Genomic Variation-Mediating Fluconazole Resistance in Yeast
Abstract
:1. Introduction
2. Antifungal Resistances Mediated by Genetic Mutations
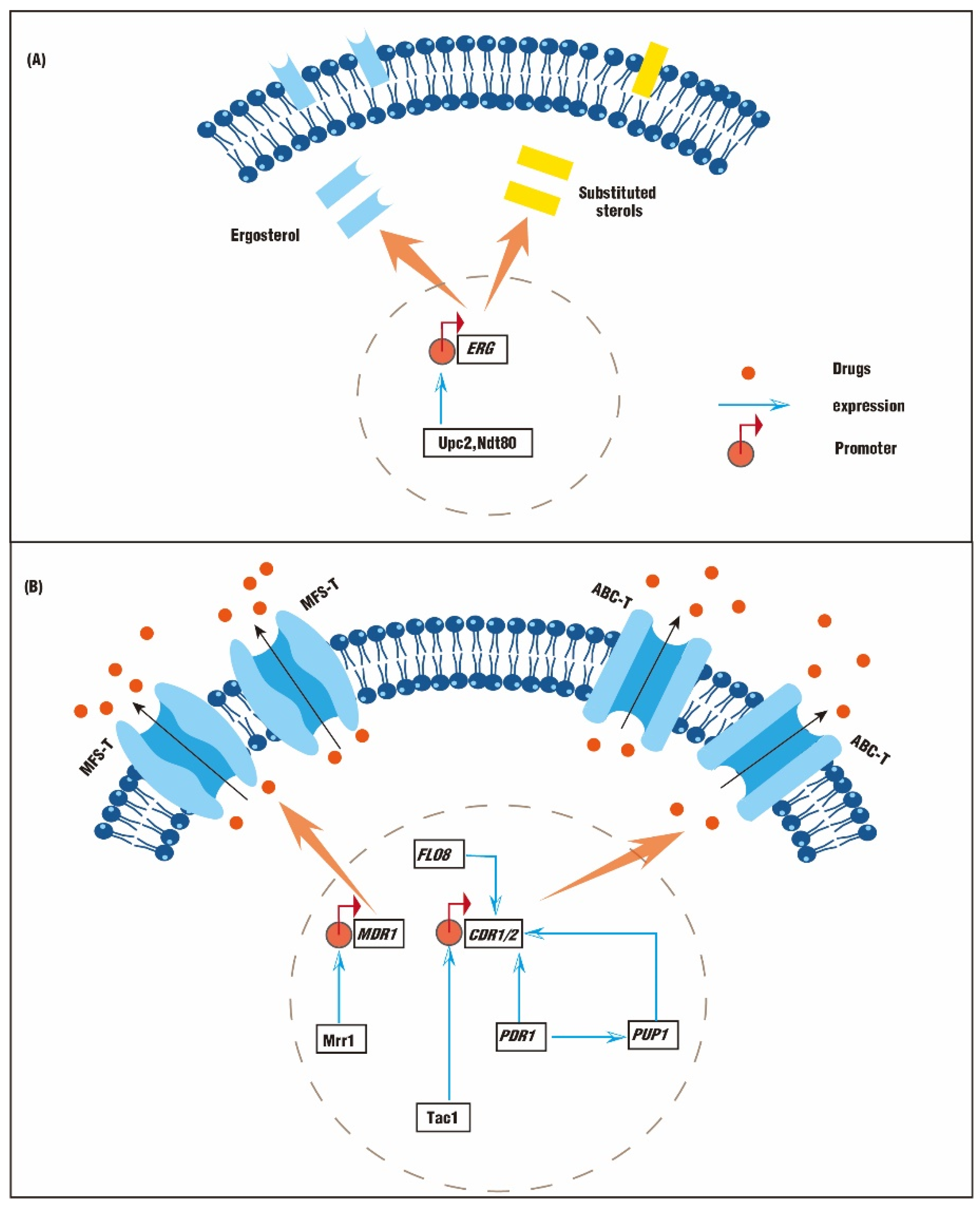
2.1. Genetic Mutations Lead to Abnormal Synthesis of Sterols
2.2. Genetic Mutations Lead to An Abnormal Efflux Pump
3. Antifungal Resistances Mediated by Aneuploidy
4. Antifungal Resistances Mediated by LOH
4.1. Chromosomal Regions of LOH Associated with Drug Resistance
4.2. Different Types of LOH Associated with Drug Resistance
4.3. Factors That Could Affect LOHs
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Almeida, F.; Rodrigues, M.L.; Coelho, C. The still underestimated problem of fungal diseases worldwide. Front. Microbiol. 2019, 10, 214. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and mechanisms of antifungal resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef]
- Costa-de-Oliveira, S.; Rodrigues, A.G. Candida albicans antifungal resistance and tolerance in bloodstream infections: The triad yeast-host-antifungal. Microorganisms 2020, 8, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, R.S.; Robbins, N.; Cowen, L.E. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev. 2011, 75, 213–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berkow, E.L.; Lockhart, S.R. Fluconazole resistance in Candida species: A current perspective. Infect. Drug Resist. 2017, 10, 237–245. [Google Scholar] [CrossRef] [Green Version]
- Cowen, L.E.; Sanglard, D.; Howard, S.J.; Rogers, P.D.; Perlin, D.S. Mechanisms of antifungal drug resistance. Cold Spring Harb. Perspect. Med. 2015, 5, a019752. [Google Scholar] [CrossRef]
- Rybak, J.M.; Sharma, C.; Doorley, L.A.; Barker, K.S.; Palmer, G.E.; Rogers, P.D. Delineation of the direct contribution of Candida auris ERG11 mutations to clinical triazole resistance. Microbiol. Spectr. 2021, 9, e01585-21. [Google Scholar] [CrossRef]
- Vale-Silva, L.A.; Coste, A.T.; Ischer, F.; Parker, J.E.; Kelly, S.L.; Pinto, E.; Sanglard, D. Azole resistance by loss of function of the sterol delta(5,6)-desaturase gene (ERG3) in Candida albicans does not necessarily decrease virulence. Antimicrob. Agents Chemother. 2012, 56, 1960–1968. [Google Scholar] [CrossRef] [Green Version]
- Martel, C.M.; Parker, J.E.; Bader, O.; Weig, M.; Gross, U.; Warrilow, A.G.S.; Kelly, D.E.; Kelly, S.L. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14 alpha-demethylase) and ERG5 (encoding c22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob. Agents Chemother. 2010, 54, 3578–3583. [Google Scholar] [CrossRef] [Green Version]
- Branco, J.; Ola, M.; Silva, R.M.; Fonseca, E.; Gomes, N.C.; Martins-Cruz, C.; Silva, A.P.; Silva-Dias, A.; Pina-Vaz, C.; Erraught, C.; et al. Impact of ERG3 mutations and expression of ergosterol genes controlled by UPC2 and NDT80 in Candida parapsilosis azole resistance. Clin. Microbiol. Infect. 2017, 23, 575.e1–575.e8. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.L.; Yang, J.; Ma, Y.; Xi, Z.Q.; Zhao, X.Q.; Zhao, X.X.; Zhao, M. The effects of secreted aspartyl proteinase inhibitor ritonavir on azoles-resistant strains of Candida albicans as well as regulatory role of SAP2 and ERG11. Immun. Inflamm. Dis. 2021, 9, 667–680. [Google Scholar] [CrossRef]
- Hervay, N.T.; Bencova, A.; Valachovic, M.; Morvova, M.; Gbelska, Y. UPC2 gene deletion modifies sterol homeostasis and susceptibility to metabolic inhibitors in Kluyveromyces lactis. Yeast 2020, 37, 647–657. [Google Scholar] [CrossRef]
- Tian, Y.; Gao, N.; Ni, Q.; Mao, Y.; Dong, D.; Huang, X.; Jiang, C.; Li, Z.; Zhang, L.; Wang, X.; et al. Sequence modification of the master regulator Pdr1 interferes with its transcriptional autoregulation and confers altered azole resistance in Candida glabrata. FEMS Yeast Res. 2018, 18, foy038. [Google Scholar] [CrossRef]
- Li, W.J.; Liu, J.Y.; Shi, C.; Zhao, Y.; Meng, L.N.; Wu, F.; Xiang, M.J. FLO8 deletion leads to azole resistance by upregulating CDR1 and CDR2 in Candida albicans. Res. Microbiol. 2019, 170, 272–279. [Google Scholar] [CrossRef]
- Ni, Q.; Wang, C.; Tian, Y.; Dong, D.F.; Jiang, C.; Mao, E.Q.; Peng, Y.B. CgPDR1 gain-of-function mutations lead to azole-resistance and increased adhesion in clinical Candida glabrata strains. Mycoses 2018, 61, 430–440. [Google Scholar] [CrossRef]
- Lohberger, A.; Coste, A.T.; Sanglard, D. Distinct roles of Candida albicans drug resistance transcription factors TAC1, MRR1, and UPC2 in virulence. Eukaryot. Cell 2014, 13, 127–142. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.L.; Rossi, J.M.; Myers, L.C. Candida albicans zn cluster transcription factors Tac1 and Znc1 are activated by farnesol to upregulate a transcriptional program including the multidrug efflux pump CDR1. Antimicrob. Agents Chemother. 2018, 62, e00968-18. [Google Scholar] [CrossRef] [Green Version]
- Selmecki, A.; Forche, A.; Berman, J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 2006, 313, 367–370. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Teoh, F.; Tan, A.S.M.; Cao, Y.B.; Pavelka, N.; Berman, J. Aneuploidy enables cross-adaptation to unrelated drugs. Mol. Biol. Evol. 2019, 36, 1768–1782. [Google Scholar] [CrossRef]
- Lee, Y.; Puumala, E.; Robbins, N.; Cowen, L.E. Antifungal drug resistance: Molecular mechanisms in Candida albicans and beyond. Chem. Rev. 2021, 121, 3390–3411. [Google Scholar] [CrossRef] [PubMed]
- vanden Bossche, H.; Marichal, P.; Odds, F.C.; Le Jeune, L.; Coene, M.C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 1992, 36, 2602–2610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon-Chung, K.J.; Chang, Y.C. Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog. 2012, 8, e1003022. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.Z.; Saha, A.; Haseeb, A.; Bennett, R.J. A chromosome 4 trisomy contributes to increased fluconazole resistance in a clinical isolate of Candida albicans. Microbiology 2017, 163, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Selmecki, A.M.; Dulmage, K.; Cowen, L.E.; Anderson, J.B.; Berman, J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 2009, 5, e1000705. [Google Scholar] [CrossRef]
- Dunkel, N.; Blass, J.; Rogers, P.D.; Morschhauser, J. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 2008, 69, 827–840. [Google Scholar] [CrossRef] [Green Version]
- Akins, R.A. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 2005, 43, 285–318. [Google Scholar] [CrossRef] [Green Version]
- Morschhauser, J.; Barker, K.S.; Liu, T.T.; Blass-Warmuth, J.; Homayouni, R.; Rogers, P.D. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 2007, 3, 1603–1616. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Gritsenko, V.; Lu, H.; Zhen, C.; Gao, L.; Berman, J.; Jiang, Y.Y. Adaptation to fluconazole via aneuploidy enables cross-adaptation to amphotericin B and flucytosine in Cryptococcus neoformans. Microbiol. Spectr. 2021, 9, e00723-21. [Google Scholar] [CrossRef]
- Harrison, B.D.; Hashemi, J.; Bibi, M.; Pulver, R.; Bavli, D.; Nahmias, Y.; Wellington, M.; Sapiro, G.; Berman, J. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 2014, 12, e1001815. [Google Scholar] [CrossRef]
- Smith, A.C.; Hickman, M.A. Host-Induced genome instability rapidly generates phenotypic variation across Candida albicans strains and ploidy states. mSphere 2020, 5, e00433-20. [Google Scholar] [CrossRef] [PubMed]
- Avramovska, O.; Hickman, M.A. The magnitude of Candida albicans stress-induced genome instability results from an interaction between ploidy and antifungal drugs. Genes Genomes Genet. 2019, 9, 4019–4027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forche, A.; Abbey, D.; Pisithkul, T.; Weinzierl, M.A.; Ringstrom, T.; Bruck, D.; Petersen, K.; Berman, J. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio 2011, 2, e00129-11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.B.; Bradford, W.D.; Seidel, C.W.; Li, R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 2012, 482, 246–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morschhauser, J. The development of fluconazole resistance in Candida albicans—An example of microevolution of a fungal pathogen. J. Microbiol. 2016, 54, 192–201. [Google Scholar] [CrossRef]
- Sasse, C.; Dunkel, N.; Schafer, T.; Schneider, S.; Dierolf, F.; Ohlsen, K.; Morschhauser, J. The stepwise acquisition of fluconazole resistance mutations causes a gradual loss of fitness in Candida albicans. Mol. Microbiol. 2012, 86, 539–556. [Google Scholar] [CrossRef]
- Ford, C.B.; Funt, J.M.; Abbey, D.; Issi, L.; Guiducci, C.; Martinez, D.A.; Delorey, T.; Li, B.Y.; White, T.C.; Cuomo, C.; et al. The evolution of drug resistance in clinical isolates of Candida albicans. eLife 2015, 4, e00662. [Google Scholar] [CrossRef] [Green Version]
- Popp, C.; Ramirez-Zavala, B.; Schwanfelder, S.; Kruger, I.; Morschhauser, J. Evolution of fluconazole-resistant Candida albicans strains by drug-induced mating competence and parasexual recombination. mBio 2019, 10, e02740-18. [Google Scholar] [CrossRef] [Green Version]
- Coste, A.; Selmecki, A.; Forche, A.; Diogo, D.; Bougnoux, M.E.; d’Enfert, C.; Berman, J.; Sanglard, D. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell 2007, 6, 1889–1904. [Google Scholar] [CrossRef] [Green Version]
- Manoharlal, R.; Gorantala, J.; Sharma, M.; Sanglard, D.; Prasad, R. PAP1 poly(A) polymerase 1 homozygosity and hyperadenylation are major determinants of increased mRNA stability of CDR1 in azole-resistant clinical isolates of Candida albicans. Microbiology 2010, 156, 313–326. [Google Scholar] [CrossRef]
- Sui, Y.; Qi, L.; Wu, J.K.; Wen, X.P.; Tang, X.X.; Ma, Z.J.; Wu, X.C.; Zhang, K.; Kokoska, R.J.; Zheng, D.Q.; et al. Genome-wide mapping of spontaneous genetic alterations in diploid yeast cells. Proc. Natl. Acad. Sci. USA 2020, 117, 28191–28200. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.H.; Bennett, R.J. The impact of gene dosage and heterozygosity on the diploid pathobiont Candida albicans. J. Fungi 2020, 6, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rustad, T.R.; Stevens, D.A.; Pfaller, M.A.; White, T.C. Homozygosity at the Candida albicans MTL locus associated with azole resistance. Microbiology 2002, 148, 1061–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsit, S.; Leducq, J.B.; Durand, E.; Marchant, A.; Filteau, M.; Landry, C.R. Evolutionary biology through the lens of budding yeast comparative genomics. Nat. Rev. Genet. 2017, 18, 581–598. [Google Scholar] [CrossRef]
- Hiller, D.; Sanglard, D.; Morschhauser, J. Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans. Antimicrob. Agents Chemother. 2006, 50, 1365–1371. [Google Scholar] [CrossRef] [Green Version]
- Hickman, M.A.; Zeng, G.S.; Forche, A.; Hirakawa, M.P.; Abbey, D.; Harrison, B.D.; Wang, Y.M.; Su, C.H.; Bennett, R.J.; Wang, Y.; et al. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature 2016, 530, 242. [Google Scholar] [CrossRef]
- Bennett, R.J.; Forche, A.; Berman, J. Rapid mechanisms for generating genome diversity: Whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb. Perspect. Med. 2014, 4, a019604. [Google Scholar] [CrossRef]
- Smith, A.C.; Morran, L.T.; Hickman, M.A. Host defense mechanisms induce genome instability leading to rapid evolution in an opportunistic fungal pathogen. Infect. Immun. 2022, 90, 12. [Google Scholar] [CrossRef]
- Weil, T.; Santamaria, R.; Lee, W.; Rung, J.; Tocci, N.; Abbey, D.; Bezerra, A.R.; Carreto, L.; Moura, G.R.; Bayes, M.; et al. Adaptive mistranslation accelerates the evolution of fluconazole resistance and induces major genomic and gene expression alterations in Candida albicans. mSphere 2017, 2, e00167-17. [Google Scholar] [CrossRef] [Green Version]
- St Charles, J.; Hazkani-Covo, E.; Yin, Y.; Andersen, S.L.; Dietrich, F.S.; Greenwell, P.W.; Malc, E.; Mieczkowski, P.; Petes, T.D. High-resolution genome-wide analysis of irradiated (uv and gamma-rays) diploid yeast cells reveals a high frequency of genomic loss of heterozygosity (LOH) events. Genetics 2012, 190, 1267–1284. [Google Scholar] [CrossRef] [Green Version]
- de Vries, A.R.G.; Couwenberg, L.G.F.; van den Broek, M.; Cortes, P.D.; ter Horst, J.; Pronk, J.T.; Daran, J.M.G. Allele-specific genome editing using CRISPR-Cas9 is associated with loss of heterozygosity in diploid yeast. Nucleic Acids Res. 2019, 47, 1362–1372. [Google Scholar]
- Li, Y.X.; Wu, Y.; Ma, L.; Guo, Z.; Xiao, W.H.; Yuan, Y.J. Loss of heterozygosity by SCRaMbLEing. Sci. China Life Sci. 2019, 62, 381–393. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.-Y.; Cai, H.-Q.; Qu, S.-Y.; Lin, W.-H.; Liang, C.-C.; Liu, H.; Xie, Z.-X.; Yuan, Y.-J. Genomic Variation-Mediating Fluconazole Resistance in Yeast. Biomolecules 2022, 12, 845. https://doi.org/10.3390/biom12060845
Wang W-Y, Cai H-Q, Qu S-Y, Lin W-H, Liang C-C, Liu H, Xie Z-X, Yuan Y-J. Genomic Variation-Mediating Fluconazole Resistance in Yeast. Biomolecules. 2022; 12(6):845. https://doi.org/10.3390/biom12060845
Chicago/Turabian StyleWang, Wen-Yao, Hong-Qing Cai, Si-Yuan Qu, Wei-Hao Lin, Cheng-Cheng Liang, Hao Liu, Ze-Xiong Xie, and Ying-Jin Yuan. 2022. "Genomic Variation-Mediating Fluconazole Resistance in Yeast" Biomolecules 12, no. 6: 845. https://doi.org/10.3390/biom12060845
APA StyleWang, W.-Y., Cai, H.-Q., Qu, S.-Y., Lin, W.-H., Liang, C.-C., Liu, H., Xie, Z.-X., & Yuan, Y.-J. (2022). Genomic Variation-Mediating Fluconazole Resistance in Yeast. Biomolecules, 12(6), 845. https://doi.org/10.3390/biom12060845




