld, M.T.M.; Helleman, J.; Boersma, A.W.M.; Van Kuijk, P.F.; Van Ijcken, W.F.; Despierre, E.; Vergote, I.; Mathijssen, R.H.J.; Berns, E.M.J.J.; Verweij, J.; et al. MiR-141 Regulates KEAP1 and Modulates Cisplatin Sensitivity in Ovarian Cancer Cells. Oncogene 2012, 32, 4284–4293. [Google Scholar] [CrossRef]
- Eades, G.; Yang, M.; Yao, Y.; Zhang, Y.; Zhou, Q. MiR-200a Regulates Nrf2 Activation by Targeting Keap1 MRNA in Breast Cancer Cells. J. Biol. Chem. 2011, 286, 40725–40733. [Google Scholar] [CrossRef]
- Cavallari, I.; Ciccarese, F.; Sharova, E.; Urso, L.; Raimondi, V.; Silic-Benussi, M.; D’Agostino, D.M.; Ciminale, V. The MiR-200 Family of MicroRNAs: Fine Tuners of Epithelial-Mesenchymal Transition and Circulating Cancer Biomarkers. Cancers 2021, 13, 5874. [Google Scholar] [CrossRef]
- Chen, J.; Tian, W.; Cai, H.; He, H.; Deng, Y. Down-Regulation of MicroRNA-200c Is Associated with Drug Resistance in Human Breast Cancer. Med. Oncol. 2012, 29, 2527–2534. [Google Scholar] [CrossRef]
- Men, D.; Liang, Y.; Chen, L. Decreased Expression of MicroRNA-200b Is an Independent Unfavorable Prognostic Factor for Glioma Patients. Cancer Epidemiol. 2014, 38, 152–156. [Google Scholar] [CrossRef]
- Thi Chung Duong, T.; Nguyen, T.H.N.; Thi Ngoc Nguyen, T.; Huynh, L.H.; Ngo, H.P.; Thi Nguyen, H. Diagnostic and Prognostic Value of MiR-200 Family in Breast Cancer: A Meta-Analysis and Systematic Review. Cancer Epidemiol. 2022, 77, 102097. [Google Scholar] [CrossRef]
- Mei, Y.; Zheng, J.; Xiang, P.; Liu, C.; Fan, Y. Prognostic Value of the MiR-200 Family in Bladder Cancer: A Systematic Review and Meta-Analysis. Medicine 2020, 99, e22891. [Google Scholar] [CrossRef]
- Madhavan, D.; Zucknick, M.; Wallwiener, M.; Cuk, K.; Modugno, C.; Scharpff, M.; Schott, S.; Heil, J.; Turchinovich, A.; Yang, R.; et al. Circulating MiRNAs as Surrogate Markers for Circulating Tumor Cells and Prognostic Markers in Metastatic Breast Cancer. Clin. Cancer Res. 2012, 18, 5972–5982. [Google Scholar] [CrossRef]
- Madhavan, D.; Peng, C.; Wallwiener, M.; Zucknick, M.; Nees, J.; Schott, S.; Rudolph, A.; Riethdorf, S.; Trumpp, A.; Pantel, K.; et al. Circulating MiRNAs with Prognostic Value in Metastatic Breast Cancer and for Early Detection of Metastasis. Carcinogenesis 2016, 37, 461–470. [Google Scholar] [CrossRef]
- Maierthaler, M.; Benner, A.; Hoffmeister, M.; Surowy, H.; Jansen, L.; Knebel, P.; Chang-Claude, J.; Brenner, H.; Burwinkel, B. Plasma MiR-122 and MiR-200 Family Are Prognostic Markers in Colorectal Cancer. Int. J. Cancer Res. 2017, 140, 176–187. [Google Scholar] [CrossRef]
- Santasusagna, S.; Moreno, I.; Navarro, A.; Martinez Rodenas, F.; Hernández, R.; Castellano, J.J.; Muñoz, C.; Monzo, M. Prognostic Impact of MiR-200 Family Members in Plasma and Exosomes from Tumor-Draining versus Peripheral Veins of Colon Cancer Patients. Oncology 2018, 95, 309–318. [Google Scholar] [CrossRef]
- Reese, M.; Flammang, I.; Yang, Z.; Dhayat, S.A. Potential of Exosomal MicroRNA-200b as Liquid Biopsy Marker in Pancreatic Ductal Adenocarcinoma. Cancers 2020, 12, 197. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, L.; Cogdell, D.E.; Zheng, H.; Schetter, A.J.; Nykter, M.; Harris, C.C.; Chen, K.; Hamilton, S.R.; Zhang, W. Circulating Plasma MiR-141 Is a Novel Biomarker for Metastatic Colon Cancer and Predicts Poor Prognosis. PLoS ONE 2011, 6, e17745. [Google Scholar] [CrossRef]
- Tejero, R.; Navarro, A.; Campayo, M.; Viñolas, N.; Marrades, R.M.; Cordeiro, A.; Ruíz-Martínez, M.; Santasusagna, S.; Molins, L.; Ramirez, J.; et al. MiR-141 and MiR-200c as Markers of Overall Survival in Early Stage Non-Small Cell Lung Cancer Adenocarcinoma. PLoS ONE 2014, 9, e101899. [Google Scholar] [CrossRef]
- Záveský, L.; Jandáková, E.; Weinberger, V.; Minář, L.; Hanzíková, V.; Dušková, D.; Drábková, L.Z.; Svobodová, I.; Hořínek, A. Ascites-Derived Extracellular MicroRNAs as Potential Biomarkers for Ovarian Cancer. Reprod. Sci. 2019, 26, 510–522. [Google Scholar] [CrossRef]
- Márton, É.; Lukács, J.; Penyige, A.; Janka, E.; Hegedüs, L.; Soltész, B.; Méhes, G.; Póka, R.; Nagy, B.; Szilágyi, M. Circulating Epithelial-Mesenchymal Transition-Associated MiRNAs Are Promising Biomarkers in Ovarian Cancer. J. Biotechnol. 2019, 297, 58–65. [Google Scholar] [CrossRef]
- Meng, X.; Müller, V.; Milde-Langosch, K.; Trillsch, F.; Pantel, K.; Schwarzenbach, H. Diagnostic and Prognostic Relevance of Circulating Exosomal MiR-373, MiR-200a, MiR-200b and MiR-200c in Patients with Epithelial Ovarian Cancer. Oncotarget 2016, 7, 16923–16935. [Google Scholar] [CrossRef]
- Choi, P.-W.; Bahrampour, A.; Ng, S.-K.; Liu, S.K.; Qiu, W.; Xie, F.; Kuo, W.P.; Kwong, J.; Hales, K.H.; Hales, D.B.; et al. Characterization of MiR-200 Family Members as Blood Biomarkers for Human and Laying Hen Ovarian Cancer. Sci. Rep. 2020, 10, 20071. [Google Scholar] [CrossRef]
- Savolainen, K.; Scaravilli, M.; Ilvesmäki, A.; Staff, S.; Tolonen, T.; Mäenpää, J.U.; Visakorpi, T.; Auranen, A. Expression of the MiR-200 Family in Tumor Tissue, Plasma and Urine of Epithelial Ovarian Cancer Patients in Comparison to Benign Counterparts. BMC Res. Notes 2020, 13, 311. [Google Scholar] [CrossRef] [PubMed]
- Zuberi, M.; Mir, R.; Das, J.; Ahmad, I.; Javid, J.; Yadav, P.; Masroor, M.; Ahmad, S.; Ray, P.C.; Saxena, A. Expression of Serum MiR-200a, MiR-200b, and MiR-200c as Candidate Biomarkers in Epithelial Ovarian Cancer and Their Association with Clinicopathological Features. Clin. Transl. Oncol. 2015, 17, 779–787. [Google Scholar] [CrossRef]
- Kan, C.W.S.; Hahn, M.A.; Gard, G.B.; Maidens, J.; Huh, J.Y.; Marsh, D.J.; Howell, V.M. Elevated Levels of Circulating MicroRNA-200 Family Members Correlate with Serous Epithelial Ovarian Cancer. BMC Cancer 2012, 12, 627. [Google Scholar] [CrossRef]
- Oliveira, D.N.P.; Carlsen, A.L.; Heegaard, N.H.H.; Prahm, K.P.; Christensen, I.J.; Høgdall, C.K.; Høgdall, E.V. Diagnostic Plasma MiRNA-Profiles for Ovarian Cancer in Patients with Pelvic Mass. PLoS ONE 2019, 14, e0225249. [Google Scholar] [CrossRef] [PubMed]
- Toiyama, Y.; Hur, K.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum MiR-200c Is a Novel Prognostic and Metastasis-Predictive Biomarker in Patients with Colorectal Cancer. Ann. Surg. 2014, 259, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Stevic, I.; Müller, V.; Ni, Q.; Oliveira-Ferrer, L.; Pantel, K.; Schwarzenbach, H. Exosomal MicroRNAs as Tumor Markers in Epithelial Ovarian Cancer. Mol. Oncol. 2018, 12, 1935–1948. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Choi, M.C.; Jeong, J.-Y.; Hwang, S.; Jung, S.G.; Joo, W.D.; Park, H.; Song, S.H.; Lee, C.; Kim, T.H.; et al. Serum Exosomal MiRNA-145 and MiRNA-200c as Promising Biomarkers for Preoperative Diagnosis of Ovarian Carcinomas. J. Cancer 2019, 10, 1958–1967. [Google Scholar] [CrossRef]
- Shen, L.; Chen, G.; Xia, Q.; Shao, S.; Fang, H. Exosomal MiR-200 Family as Serum Biomarkers for Early Detection and Prognostic Prediction of Cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2019, 12, 3870–3876. [Google Scholar]
- Hulstaert, E.; Morlion, A.; Levanon, K.; Vandesompele, J.; Mestdagh, P. Candidate RNA Biomarkers in Biofluids for Early Diagnosis of Ovarian Cancer: A Systematic Review. Gynecol. Oncol. 2021, 160, 633–642. [Google Scholar] [CrossRef]
- Hydbring, P.; De Petris, L.; Zhang, Y.; Brandén, E.; Koyi, H.; Novak, M.; Kanter, L.; Hååg, P.; Hurley, J.; Tadigotla, V.; et al. Exosomal RNA-Profiling of Pleural Effusions Identifies Adenocarcinoma Patients through Elevated MiR-200 and LCN2 Expression. Lung Cancer 2018, 124, 45–52. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of MicroRNA Function in Animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Zhao, Y.; Qin, F.; Han, S.; Li, S.; Zhao, Y.; Wang, H.; Tian, J.; Cen, X. MicroRNAs in Drug Addiction: Current Status and Future Perspectives. Pharmacol. Ther. 2022, 108215. [Google Scholar] [CrossRef]
- Jouza, M.; Bohosova, J.; Stanikova, A.; Pecl, J.; Slaby, O.; Jabandziev, P. MicroRNA as an Early Biomarker of Neonatal Sepsis. Front. Pediatr. 2022, 10, 854324. [Google Scholar] [CrossRef]
- Gao, Y.-N.; Zhang, Y.-Q.; Wang, H.; Deng, Y.-L.; Li, N.-M. A New Player in Depression: MiRNAs as Modulators of Altered Synaptic Plasticity. Int. J. Mol. Sci. 2022, 23, 4555. [Google Scholar] [CrossRef]
- Żurawek, D.; Turecki, G. The MiRNome of Depression. Int. J. Mol. Sci. 2021, 22, 11312. [Google Scholar] [CrossRef]
- Hu, C.; Liang, X.; Fang, S.; Xu, L.; Gong, M.; Wang, Y.; Bi, Y.; Hong, S.; He, Y. ATRA Induces the Differentiation of Hepatic Progenitor Cells by Upregulating MicroRNA-200a. In Vitro Cell Dev. Biol. Anim. 2019, 55, 713–722. [Google Scholar] [CrossRef]
- Milevskiy, M.J.G.; Gujral, U.; Del Lama Marques, C.; Stone, A.; Northwood, K.; Burke, L.J.; Gee, J.M.W.; Nephew, K.; Clark, S.; Brown, M.A. MicroRNA-196a Is Regulated by ER and Is a Prognostic Biomarker in ER+ Breast Cancer. Br. J. Cancer 2019, 120, 621–632. [Google Scholar] [CrossRef]
- Nuñez-Olvera, S.I.; Puente-Rivera, J.; Ramos-Payán, R.; Pérez-Plasencia, C.; Salinas-Vera, Y.M.; Aguilar-Arnal, L.; López-Camarillo, C. Three-Dimensional Genome Organization in Breast and Gynecological Cancers: How Chromatin Folding Influences Tumorigenic Transcriptional Programs. Cells 2021, 11, 75. [Google Scholar] [CrossRef]
- Kang, S.Y.; Kim, D.G.; Kim, H.; Cho, Y.A.; Ha, S.Y.; Kwon, G.Y.; Jang, K.-T.; Kim, K.-M. Direct Comparison of the Next-Generation Sequencing and ITERT PCR Methods for the Diagnosis of TERT Hotspot Mutations in Advanced Solid Cancers. BMC Med. Genom. 2022, 15, 25. [Google Scholar] [CrossRef]
- Lim, G.X.Y.; Yeo, M.; Koh, Y.Y.; Winarni, T.I.; Rajan-Babu, I.-S.; Chong, S.S.; Faradz, S.M.H.; Guan, M. Validation of a Commercially Available Test That Enables the Quantification of the Numbers of CGG Trinucleotide Repeat Expansion in FMR1 Gene. PLoS ONE 2017, 12, e0173279. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).