The Mitochondrial HSP90 Paralog TRAP1: Structural Dynamics, Interactome, Role in Metabolic Regulation, and Inhibitors
Abstract
:1. Introduction
2. TRAP1: Cytoprotective or Pro-Neoplastic?
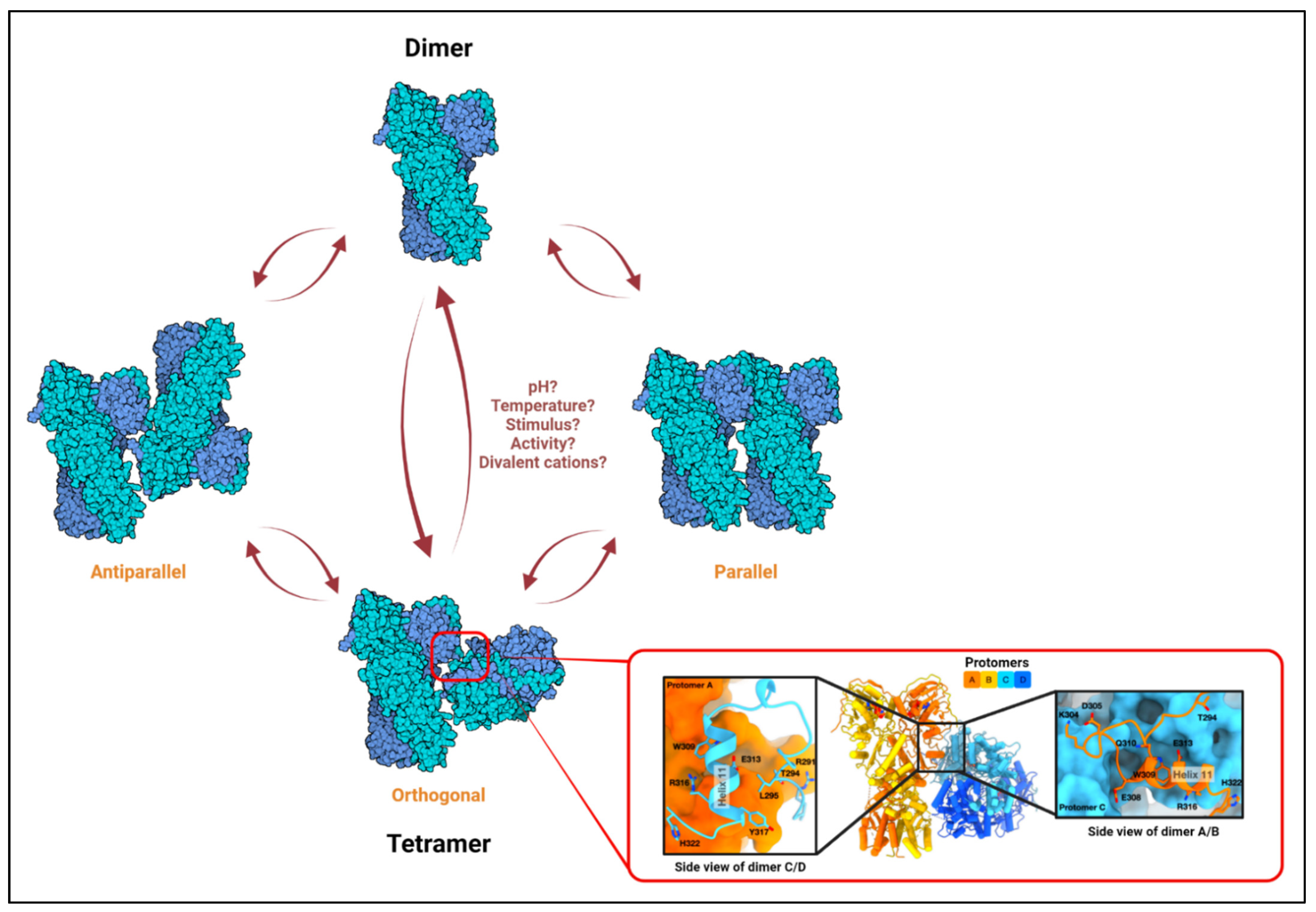
3. Structure, ATPase Cycle, Dimers, and Tetramers
4. Cancer and Metabolic Rewiring
5. Defining a TRAP1 Interactome
6. TRAP1 Inhibitors
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Hartl, F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Carroni, M.; Nussbaum-Krammer, C.; Mogk, A.; Nillegoda, N.B.; Szlachcic, A.; Guilbride, D.L.; Saibil, H.R.; Mayer, M.P.; Bukau, B. Human Hsp70 Disaggregase Reverses Parkinson’s-Linked α-Synuclein Amyloid Fibrils. Mol. Cell 2015, 59, 781–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nillegoda, N.B.; Bukau, B. Metazoan Hsp70-based protein disaggregases: Emergence and mechanisms. Front. Mol. Biosci. 2015, 2, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Funez, P.; Sanchez-Garcia, J.; de Mena, L.; Zhang, Y.; Levites, Y.; Khare, S.; Golde, T.E.; Rincon-Limas, D.E. Holdase activity of secreted Hsp70 masks amyloid-β42 neurotoxicity in Drosophila. Proc. Natl. Acad. Sci. USA 2016, 113, E5212–E5221. [Google Scholar] [CrossRef] [Green Version]
- Thoma, J.; Burmann, B.M.; Hiller, S.; Müller, D.J. Impact of holdase chaperones Skp and SurA on the folding of β-barrel outer-membrane proteins. Nat. Struct. Mol. Biol. 2015, 22, 795–802. [Google Scholar] [CrossRef]
- Demand, J.; Alberti, S.; Patterson, C.; Höhfeld, J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 2001, 11, 1569–1577. [Google Scholar] [CrossRef] [Green Version]
- Gamerdinger, M.; Hajieva, P.; Kaya, A.M.; Wolfrum, U.; Hartl, F.U.; Behl, C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009, 28, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Hartmann-Petersen, R.; Seeger, M.; Gordon, C. Transferring substrates to the 26S proteasome. Trends Biochem. Sci. 2003, 28, 26–31. [Google Scholar] [CrossRef]
- Csermely, P.; Schnaider, T.; Soti, C.; Prohászka, Z.; Nardai, G. The 90-kDa molecular chaperone family: Structure, function, and clinical applications. A comprehensive review. Pharmacol. Ther. 1998, 79, 129–168. [Google Scholar] [CrossRef]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Radli, M.; Rüdiger, S.G.D. Dancing with the Diva: Hsp90–Client Interactions. J. Mol. Biol. 2018, 430, 3029–3040. [Google Scholar] [CrossRef] [PubMed]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Zuehlke, A.D.; Beebe, K.; Neckers, L.; Prince, T. Regulation and function of the human HSP90AA1 gene. Gene 2015, 570, 8–16. [Google Scholar] [CrossRef] [Green Version]
- Marzec, M.; Eletto, D.; Argon, Y. GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim. Biophys. Acta 2012, 1823, 774–787. [Google Scholar] [CrossRef] [Green Version]
- Felts, S.J.; Owen, B.A.; Nguyen, P.; Trepel, J.; Donner, D.B.; Toft, D.O. The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J. Biol. Chem. 2000, 275, 3305–3312. [Google Scholar] [CrossRef] [Green Version]
- Cechetto, J.D.; Gupta, R.S. Immunoelectron microscopy provides evidence that tumor necrosis factor receptor-associated protein 1 (TRAP-1) is a mitochondrial protein which also localizes at specific extramitochondrial sites. Exp. Cell Res. 2000, 260, 30–39. [Google Scholar] [CrossRef]
- Yoshida, S.; Tsutsumi, S.; Muhlebach, G.; Sourbier, C.; Lee, M.-J.; Lee, S.; Vartholomaiou, E.; Tatokoro, M.; Beebe, K.; Miyajima, N.; et al. Molecular chaperone TRAP1 regulates a metabolic switch between mitochondrial respiration and aerobic glycolysis. Proc. Natl. Acad. Sci. USA 2013, 110, E1604–E1612. [Google Scholar] [CrossRef] [Green Version]
- Sciacovelli, M.; Guzzo, G.; Morello, V.; Frezza, C.; Zheng, L.; Nannini, N.; Calabrese, F.; Laudiero, G.; Esposito, F.; Landriscina, M.; et al. The Mitochondrial Chaperone TRAP1 Promotes Neoplastic Growth by Inhibiting Succinate Dehydrogenase. Cell Metab. 2013, 17, 988–999. [Google Scholar] [CrossRef] [Green Version]
- Chae, Y.C.; Angelin, A.; Lisanti, S.; Kossenkov, A.V.; Speicher, K.D.; Wang, H.; Powers, J.F.; Tischler, A.S.; Pacak, K.; Fliedner, S.; et al. Landscape of the mitochondrial Hsp90 metabolome in tumours. Nat. Commun. 2013, 4, 2139. [Google Scholar] [CrossRef] [Green Version]
- Joshi, A.; Dai, L.; Liu, Y.; Lee, J.; Ghahhari, N.M.; Segala, G.; Beebe, K.; Jenkins, L.M.; Lyons, G.C.; Bernasconi, L.; et al. The mitochondrial HSP90 paralog TRAP1 forms an OXPHOS-regulated tetramer and is involved in mitochondrial metabolic homeostasis. BMC Biol. 2020, 18, 10. [Google Scholar] [CrossRef] [Green Version]
- Park, H.K.; Hong, J.H.; Oh, Y.T.; Kim, S.S.; Yin, J.; Lee, A.J.; Chae, Y.C.; Kim, J.H.; Park, S.H.; Park, C.K.; et al. Interplay between TRAP1 and Sirtuin-3 Modulates Mitochondrial Respiration and Oxidative Stress to Maintain Stemness of Glioma Stem Cells. Cancer Res. 2019, 79, 1369–1382. [Google Scholar] [CrossRef] [Green Version]
- Rasola, A.; Neckers, L.; Picard, D. Mitochondrial oxidative phosphorylation TRAP(1)ped in tumor cells. Trends Cell Biol. 2014, 24, 455–463. [Google Scholar] [CrossRef]
- Masgras, I.; Laquatra, C.; Cannino, G.; Serapian, S.A.; Colombo, G.; Rasola, A. The molecular chaperone TRAP1 in cancer: From the basics of biology to pharmacological targeting. Semin. Cancer Biol. 2021, 76, 45–53. [Google Scholar] [CrossRef]
- Takamura, H.; Koyama, Y.; Matsuzaki, S.; Yamada, K.; Hattori, T.; Miyata, S.; Takemoto, K.; Tohyama, M.; Katayama, T. TRAP1 controls mitochondrial fusion/fission balance through Drp1 and Mff expression. PLoS ONE 2012, 7, e51912. [Google Scholar] [CrossRef]
- Costa, A.C.; Loh, S.H.Y.; Martins, L.M. Drosophila Trap1 protects against mitochondrial dysfunction in a PINK1/parkin model of Parkinson’s disease. Cell Death Dis. 2013, 4, e467. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Karsten, P.; Hamm, S.; Pogson, J.H.; Müller-Rischart, A.K.; Exner, N.; Haass, C.; Whitworth, A.J.; Winklhofer, K.F.; Schulz, J.B.; et al. TRAP1 rescues PINK1 loss-of-function phenotypes. Hum. Mol. Genet. 2013, 22, 2829–2841. [Google Scholar] [CrossRef] [Green Version]
- Hua, G.; Zhang, Q.; Fan, Z. Heat Shock Protein 75 (TRAP1) Antagonizes Reactive Oxygen Species Generation and Protects Cells from Granzyme M-mediated Apoptosis. J. Biol. Chem. 2007, 282, 20553–20560. [Google Scholar] [CrossRef] [Green Version]
- Montesano Gesualdi, N.; Chirico, G.; Pirozzi, G.; Costantino, E.; Landriscina, M.; Esposito, F. Tumor necrosis factor-associated protein 1 (TRAP-1) protects cells from oxidative stress and apoptosis. Stress 2007, 10, 342–350. [Google Scholar] [CrossRef]
- Pridgeon, J.W.; Olzmann, J.A.; Chin, L.-S.; Li, L. PINK1 Protects against Oxidative Stress by Phosphorylating Mitochondrial Chaperone TRAP1. PLoS Biol. 2007, 5, e172. [Google Scholar] [CrossRef]
- Ramos Rego, I.; Santos Cruz, B.; Ambrósio, A.F.; Alves, C.H. TRAP1 in Oxidative Stress and Neurodegeneration. Antioxidants 2021, 10, 1829. [Google Scholar] [CrossRef]
- Kang, B.H.; Plescia, J.; Dohi, T.; Rosa, J.; Doxsey, S.J.; Altieri, D.C. Regulation of Tumor Cell Mitochondrial Homeostasis by an Organelle-Specific Hsp90 Chaperone Network. Cell 2007, 131, 257–270. [Google Scholar] [CrossRef]
- Xu, L.; Voloboueva, L.A.; Ouyang, Y.; Emery, J.F.; Giffard, R.G. Overexpression of mitochondrial Hsp70/Hsp75 in rat brain protects mitochondria, reduces oxidative stress, and protects from focal ischemia. J. Cereb. Blood Flow Metab. 2009, 29, 365–374. [Google Scholar] [CrossRef]
- Xiang, F.; Huang, Y.S.; Shi, X.H.; Zhang, Q. Mitochondrial chaperone tumour necrosis factor receptor-associated protein 1 protects cardiomyocytes from hypoxic injury by regulating mitochondrial permeability transition pore opening. FEBS J. 2010, 277, 1929–1938. [Google Scholar] [CrossRef]
- Zhang, P.; Lu, Y.; Yu, D.; Zhang, D.; Hu, W. TRAP1 Provides Protection Against Myocardial Ischemia-Reperfusion Injury by Ameliorating Mitochondrial Dysfunction. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2015, 36, 2072–2082. [Google Scholar] [CrossRef]
- Clarke, B.E.; Kalmar, B.; Greensmith, L. Enhanced Expression of TRAP1 Protects Mitochondrial Function in Motor Neurons under Conditions of Oxidative Stress. Int. J. Mol. Sci. 2022, 23, 1789. [Google Scholar] [CrossRef]
- Zhang, L.; Su, N.; Luo, Y.; Chen, S.; Zhao, T. TRAP1 inhibits MIC60 ubiquitination to mitigate the injury of cardiomyocytes and protect mitochondria in extracellular acidosis. Cell Death Discov. 2021, 7, 389. [Google Scholar] [CrossRef]
- Butler, E.K.; Voigt, A.; Lutz, A.K.; Toegel, J.P.; Gerhardt, E.; Karsten, P.; Falkenburger, B.; Reinartz, A.; Winklhofer, K.F.; Schulz, J.B. The mitochondrial chaperone protein TRAP1 mitigates α-Synuclein toxicity. PLoS Genet. 2012, 8, e1002488. [Google Scholar] [CrossRef] [Green Version]
- Malpartida, A.B.; Williamson, M.; Narendra, D.P.; Wade-Martins, R.; Ryan, B.J. Mitochondrial Dysfunction and Mitophagy in Parkinson’s Disease: From Mechanism to Therapy. Trends Biochem. Sci. 2021, 46, 329–343. [Google Scholar] [CrossRef]
- Chen, J.F.; Wu, Q.S.; Xie, Y.X.; Si, B.L.; Yang, P.P.; Wang, W.Y.; Hua, Q.; He, Q. TRAP1 ameliorates renal tubulointerstitial fibrosis in mice with unilateral ureteral obstruction by protecting renal tubular epithelial cell mitochondria. FASEB J. 2017, 31, 4503–4514. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, E.; Demidova, E.V.; Iqbal, W.; Serebriiskii, I.G.; Vlasenkova, R.; Ghatalia, P.; Zhou, Y.; Rainey, K.; Forman, A.F.; Dunbrack, R.L., Jr.; et al. Interaction of germline variants in a family with a history of early-onset clear cell renal cell carcinoma. Mol. Genet. Genom. Med. 2019, 7, e556. [Google Scholar] [CrossRef]
- Fitzgerald, J.C.; Zimprich, A.; Carvajal Berrio, D.A.; Schindler, K.M.; Maurer, B.; Schulte, C.; Bus, C.; Hauser, A.K.; Kübler, M.; Lewin, R.; et al. Metformin reverses TRAP1 mutation-associated alterations in mitochondrial function in Parkinson’s disease. Brain J. Neurol. 2017, 140, 2444–2459. [Google Scholar] [CrossRef]
- Skinner, S.J.; Doonanco, K.R.; Boles, R.G.; Chan, A.K.J. Homozygous TRAP1 sequence variant in a child with Leigh syndrome and normal kidneys. Kidney Int. 2014, 86, 860. [Google Scholar] [CrossRef] [Green Version]
- Boles, R.G.; Hornung, H.A.; Moody, A.E.; Ortiz, T.B.; Wong, S.A.; Eggington, J.M.; Stanley, C.M.; Gao, M.; Zhou, H.; McLaughlin, S.; et al. Hurt, tired and queasy: Specific variants in the ATPase domain of the TRAP1 mitochondrial chaperone are associated with common, chronic “functional” symptomatology including pain, fatigue and gastrointestinal dysmotility. Mitochondrion 2015, 23, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Saisawat, P.; Kohl, S.; Hilger, A.C.; Hwang, D.Y.; Yung Gee, H.; Dworschak, G.C.; Tasic, V.; Pennimpede, T.; Natarajan, S.; Sperry, E.; et al. Whole-exome resequencing reveals recessive mutations in TRAP1 in individuals with CAKUT and VACTERL association. Kidney Int. 2014, 85, 1310–1317. [Google Scholar] [CrossRef] [Green Version]
- Megger, D.A.; Bracht, T.; Kohl, M.; Ahrens, M.; Naboulsi, W.; Weber, F.; Hoffmann, A.-C.; Stephan, C.; Kuhlmann, K.; Eisenacher, M.; et al. Proteomic differences between hepatocellular carcinoma and nontumorous liver tissue investigated by a combined gel-based and label-free quantitative proteomics study. Mol. Cell. Proteom. 2013, 12, 2006–2020. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Wang, J.; Huang, Z.; Wei, P.; Liu, Y.; Hao, J.; Zhao, L.; Zhang, F.; Tu, Y.; Wei, T. Aberrantly upregulated TRAP1 is required for tumorigenesis of breast cancer. Oncotarget 2015, 6, 44495. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Lv, Q.; Sun, H.; Xue, Y.; Wang, P.; Liu, L.; Li, Z.; Li, Z.; Tian, X.; Liu, Y.H. Expression of TRAP1 predicts poor survival of malignant glioma patients. J. Mol. Neurosci. 2015, 55, 62–68. [Google Scholar] [CrossRef]
- Lee, J.H.; Kang, K.W.; Kim, J.-E.; Hwang, S.W.; Park, J.H.; Kim, S.-H.; Ji, J.H.; Kim, T.G.; Nam, H.-Y.; Roh, M.S.; et al. Differential expression of heat shock protein 90 isoforms in small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 9487–9493. [Google Scholar]
- Gao, C.; Li, M.; Jiang, A.-L.; Sun, R.; Jin, H.-L.; Gui, H.-W.; Xiao, F.; Ding, X.-W.; Fu, Z.-M.; Feng, J.-P. Overexpression of the mitochondrial chaperone tumor necrosis factor receptor-associated protein 1 is associated with the poor prognosis of patients with colorectal cancer. Oncol. Lett. 2018, 15, 5451–5458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.-Y.; Song, B.-R.; Peng, J.-J.; Lu, Y.-M. Correlation between mitochondrial TRAP-1 expression and lymph node metastasis in colorectal cancer. World J. Gastroenterol. 2012, 18, 5965–5971. [Google Scholar] [CrossRef] [PubMed]
- Leav, I.; Plescia, J.; Goel, H.L.; Li, J.; Jiang, Z.; Cohen, R.J.; Languino, L.R.; Altieri, D.C. Cytoprotective mitochondrial chaperone TRAP-1 as a novel molecular target in localized and metastatic prostate cancer. Am. J. Pathol. 2010, 176, 393–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Q.; Sun, H.; Cao, C.; Gao, B.; Qi, Y. Overexpression of tumor necrosis factor receptor-associated protein 1 (TRAP1) are associated with poor prognosis of epithelial ovarian cancer. Tumor Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 2721–2727. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Liu, L.; Xue, L.; Zhou, W.; Zhao, Z.; Xu, B.; Song, Y.; Zhan, Q. TRAP1 shows clinical significance and promotes cellular migration and invasion through STAT3/MMP2 pathway in human esophageal squamous cell cancer. J. Genet. Genom. Yi Chuan Xue Bao 2014, 41, 529–537. [Google Scholar] [CrossRef]
- Pak, M.G.; Koh, H.J.; Roh, M.S. Clinicopathologic significance of TRAP1 expression in colorectal cancer: A large scale study of human colorectal adenocarcinoma tissues. Diagn. Pathol. 2017, 12, 6. [Google Scholar] [CrossRef] [Green Version]
- Si, T.; Yang, G.; Qiu, X.; Luo, Y.; Liu, B.; Wang, B. Expression of tumor necrosis factor receptor-associated protein 1 and its clinical significance in kidney cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 13090–13095. [Google Scholar]
- Chen, R.; Pan, S.; Lai, K.; Lai, L.A.; Crispin, D.A.; Bronner, M.P.; Brentnall, T.A. Up-regulation of mitochondrial chaperone TRAP1 in ulcerative colitis associated colorectal cancer. World J. Gastroenterol. 2014, 20, 17037–17048. [Google Scholar] [CrossRef]
- Kowalik, M.A.; Guzzo, G.; Morandi, A.; Perra, A.; Menegon, S.; Masgras, I.; Trevisan, E.; Angioni, M.M.; Fornari, F.; Quagliata, L.; et al. Metabolic reprogramming identifies the most aggressive lesions at early phases of hepatic carcinogenesis. Oncotarget 2016, 7, 32375–32393. [Google Scholar] [CrossRef]
- Wallace, D.C. Mitochondria and cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef] [Green Version]
- Vartholomaiou, E.; Madon-Simon, M.; Hagmann, S.; Mühlebach, G.; Wurst, W.; Floss, T.; Picard, D. Cytosolic Hsp90α and its mitochondrial isoform Trap1 are differentially required in a breast cancer model. Oncotarget 2017, 8, 17428. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Piel, W.H.; Gui, L.; Bruford, E.; Monteiro, A. The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 2005, 86, 627–637. [Google Scholar] [CrossRef]
- Lisanti, S.; Tavecchio, M.; Chae, Y.C.; Liu, Q.; Brice, A.K.; Thakur, M.L.; Languino, L.R.; Altieri, D.C. Deletion of the Mitochondrial Chaperone TRAP-1 Uncovers Global Reprogramming of Metabolic Networks. Cell Rep. 2014, 8, 671–677. [Google Scholar] [CrossRef]
- Jackson, S.E. Hsp90: Structure and function. Top. Curr. Chem. 2013, 328, 155–240. [Google Scholar] [CrossRef]
- Lavery, L.A.; Partridge, J.R.; Ramelot, T.A.; Elnatan, D.; Kennedy, M.A.; Agard, D.A. Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism. Mol. Cell 2014, 53, 330–343. [Google Scholar] [CrossRef] [Green Version]
- Kang, B.H. TRAP1 regulation of mitochondrial life or death decision in cancer cells and mitochondria-targeted TRAP1 inhibitors. BMB Rep. 2012, 45, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Tsutsumi, S.; Mollapour, M.; Prodromou, C.; Lee, C.T.; Panaretou, B.; Yoshida, S.; Mayer, M.P.; Neckers, L.M. Charged linker sequence modulates eukaryotic heat shock protein 90 (Hsp90) chaperone activity. Proc. Natl. Acad. Sci. USA 2012, 109, 2937–2942. [Google Scholar] [CrossRef] [Green Version]
- Partridge, J.R.; Lavery, L.A.; Elnatan, D.; Naber, N.; Cooke, R.; Agard, D.A. A novel N-terminal extension in mitochondrial TRAP1 serves as a thermal regulator of chaperone activity. eLife 2014, 3, e03487. [Google Scholar] [CrossRef]
- Sung, N.; Lee, J.; Kim, J.-H.; Chang, C.; Joachimiak, A.; Lee, S.; Tsai, F.T.F. Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate. Proc. Natl. Acad. Sci. USA 2016, 113, 2952–2957. [Google Scholar] [CrossRef] [Green Version]
- Leskovar, A.; Wegele, H.; Werbeck, N.D.; Buchner, J.; Reinstein, J. The ATPase cycle of the mitochondrial Hsp90 analog Trap1. J. Biol. Chem. 2008, 283, 11677–11688. [Google Scholar] [CrossRef] [Green Version]
- Elnatan, D.; Betegon, M.; Liu, Y.; Ramelot, T.; Kennedy, M.A.; Agard, D.A. Symmetry broken and rebroken during the ATP hydrolysis cycle of the mitochondrial Hsp90 TRAP1. eLife 2017, 6, e25235. [Google Scholar] [CrossRef]
- Elnatan, D.; Agard, D.A. Calcium binding to a remote site can replace magnesium as cofactor for mitochondrial Hsp90 (TRAP1) ATPase activity. J. Biol. Chem. 2018, 293, 13717–13724. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Sun, M.; Elnatan, D.; Larson, A.G.; Agard, D.A. Cryo-EM analysis of human mitochondrial Hsp90 in multiple tetrameric states. bioRxiv 2020. [Google Scholar] [CrossRef]
- Shiau, A.K.; Harris, S.F.; Southworth, D.R.; Agard, D.A. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell 2006, 127, 329–340. [Google Scholar] [CrossRef] [Green Version]
- Nemoto, T.; Sato, N. Oligomeric forms of the 90-kDa heat shock protein. Biochem. J. 1998, 330, 989–995. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-C.; Lin, T.-W.; Ko, T.-P.; Wang, A.H.J. The Hexameric Structures of Human Heat Shock Protein 90. PLoS ONE 2011, 6, e19961. [Google Scholar] [CrossRef] [Green Version]
- Chadli, A.; Ladjimi, M.M.; Baulieu, E.-E.; Catelli, M.G. Heat-induced Oligomerization of the Molecular Chaperone Hsp90: Inhibition by atp and geldanamycin and activation by transition metal oxyanions. J. Biol. Chem. 1999, 274, 4133–4139. [Google Scholar] [CrossRef] [Green Version]
- Moullintraffort, L.; Bruneaux, M.; Nazabal, A.; Allegro, D.; Giudice, E.; Zal, F.; Peyrot, V.; Barbier, P.; Thomas, D.; Garnier, C. Biochemical and Biophysical Characterization of the Mg2+-induced 90-kDa Heat Shock Protein Oligomers. J. Biol. Chem. 2010, 285, 15100–15110. [Google Scholar] [CrossRef] [Green Version]
- Yonehara, M.; Minami, Y.; Kawata, Y.; Nagai, J.; Yahara, I. Heat-induced Chaperone Activity of HSP90. J. Biol. Chem. 1996, 271, 2641–2645. [Google Scholar] [CrossRef] [Green Version]
- Jakob, U.; Meyer, I.; Bügl, H.; André, S.; Bardwell, J.C.A.; Buchner, J. Structural Organization of Procaryotic and Eucaryotic Hsp90. Influence of divalent cations on structure and function. J. Biol. Chem. 1995, 270, 14412–14419. [Google Scholar] [CrossRef] [Green Version]
- Chrétien, D.; Bénit, P.; Ha, H.-H.; Keipert, S.; El-Khoury, R.; Chang, Y.-T.; Jastroch, M.; Jacobs, H.T.; Rustin, P.; Rak, M. Mitochondria are physiologically maintained at close to 50 °C. PLoS Biol. 2018, 16, e2003992. [Google Scholar] [CrossRef] [Green Version]
- Lepvrier, E.; Thomas, D.; Garnier, C. Hsp90 Quaternary Structures and the Chaperone Cycle: Highly Flexible Dimeric and Oligomeric Structures and Their Regulation by Co-Chaperones. Curr. Proteom. 2019, 16, 5–11. [Google Scholar] [CrossRef]
- Hsu, P.P.; Sabatini, D.M. Cancer cell metabolism: Warburg and beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [Green Version]
- Chae, Y.C.; Caino, M.C.; Lisanti, S.; Ghosh, J.C.; Dohi, T.; Danial, N.N.; Villanueva, J.; Ferrero, S.; Vaira, V.; Santambrogio, L.; et al. Control of tumor bioenergetics and survival stress signaling by mitochondrial HSP90s. Cancer Cell 2012, 22, 331–344. [Google Scholar] [CrossRef] [Green Version]
- King, A.; Selak, M.A.; Gottlieb, E. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene 2006, 25, 4675–4682. [Google Scholar] [CrossRef] [Green Version]
- Tretter, L.; Patocs, A.; Chinopoulos, C. Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim. Biophys. Acta (BBA)-Bioenerg. 2016, 1857, 1086–1101. [Google Scholar] [CrossRef]
- Hayashi, Y.; Yokota, A.; Harada, H.; Huang, G. Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1α in cancer. Cancer Sci. 2019, 110, 1510–1517. [Google Scholar] [CrossRef] [Green Version]
- Selak, M.A.; Armour, S.M.; MacKenzie, E.D.; Boulahbel, H.; Watson, D.G.; Mansfield, K.D.; Pan, Y.; Simon, M.C.; Thompson, C.B.; Gottlieb, E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 2005, 7, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Mao, H.; Yang, A.; Zhao, Y.; Lei, L.; Li, H. Succinate Supplement Elicited “Pseudohypoxia” Condition to Promote Proliferation, Migration, and Osteogenesis of Periodontal Ligament Cells. Stem Cells Int. 2020, 2020, 2016809. [Google Scholar] [CrossRef] [Green Version]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-Modified Cancer Cell Metabolism. Front. Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Cannino, G.; Urbani, A.; Gaspari, M.; Varano, M.; Negro, A.; Filippi, A.; Ciscato, F.; Masgras, I.; Gerle, C.; Tibaldi, E.; et al. The mitochondrial chaperone TRAP1 regulates F-ATP synthase channel formation. Cell Death Differ. 2022. [Google Scholar] [CrossRef]
- Martins, F.; Gonçalves, L.G.; Pojo, M.; Serpa, J. Take Advantage of Glutamine Anaplerosis, the Kernel of the Metabolic Rewiring in Malignant Gliomas. Biomolecules 2020, 10, 1370. [Google Scholar] [CrossRef]
- Wise, D.R.; Thompson, C.B. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 2010, 35, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef] [Green Version]
- Cluntun, A.A.; Lukey, M.J.; Cerione, R.A.; Locasale, J.W. Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer 2017, 3, 169–180. [Google Scholar] [CrossRef] [Green Version]
- Bruno, G.; Li Bergolis, V.; Piscazzi, A.; Crispo, F.; Condelli, V.; Zoppoli, P.; Maddalena, F.; Pietrafesa, M.; Giordano, G.; Matassa, D.S.; et al. TRAP1 regulates the response of colorectal cancer cells to hypoxia and inhibits ribosome biogenesis under conditions of oxygen deprivation. Int. J. Oncol. 2022, 60, 79. [Google Scholar] [CrossRef]
- Wang, X.; Venable, J.; LaPointe, P.; Hutt, D.M.; Koulov, A.V.; Coppinger, J.; Gurkan, C.; Kellner, W.; Matteson, J.; Plutner, H.; et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 2006, 127, 803–815. [Google Scholar] [CrossRef] [Green Version]
- Prince, T.L.; Kijima, T.; Tatokoro, M.; Lee, S.; Tsutsumi, S.; Yim, K.; Rivas, C.; Alarcon, S.; Schwartz, H.; Khamit-Kush, K.; et al. Client Proteins and Small Molecule Inhibitors Display Distinct Binding Preferences for Constitutive and Stress-Induced HSP90 Isoforms and Their Conformationally Restricted Mutants. PLoS ONE 2015, 10, e0141786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kijima, T.; Prince, T.L.; Tigue, M.L.; Yim, K.H.; Schwartz, H.; Beebe, K.; Lee, S.; Budzynski, M.A.; Williams, H.; Trepel, J.B.; et al. HSP90 inhibitors disrupt a transient HSP90-HSF1 interaction and identify a noncanonical model of HSP90-mediated HSF1 regulation. Sci. Rep. 2018, 8, 6976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, B.H.; Plescia, J.; Song, H.Y.; Meli, M.; Colombo, G.; Beebe, K.; Scroggins, B.; Neckers, L.; Altieri, D.C. Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. J. Clin. Investig. 2009, 119, 454–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoye, A.T.; Davoren, J.E.; Wipf, P.; Fink, M.P.; Kagan, V.E. Targeting mitochondria. Acc. Chem. Res. 2008, 41, 87–97. [Google Scholar] [CrossRef]
- Kang, B.H.; Siegelin, M.D.; Plescia, J.; Raskett, C.M.; Garlick, D.S.; Dohi, T.; Lian, J.B.; Stein, G.S.; Languino, L.R.; Altieri, D.C. Preclinical characterization of mitochondria-targeted small molecule hsp90 inhibitors, gamitrinibs, in advanced prostate cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 4779–4788. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Park, H.K.; Jeong, H.; Lim, J.; Lee, A.J.; Cheon, K.Y.; Kim, C.S.; Thomas, A.P.; Bae, B.; Kim, N.D.; et al. Development of a mitochondria-targeted Hsp90 inhibitor based on the crystal structures of human TRAP1. J. Am. Chem. Soc. 2015, 137, 4358–4367. [Google Scholar] [CrossRef]
- Bryant, K.G.; Chae, Y.C.; Martinez, R.L.; Gordon, J.C.; Elokely, K.M.; Kossenkov, A.V.; Grant, S.; Childers, W.E.; Abou-Gharbia, M.; Altieri, D.C. A Mitochondrial-targeted purine-based HSP90 antagonist for leukemia therapy. Oncotarget 2017, 8, 112184–112198. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.; Ferraro, M.; Thomas, A.P.; Chung, J.M.; Yoon, N.G.; Seol, J.-H.; Kim, S.; Kim, H.-u.; An, M.Y.; Ok, H.; et al. Dual Binding to Orthosteric and Allosteric Sites Enhances the Anticancer Activity of a TRAP1-Targeting Drug. J. Med. Chem. 2020, 63, 2930–2940. [Google Scholar] [CrossRef]
- Trnka, J.; Elkalaf, M.; Anděl, M. Lipophilic triphenylphosphonium cations inhibit mitochondrial electron transport chain and induce mitochondrial proton leak. PLoS ONE 2015, 10, e0121837. [Google Scholar] [CrossRef] [Green Version]
- Park, H.-K.; Jeong, H.; Ko, E.; Lee, G.; Lee, J.-E.; Lee, S.K.; Lee, A.-J.; Im, J.Y.; Hu, S.; Kim, S.H.; et al. Paralog Specificity Determines Subcellular Distribution, Action Mechanism, and Anticancer Activity of TRAP1 Inhibitors. J. Med. Chem. 2017, 60, 7569–7578. [Google Scholar] [CrossRef]
- Lundgren, K.; Zhang, H.; Brekken, J.; Huser, N.; Powell, R.E.; Timple, N.; Busch, D.J.; Neely, L.; Sensintaffar, J.L.; Yang, Y.C.; et al. BIIB021, an orally available, fully synthetic small-molecule inhibitor of the heat shock protein Hsp90. Mol. Cancer Ther. 2009, 8, 921–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moroni, E.; Agard, D.A.; Colombo, G. The Structural Asymmetry of Mitochondrial Hsp90 (Trap1) Determines Fine Tuning of Functional Dynamics. J. Chem. Theory Comput. 2018, 14, 1033–1044. [Google Scholar] [CrossRef]
- Montefiori, M.; Pilotto, S.; Marabelli, C.; Moroni, E.; Ferraro, M.; Serapian, S.A.; Mattevi, A.; Colombo, G. Impact of Mutations on NPAC Structural Dynamics: Mechanistic Insights from MD Simulations. J. Chem. Inf. Modeling 2019, 59, 3927–3937. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Martin, C.; Moroni, E.; Ferraro, M.; Laquatra, C.; Cannino, G.; Masgras, I.; Negro, A.; Quadrelli, P.; Rasola, A.; Colombo, G. Rational Design of Allosteric and Selective Inhibitors of the Molecular Chaperone TRAP1. Cell Rep. 2020, 31, 107531. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Martin, C.; Menon, D.; Moroni, E.; Ferraro, M.; Masgras, I.; Elsey, J.; Arbiser, J.L.; Colombo, G.; Rasola, A. Honokiol Bis-Dichloroacetate Is a Selective Allosteric Inhibitor of the Mitochondrial Chaperone TRAP1. Antioxid. Redox Signal. 2021, 34, 505–516. [Google Scholar] [CrossRef]
- Zhang, G.; Frederick, D.T.; Wu, L.; Wei, Z.; Krepler, C.; Srinivasan, S.; Chae, Y.C.; Xu, X.; Choi, H.; Dimwamwa, E.; et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Investig. 2016, 126, 1834–1856. [Google Scholar] [CrossRef]
- Ishida, C.T.; Shu, C.; Halatsch, M.-E.; Westhoff, M.-A.; Altieri, D.C.; Karpel-Massler, G.; Siegelin, M.D. Mitochondrial matrix chaperone and c-myc inhibition causes enhanced lethality in glioblastoma. Oncotarget 2017, 8, 37140–37153. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.T.T.; Zhang, Y.; Shang, E.; Shu, C.; Quinzii, C.M.; Westhoff, M.A.; Karpel-Massler, G.; Siegelin, M.D. Inhibition of HDAC1/2 Along with TRAP1 Causes Synthetic Lethality in Glioblastoma Model Systems. Cells 2020, 9, 1661. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joshi, A.; Ito, T.; Picard, D.; Neckers, L. The Mitochondrial HSP90 Paralog TRAP1: Structural Dynamics, Interactome, Role in Metabolic Regulation, and Inhibitors. Biomolecules 2022, 12, 880. https://doi.org/10.3390/biom12070880
Joshi A, Ito T, Picard D, Neckers L. The Mitochondrial HSP90 Paralog TRAP1: Structural Dynamics, Interactome, Role in Metabolic Regulation, and Inhibitors. Biomolecules. 2022; 12(7):880. https://doi.org/10.3390/biom12070880
Chicago/Turabian StyleJoshi, Abhinav, Takeshi Ito, Didier Picard, and Len Neckers. 2022. "The Mitochondrial HSP90 Paralog TRAP1: Structural Dynamics, Interactome, Role in Metabolic Regulation, and Inhibitors" Biomolecules 12, no. 7: 880. https://doi.org/10.3390/biom12070880
APA StyleJoshi, A., Ito, T., Picard, D., & Neckers, L. (2022). The Mitochondrial HSP90 Paralog TRAP1: Structural Dynamics, Interactome, Role in Metabolic Regulation, and Inhibitors. Biomolecules, 12(7), 880. https://doi.org/10.3390/biom12070880






