Selenium Metabolism and Selenoproteins in Prokaryotes: A Bioinformatics Perspective
Abstract
1. Introduction
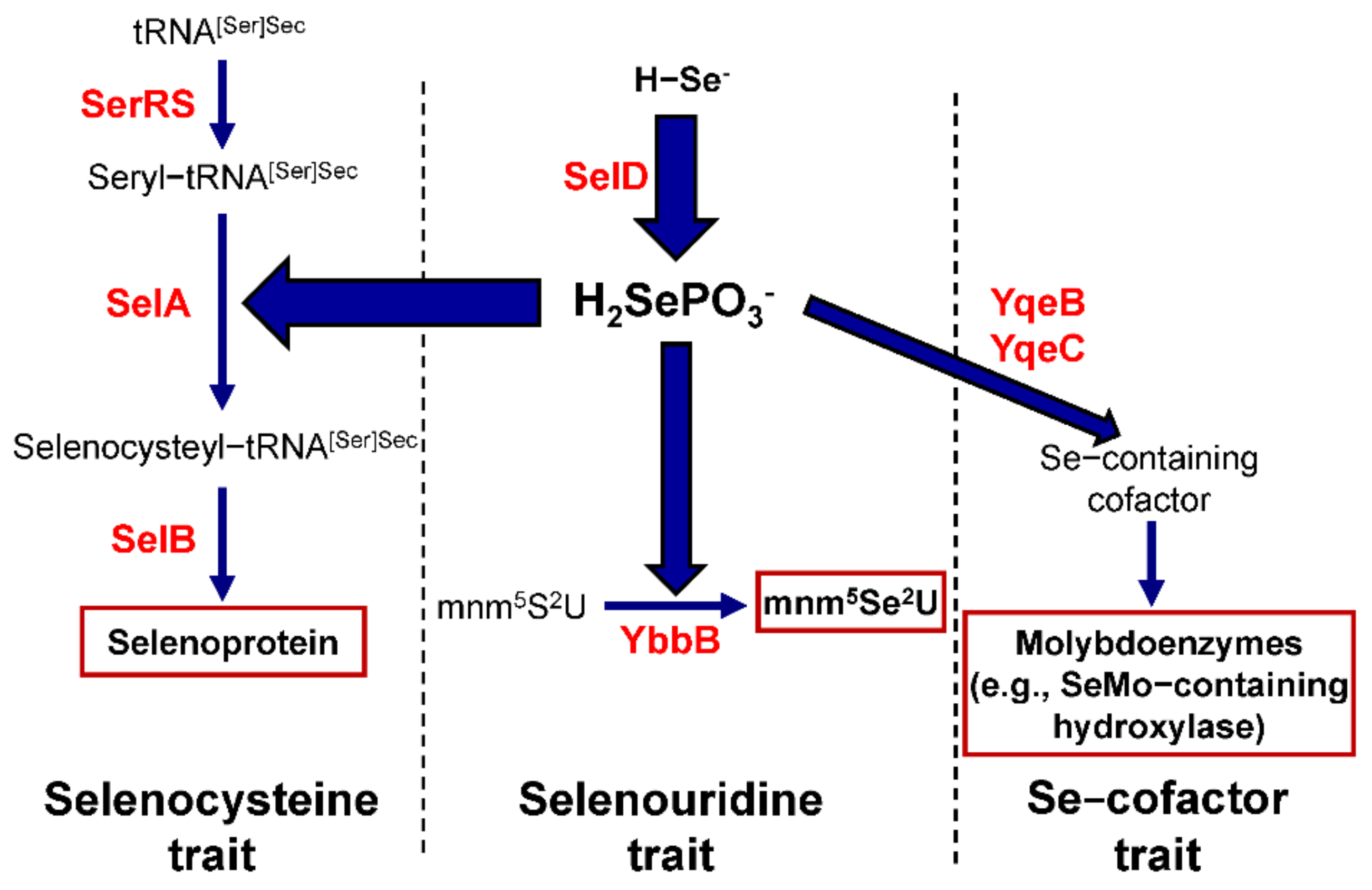
2. An Overview of Selenium Metabolism in Prokaryotes
3. Computational Identification and Classification of Selenoproteins in Prokaryotes
4. Comparative Genomics of Selenium Utilization in Prokaryotes
5. Comparative Metagenomics of Selenium Utilization
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Roman, M.; Jitaru, P.; Barbante, C. Selenium biochemistry and its role for human health. Metallomics 2014, 6, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Mangiapane, E.; Pessione, A.; Pessione, E. Selenium and selenoproteins: An overview on different biological systems. Curr. Protein Pept. Sci. 2014, 15, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Copeland, P.R. Regulation of gene expression by stop codon recoding: Selenocysteine. Gene 2003, 312, 17–25. [Google Scholar] [CrossRef]
- Hariharan, S.; Dharmaraj, S. Selenium and selenoproteins: It’s role in regulation of inflammation. Inflammopharmacology 2020, 28, 667–695. [Google Scholar] [CrossRef] [PubMed]
- Avery, J.C.; Hoffmann, P.R. Selenium, Selenoproteins, and Immunity. Nutrients 2018, 10, 1203. [Google Scholar] [CrossRef]
- Fairweather-Tait, S.J.; Bao, Y.; Broadley, M.R.; Collings, R.; Ford, D.; Hesketh, J.E.; Hurst, R. Selenium in human health and disease. Antioxid. Redox Signal. 2011, 14, 1337–1383. [Google Scholar] [CrossRef]
- Gonzalez-Flores, J.N.; Shetty, S.P.; Dubey, A.; Copeland, P.R. The molecular biology of selenocysteine. Biomol. Concepts 2013, 4, 349–365. [Google Scholar] [CrossRef]
- Bulteau, A.L.; Chavatte, L. Update on selenoprotein biosynthesis. Antioxid. Redox Signal. 2015, 23, 775–794. [Google Scholar] [CrossRef]
- Mariotti, M. SECISearch3 and Seblastian: In-Silico Tools to Predict SECIS Elements and Selenoproteins. Methods Mol. Biol. 2018, 1661, 3–16. [Google Scholar]
- Santesmasses, D.; Mariotti, M.; Guigó, R. Selenoprofiles: A Computational Pipeline for Annotation of Selenoproteins. Methods Mol. Biol. 2018, 1661, 17–28. [Google Scholar]
- Zhang, Y.; Gladyshev, V.N. An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics 2005, 21, 2580–2589. [Google Scholar] [CrossRef] [PubMed]
- Kryukov, G.V.; Gladyshev, V.N. The prokaryotic selenoproteome. EMBO Rep. 2004, 5, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigó, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, H.; Speckmann, B.; Klotz, L.O. Selenoproteins: Antioxidant selenoenzymes and beyond. Arch. Biochem. Biophys. 2016, 595, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, A.J.; Stadtman, T.C. Biosynthesis of 5-methylaminomethyl-2-selenouridine, a naturally occurring nucleoside in Escherichia coli tRNA. Arch. Biochem. Biophys. 1986, 248, 540–550. [Google Scholar] [CrossRef]
- Self, W.T.; Wolfe, M.D.; Stadtman, T.C. Cofactor determination and spectroscopic characterization of the selenium-dependent purine hydroxylase from Clostridium purinolyticum. Biochemistry 2003, 42, 11382–11390. [Google Scholar] [CrossRef]
- Caton-Williams, J.; Huang, Z. Biochemistry of selenium-derivatized naturally occurring and unnatural nucleic acids. Chem. Biodivers. 2008, 5, 396–407. [Google Scholar] [CrossRef]
- Srivastava, M.; Mallard, C.; Barke, T.; Hancock, L.E.; Self, W.T. A selenium-dependent xanthine dehydrogenase triggers biofilm proliferation in Enterococcus faecalis through oxidant production. J. Bacteriol. 2011, 193, 1643–1652. [Google Scholar] [CrossRef]
- Aguilar-Barajas, E.; Díaz-Pérez, C.; Ramírez-Díaz, M.I.; Riveros-Rosas, H.; Cervantes, C. Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 2011, 24, 687–707. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, B.; Li, W.; Che, R.; Deng, K.; Li, H.; Yu, F.; Ling, H.; Li, Y.; Chu, C. OsPT2, a phosphate transporter, is involved in the active uptake of selenite in rice. New Phytol. 2014, 201, 1183–1191. [Google Scholar] [CrossRef]
- Lazard, M.; Blanquet, S.; Fisicaro, P.; Labarraque, G.; Plateau, P. Uptake of selenite by Saccharomyces cerevisiae involves the high and low affinity orthophosphate transporters. J. Biol. Chem. 2010, 285, 32029–32037. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.T.; Tian, L.J.; Yu, H.Q. Phosphate-Suppressed Selenite Biotransformation by Escherichia coli. Environ. Sci. Technol. 2020, 54, 10713–10721. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.; Basu, P.; Stolz, J.F. The physiology and evolution of microbial selenium metabolism. Metallomics 2021, 13, mfab024. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, S.; Böck, A. The many levels of control on bacterial selenoprotein synthesis. Biochim. Biophys. Acta 2009, 1790, 1404–1414. [Google Scholar] [CrossRef] [PubMed]
- Rother, M.; Resch, A.; Wilting, R.; Böck, A. Selenoprotein synthesis in archaea. Biofactors 2001, 14, 75–83. [Google Scholar] [CrossRef]
- Fu, X.; Söll, D.; Sevostyanova, A. Challenges of site-specific selenocysteine incorporation into proteins by Escherichia coli. RNA Biol. 2018, 15, 461–470. [Google Scholar] [CrossRef]
- Rother, M.; Quitzke, V. Selenoprotein synthesis and regulation in Archaea. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2451–2462. [Google Scholar] [CrossRef]
- Xu, X.M.; Carlson, B.A.; Zhang, Y.; Mix, H.; Kryukov, G.V.; Glass, R.S.; Berry, M.J.; Gladyshev, V.N.; Hatfield, D.L. New developments in selenium biochemistry: Selenocysteine biosynthesis in eukaryotes and archaea. Biol. Trace Elem. Res. 2007, 119, 234–241. [Google Scholar] [CrossRef]
- Wilting, R.; Schorling, S.; Persson, B.C.; Böck, A. Selenoprotein synthesis in archaea: Identification of an mRNA element of Methanococcus jannaschii probably directing selenocysteine insertion. J. Mol. Biol. 1997, 266, 637–641. [Google Scholar] [CrossRef][Green Version]
- Sierant, M.; Leszczynska, G.; Sadowska, K.; Komar, P.; Radzikowska-Cieciura, E.; Sochacka, E.; Nawrot, B. Escherichia coli tRNA 2-selenouridine synthase (SelU) converts S2U-RNA to Se2U-RNA via S-geranylated-intermediate. FEBS Lett. 2018, 592, 2248–2258. [Google Scholar] [CrossRef]
- Zhang, Y.; Turanov, A.A.; Hatfield, D.L.; Gladyshev, V.N. In silico identification of genes involved in selenium metabolism: Evidence for a third selenium utilization trait. BMC Genomics 2008, 9, 251. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; Self, W.T. Orphan SelD proteins and selenium-dependent molybdenum hydroxylases. Biol. Direct 2008, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Tobe, R.; Mihara, H. Delivery of selenium to selenophosphate synthetase for selenoprotein biosynthesis. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2433–2440. [Google Scholar] [CrossRef] [PubMed]
- Seale, L.A. Selenocysteine β-Lyase: Biochemistry, Regulation and Physiological Role of the Selenocysteine Decomposition Enzyme. Antioxidants 2019, 8, 357. [Google Scholar] [CrossRef]
- Yee, N.; Choi, J.; Porter, A.W.; Carey, S.; Rauschenbach, I.; Harel, A. Selenate reductase activity in Escherichia coli requires Isc iron-sulfur cluster biosynthesis genes. FEMS Microbiol. Lett. 2014, 361, 138–143. [Google Scholar] [CrossRef]
- Kessi, J.; Hanselmann, K.W. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J. Biol. Chem. 2004, 279, 50662–50669. [Google Scholar] [CrossRef]
- Huang, S.; Wang, Y.; Tang, C.; Jia, H.; Wu, L. Speeding up selenite bioremediation using the highly selenite-tolerant strain Providencia rettgeri HF16-A novel mechanism of selenite reduction based on proteomic analysis. J. Hazard Mater. 2021, 406, 124690. [Google Scholar] [CrossRef]
- Wells, M.; McGarry, J.; Gaye, M.M.; Basu, P.; Oremland, R.S.; Stolz, J.F. Respiratory Selenite Reductase from Bacillus selenitireducens Strain MLS10. J. Bacteriol. 2019, 201, e00614-18. [Google Scholar] [CrossRef]
- Gong, Y.; Wu, Y.; Khan, A.; Song, P.; Wang, Z.; Ni, H.; Ji, J.; Salama, E.S.; Liu, P.; Li, X. Improving selenium accumulation in broilers using Escherichia coli Nissle 1917 with surface-displayed selenite reductase SerV01. Food Funct. 2022, 13, 4537–4550. [Google Scholar] [CrossRef]
- Li, D.B.; Cheng, Y.Y.; Wu, C.; Li, W.W.; Li, N.; Yang, Z.C.; Tong, Z.H.; Yu, H.Q. Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm. Sci. Rep. 2014, 4, 3735. [Google Scholar] [CrossRef]
- Tamura, T.; Sato, K.; Komori, K.; Imai, T.; Kuwahara, M.; Okugochi, T.; Mihara, H.; Esaki, N.; Inagaki, K. Selenite reduction by the thioredoxin system: Kinetics and identification of protein-bound selenide. Biosci. Biotechnol. Biochem. 2011, 75, 1184–1187. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, A.; Tobem, R.; Aono, R.; Inoue, M.; Hagita, S.; Kiriyama, K.; Toyotake, Y.; Ogawa, T.; Kurihara, T.; Goto, K.; et al. Initial Step of Selenite Reduction via Thioredoxin for Bacterial Selenoprotein Biosynthesis. Int. J. Mol. Sci. 2021, 22, 10965. [Google Scholar] [CrossRef] [PubMed]
- Santesmasses, D.; Mariotti, M.; Gladyshev, V.N. Bioinformatics of Selenoproteins. Antioxid. Redox Signal. 2020, 33, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. Prokaryotic selenoproteins and selenoproteomes. In Selenium—Its Molecular Biology and Role in Human Health, 4th ed.; Hatfield, D.L., Schweizer, U., Tsuji, P.A., Gladyshev, V.N., Eds.; Springer: New York, NY, USA, 2016; pp. 141–150. [Google Scholar]
- Zhang, Y.; Fomenko, D.E.; Gladyshev, V.N. The microbial selenoproteome of the Sargasso Sea. Genome Biol. 2005, 6, R37. [Google Scholar] [CrossRef]
- Zhang, Y.; Gladyshev, V.N. General trends in trace element utilization revealed by comparative genomic analyses of Co, Cu, Mo, Ni, and Se. J. Biol. Chem. 2010, 285, 3393–3405. [Google Scholar] [CrossRef]
- Mariotti, M.; Lobanov, A.V.; Manta, B.; Santesmasses, D.; Bofill, A.; Guigó, R.; Gabaldón, T.; Gladyshev, V.N. Lokiarchaeota Marks the Transition between the Archaeal and Eukaryotic Selenocysteine Encoding Systems. Mol. Biol. Evol. 2016, 33, 2441–2453. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Z.; Pan, J.; Baker, B.J.; Gu, J.D.; Li, M. Comparative genomic inference suggests mixotrophic lifestyle for Thorarchaeota. ISME J. 2018, 12, 1021–1031. [Google Scholar] [CrossRef]
- Böck, A.; Forchhammer, K.; Heider, J.; Leinfelder, W.; Sawers, G.; Veprek, B.; Zinoni, F. Selenocysteine: The 21st amino acid. Mol. Microbiol. 1991, 5, 515–520. [Google Scholar] [CrossRef]
- Vorholt, J.A.; Vaupel, M.; Thauer, R.K. A selenium-dependent and a selenium-independent formylmethanofuran dehydrogenase and their transcriptional regulation in the hyperthermophilic Methanopyrus kandleri. Mol. Microbiol. 1997, 23, 1033–1042. [Google Scholar] [CrossRef]
- Tormay, P.; Wilting, R.; Lottspeich, F.; Mehta, P.K.; Christen, P.; Böck, A. Bacterial Selenocysteine synthase—Structural and functional properties. Eur. J. Biochem. 1998, 254, 655–661. [Google Scholar] [CrossRef]
- Rother, M.; Resch, A.; Gardner, W.L.; Whitman, W.B.; Böck, A. Heterologous expression of archaeal selenoprotein genes directed by the SECIS element located in the 3’ non-translated region. Mol. Microbiol. 2001, 40, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Halboth, S.; Klein, A. Methanococcus voltae harbors four gene clusters potentially encoding two [NiFe] and two [NiFeSe] hydrogenases, each of the cofactor F420-reducing or F420-non-reducing types. Mol. Gen. Genet. 1992, 233, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Shokes, J.E.; Duin, E.C.; Bauer, C.; Jaun, B.; Hedderich, R.; Koch, J.; Scott, R.A. Direct interaction of coenzyme M with the active-site Fe-S cluster of heterodisulfide reductase. FEBS Lett. 2005, 579, 1741–1744. [Google Scholar] [CrossRef]
- Stock, T.; Selzer, M.; Rother, M. In vivo requirement of selenophosphate for selenoprotein synthesis in archaea. Mol. Microbiol. 2010, 75, 149–160. [Google Scholar] [CrossRef]
- Kreimer, S.; Andreesen, J.R. Glycine reductase of Clostridium litorale. Cloning, sequencing, and molecular analysis of the grdAB operon that contains two in-frame TGA codons for selenium incorporation. Eur. J. Biochem. 1995, 234, 192–199. [Google Scholar] [CrossRef]
- Andreesen, J.R.; Wagner, M.; Sonntag, D.; Kohlstock, M.; Harms, C.; Gursinsky, T.; Jäger, J.; Parther, T.; Kabisch, U.; Gräntzdörffer, A.; et al. Various functions of selenols and thiols in anaerobic gram-positive, amino acids-utilizing bacteria. Biofactors 1999, 10, 263–270. [Google Scholar] [CrossRef]
- Söhling, B.; Parther, T.; Rücknagel, K.P.; Wagner, M.A.; Andreesen, J.R. A selenocysteine-containing peroxiredoxin from the strictly anaerobic organism Eubacterium acidaminophilum. Biol. Chem. 2001, 382, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Lee, B.C.; Hwang, K.Y.; Gladyshev, V.N.; Kim, H.Y. Selenium utilization in thioredoxin and catalytic advantage provided by selenocysteine. Biochem. Biophys. Res. Commun. 2015, 461, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Lee, B.C.; Jeong, J.; Lee, K.J.; Hwang, K.Y.; Gladyshev, V.N.; Kim, H.Y. Tandem use of selenocysteine: Adaptation of a selenoprotein glutaredoxin for reduction of selenoprotein methionine sulfoxide reductase. Mol. Microbiol. 2011, 79, 1194–1203. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Zhang, Y.; Lee, B.C.; Kim, J.R.; Gladyshev, V.N. The selenoproteome of Clostridium sp. OhILAs: Characterization of anaerobic bacterial selenoprotein methionine sulfoxide reductase A. Proteins 2009, 74, 1008–1017. [Google Scholar] [CrossRef]
- Fomenko, D.E.; Xing, W.; Adair, B.M.; Thomas, D.J.; Gladyshev, V.N. High-throughput identification of catalytic redox-active cysteine residues. Science 2007, 315, 387–389. [Google Scholar] [CrossRef]
- Zhang, Y.; Gladyshev, V.N. High content of proteins containing 21st and 22nd amino acids, selenocysteine and pyrrolysine, in a symbiotic deltaproteobacterium of gutless worm Olavius algarvensis. Nucleic Acids Res. 2007, 35, 4952–4963. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gladyshev, V.N. Trends in selenium utilization in marine microbial world revealed through the analysis of the global ocean sampling (GOS) project. PLoS Genet. 2008, 4, e1000095. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Lin, J.; Xu, Y.Z.; Zhang, Y. Comparative genomics reveals new evolutionary and ecological patterns of selenium utilization in bacteria. ISME J. 2016, 10, 2048–2059. [Google Scholar] [CrossRef] [PubMed]
- Fonknechten, N.; Chaussonnerie, S.; Tricot, S.; Lajus, A.; Andreesen, J.R.; Perchat, N.; Pelletier, E.; Gouyvenoux, M.; Barbe, V.; Salanoubat, M.; et al. Clostridium sticklandii, a specialist in amino acid degradation:Revisiting its metabolism through its genome sequence. BMC Genomics 2010, 11, 555. [Google Scholar] [CrossRef] [PubMed]
- Cravedi, P.; Mori, G.; Fischer, F.; Percudani, R. Evolution of the Selenoproteome in Helicobacter pylori and Epsilonproteobacteria. Genome Biol. Evol. 2015, 7, 2692–2704. [Google Scholar]
- Santesmasses, D.; Mariotti, M.; Guigó, R. Computational identification of the selenocysteine tRNA (tRNASec) in genomes. PLoS Comput. Biol. 2017, 13, e1005383. [Google Scholar] [CrossRef]
- Hardison, R.C. Comparative genomics. PLoS Biol. 2003, 1, E58. [Google Scholar] [CrossRef]
- Ellegren, H. Comparative genomics and the study of evolution by natural selection. Mol. Ecol. 2008, 17, 4586–4596. [Google Scholar] [CrossRef]
- Zhang, Y.; Ying, H.; Xu, Y. Comparative genomics and metagenomics of the metallomes. Metallomics 2019, 11, 1026–1043. [Google Scholar] [CrossRef]
- Davy, T.; Castellano, S. The genomics of selenium: Its past, present and future. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2427–2432. [Google Scholar] [CrossRef] [PubMed]
- Li, G.P.; Jiang, L.; Ni, J.Z.; Liu, Q.; Zhang, Y. Computational identification of a new SelD-like family that may participate in sulfur metabolism in hyperthermophilic sulfur-reducing archaea. BMC Genomics 2014, 15, 908. [Google Scholar] [CrossRef]
- Mariotti, M.; Santesmasses, D.; Capella-Gutierrez, S.; Mateo, A.; Arnan, C.; Johnson, R.; D’Aniello, S.; Yim, S.H.; Gladyshev, V.N.; Serras, F.; et al. Evolution of selenophosphate synthetases: Emergence and relocation of function through independent duplications and recurrent subfunctionalization. Genome Res. 2015, 25, 1256–1267. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Peng, T.; Jiang, L.; Ni, J.Z.; Liu, Q.; Chen, L.; Zhang, Y. Comparative genomics reveals new candidate genes involved in selenium metabolism in prokaryotes. Genome Biol. Evol. 2015, 7, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Quitzke, V.; Fersch, J.; Seyhan, D.; Rother, M. Selenium-dependent gene expression in Methanococcus maripaludis: Involvement of the transcriptional regulator HrsM. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2441–2450. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.G.; Yee, E.; Lopes, B.S.; Chapman, M.H.; Huynh, S.; Bono, J.L.; Parker, C.T.; Strachan, N.J.C.; Forbes, K.J. Comparative Genomic Analysis Identifies a Campylobacter Clade Deficient in Selenium Metabolism. Genome Biol. Evol. 2017, 9, 1843–1858. [Google Scholar] [CrossRef]
- Parker, C.T.; Cooper, K.K.; Schiaffino, F.; Miller, W.G.; Huynh, S.; Gray, H.K.; Olortegui, M.P.; Bardales, P.G.; Trigoso, D.R.; Penataro-Yori, P.; et al. Genomic Characterization of Campylobacter jejuni Adapted to the Guinea Pig (Cavia porcellus) Host. Front. Cell. Infect. Microbiol. 2021, 11, 607747. [Google Scholar] [CrossRef]
- Mukai, T. Bioinformatic Prediction of an tRNASec Gene Nested inside an Elongation Factor SelB Gene in Alphaproteobacteria. Int. J. Mol. Sci. 2021, 22, 4605. [Google Scholar] [CrossRef]
- Alves, L.F.; Westmann, C.A.; Lovate, G.L.; de Siqueira, G.M.V.; Borelli, T.C.; Guazzaroni, M.E. Metagenomic Approaches for Understanding New Concepts in Microbial Science. Int. J. Genomics 2018, 2018, 2312987. [Google Scholar] [CrossRef]
- Faust, K.; Lahti, L.; Gonze, D.; de Vos, W.M.; Raes, J. Metagenomics meets time series analysis: Unraveling microbial community dynamics. Curr. Opin. Microbiol. 2015, 25, 56–66. [Google Scholar] [CrossRef]
- Gwak, H.J.; Lee, S.J.; Rho, M. Application of computational approaches to analyze metagenomic data. J. Microbiol. 2021, 59, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Morel, F.M. The co-evolution of phytoplankton and trace element cycles in the oceans. Geobiology 2008, 6, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.; Stolz, J.F. Microbial selenium metabolism: A brief history, biogeochemistry and ecophysiology. FEMS Microbiol. Ecol. 2020, 96, fiaa209. [Google Scholar] [CrossRef]
- Xu, Y.; Cao, J.; Jiang, L.; Zhang, Y. Biogeographic and Evolutionary Patterns of Trace Element Utilization in Marine Microbial World. Genomics Proteomics Bioinformatics 2021, S1672-0229(21)00036-X. [Google Scholar] [CrossRef] [PubMed]
- Farukh, M. Comparative genomic analysis of selenium utilization traits in different marine environments. J. Microbiol. 2020, 58, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Kivenson, V.; Paul, B.G.; Valentine, D.L. An Ecological Basis for Dual Genetic Code Expansion in Marine Deltaproteobacteria. Front. Microbiol. 2021, 12, 680620. [Google Scholar] [CrossRef]
- Nkansah-Boadu, F.; Hatam, I.; Baldwin, S.A. Microbial consortia capable of reducing selenate in the presence of nitrate enriched from coalmining-impacted environments. Appl. Microbiol. Biotechnol. 2021, 105, 1287–1300. [Google Scholar] [CrossRef]
- Shi, L.D.; Lv, P.L.; Niu, Z.F.; Lai, C.Y.; Zhao, H.P. Why does sulfate inhibit selenate reduction: Molybdenum deprivation from Mo-dependent selenate reductase. Water Res. 2020, 178, 115832. [Google Scholar] [CrossRef]

| Selenoprotein Family or Subfamily Name | Domain ID (Name) | Sec-Related Motif | Representative Sequence (Genbank/Refseq) | Ref. |
|---|---|---|---|---|
| Experimentally verified (16) | ||||
| Formate dehydrogenase alpha subunit * | COG0243 (BisC) | - | WP_010904702.1 | [49] |
| Formylmethanofuran dehydrogenase subunit B * | COG1029 (FwdB) | - | CAA67419.1 | [50] |
| Selenophosphate synthetase * | COG0709 (SelD) | UxxK | WP_083774555.1 | [51] |
| Coenzyme F420-reducing hydrogenase alpha subunit * | COG3259 (FrhA) | UxxC | WP_083774535.1 | [52] |
| Methylviologen-reducing (or F420-nonreducing) hydrogenase alpha subunit * | COG3259 (FrhA) | UxxC | P0C1V6.2 | [27] |
| Coenzyme F420-reducing hydrogenase delta subunit * | COG1908 (FrhD) | - | WP_010870703.1 | [53] |
| Heterodisulfide reductase alpha subunit * | COG1148 (HdrA) | CxxU | WP_162484757.1 | [54] |
| HesB-like protein * | TIGR01911 (HesB_rel_seleno) | - | WP_083774540.1 | [55] |
| Glycine reductase complex selenoprotein A | pfam04723 (GRDA) | CxxU | WP_079747582.1 | [56] |
| Glycine reductase complex selenoprotein B | pfam07355 (GRDB) | UxxC | WP_246895825.1 | [56] |
| D-proline reductase | TIGR04483 (D_pro_red_PrdB) | UxxC | WP_079281142.1 | [57] |
| Peroxiredoxin (Prx) | COG1225 (Bcp) | TxxU | WP_011365628.1 | [58] |
| Thioredoxin (Trx) | pfam00085 (Thioredoxin) | UxxC | WP_010956703.1 | [59] |
| Glutaredoxin (Grx) | pfam00462 (Glutaredoxin) | UxxC | WP_010943784.1 | [60] |
| Methione sulfoxide reductase A | COG0225 (MsrA) | - | MBI4965933.1 | [61] |
| Arsenite methyltransferase | PRK11873 (arsM) | - | WP_011987699.1 | [62] |
| Predicted (71) | ||||
| Radical SAM domain protein | TIGR04167 (rSAM_SeCys) | - | AAR34688.1 | [11] |
| Rhodanese-like domain-containing protein | pfam00581 (Rhodanese) | - | WP_010941598.1 | [11] |
| Rhodanese-related sulfurtransferase COG0607 form 1 | COG0607 (PspE) | - | MBM9537886.1 | [11] |
| Rhodanese-related sulfurtransferase COG0607 form 2 | COG0607 (PspE) | CxU | TKB26178.1 | [11] |
| Prx-like thiol:disulfide oxidoreductase * | pfam00578 (AhpC-TSA) | UxxC, UxxU | WP_010940744.1 | [12] |
| Thiol:disulfide interchange protein | pfam13098 (Thioredoxin_2) | UxxC | WP_011366075.1 | [12] |
| Selenoprotein W (SELENOW)-like protein | pfam10262 (Rdx) | CxxU | AOH51717.1 | [12] |
| Glutathione peroxidase (GPX)-like protein | pfam00255 (GSHPx) | UxxT | WP_010957027.1 | [12] |
| Homolog of AhpF N-terminal domain (Grx-like domain protein) | TIGR02187 (GlrX_arch) | UxxC | ABB15282.1 | [12] |
| DsbG-like protein | pfam13462 (Thioredoxin_4) | UxxC | WP_012258530.1 ** | [12] |
| Fe-S oxidoreductase-like protein | COG0247 (GlpC) | - | WP_174406253.1 | [12] |
| DsrE-like protein | pfam02635 (DsrE) | UxxC | WP_014524487.1 | [12] |
| FAD-dependent oxidoreductase (CoA-disulfide reductase, NADH oxidase) | COG0446 (FadH2) | - | WP_011365774.1 | [12] |
| Distant Alkylhydroperoxidase (AhpD) homolog | COG0599 (YurZ) | CxxU | AAR36519.2 | [12] |
| AhpD-like protein | COG2128 (YciW) | CxxU | MCB9421940.1 | [45] |
| Arsenate reductase | COG1393 (ArsC) | UxxS | MBT3519430.1 | [45] |
| Molybdopterin-synthase adenylyltransferase MoeB | COG0476 (ThiF) | - | MBT7809913.1 | [45] |
| DsbA-like protein | pfam01323 (DSBA) | UxxC | NIP15863.1 | [45] |
| Glutathione S-transferase-like (GST-like) | COG0625 (GstA) | - | PPR62222.1 | [45] |
| Deiodinase-like protein | pfam00837 (T4_deiodinase) | UxxC | MBO99264.1 | [45] |
| Thiol-disulfide isomerase-like protein | pfam13905 (Thioredoxin_8) | UxxC | MAK15852.1 | [45] |
| Carboxymuconolactone decarboxylase(CMD)-like protein | pfam02627 (CMD) | CxxU | MBW1767730.1 | [45] |
| Hypothetical protein 1 (Sargasso Sea metagenome) | - | CxxU | MBR86424.1 | [45] |
| OsmC-like protein | COG1765 (YhfA) | UxxT | MBR72571.1 | [45] |
| Rhodanase-related sulfurtransferase | COG2897 (SseA) | - | MQG53192.1 | [45] |
| NADH:ubiquinone oxidoreductase subunit E | COG2209 (NqrE) | TxxU | - | [45] |
| Putative mercuric transport protein | pfam02411 (MerT) | - | ABB16073.1 | [63] |
| Cation-transporting ATPase, E1-E2 family | COG2217 (ZntA) | UxxC | ABB15669.1 | [63] |
| Methylated-DNA-protein-cysteine methyltransferase | COG0350 (AdaB) | - | ABB14497.1 | [63] |
| UGSC-containing protein | - | UxxC | ABI76733.1 | [63] |
| DUF3179 domain-containing protein | pfam11376 (DUF3179) | UxxC/T | MBW1804167.1 | [63] |
| YHS domain-containing protein | pfam04945 (YHS) | - | - | [63] |
| Putative redox protein | - | - | KGM38912.1 | [63] |
| DUF166 domain-containing protein | pfam02593 (DUF166) | - | - | [63] |
| DUF1573 domain-containing protein | pfam07610 (DUF1573) | UGC | CAB1076174.1 | [63] |
| Hypothetical protein OS_HP3 | - | - | SMF39960.1 | [63] |
| Putative mercuric reductase | PRK13748 (PRK13748) | UxxU | CAB1070815.1 | [63] |
| Hypothetical protein OS_HP4 | - | UxxC | - | [63] |
| Cobalamin synthesis protein CobW-like | COG0523 (YejR) | UxxC | CAB1077436.1 | [63] |
| AhpC/TSA family protein | pfam13911 (AhpC-TSA_2) | UxxS | CAB1081847.1 | [63] |
| Hypothetical protein OS_HP5 | - | - | - | [63] |
| Distant Grx-like protein 1 | TIGR02196 (GlrX_YruB) | UxxT | MBW2590879.1 | [64] |
| Arsenate reductase-like protein | COG1393 (ArsC) | UxxC | MAM02162.1 | [64] |
| Fe-S cluster domain-containing protein | PRK07118 (PRK07118) | UxxC | ABC78902.1 | [64] |
| (2Fe-2S)-binding protein (copper chaperone Copz family) form 1 | cd10141 (CopZ-like_Fer2_BFD-like) | - | WP_245779778.1 | [64] |
| (2Fe-2S)-binding protein (copper chaperone Copz family) form 2 | cd10141 (CopZ-like_Fer2_BFD-like) | - | MBF1269327.1 | [64] |
| Hypothetical protein predicted in Moorella thermoacetica | - | - | WP_155767724.1 | [64,65] |
| Alkylmercury lyase MerB-like protein | pfam03243 (MerB) | - | WP_238493467.1 | [64,65] |
| DUF1858 domain-containing protein | pfam08984 (DUF1858) | CxxU | WP_012065717.1 | [64,65] |
| Proline reductase-associated electron transfer protein PrdC form 1 | TIGR04481 (PR_assoc_PrdC) | CxxU | WP_243183503.1 | [64,66] |
| Proline reductase-associated electron transfer protein PrdC form 2 | TIGR04481 (PR_assoc_PrdC) | - | WP_245122565.1 | [64,65] |
| cytochrome c family protein | pfam13435 (Cytochrome_C554) | - | WP_013164579.1 | [64,65] |
| MtrB/PioB family outer membrane beta-barrel protein | pfam11854 (MtrB_PioB) | - | WP_005997773.1 | [64] |
| UshA-like protein | COG0737 (UshA) | CxU | WP_013162925.1 | [64] |
| C-GCAxxG-C-C family protein | pfam09719 (C_GCAxxG_C_C) | - | WP_012158890.1 | [64] |
| CO dehydrogenase/acetyl-CoA synthase gamma subunit | COG1456 (CdhE) | - | WP_012647565.1 | [64] |
| YeeE/YedE family protein | pfam04143 (Sulf_transp) | - | WP_012471001.1 | [64,65] |
| UGC-containing Prx-like protein | pfam00578 (AhpC-TSA) | UGC | MBL6689828.1 | [64] |
| Ferredoxin-thioredoxin reductase | COG4802 (FtrB) | CxU | MBG54406.1 | [64] |
| Trypsin-like serine protease | pfam00089 (Trypsin) | - | - | [64] |
| Putative regulatory protein, FmdB family | TIGR02605 (CxxC_CxxC_SSSS) | U/CxxU | - | [64] |
| PDZ domain-containing protein | pfam13899 (Thioredoxin_7) | CxxU | MBM3766709.1 | [64] |
| Hypothetical protein GOS_A | - | - | - | [64] |
| Hypothetical protein GOS_B | - | - | NBR19009.1 | [64] |
| Hypothetical protein GOS_C | cd02973 (TRX_GRX_like) | UxxC | MBI79719.1 | [64] |
| Redoxin family protein | - | UxxC | MBA3499694.1 | [64,65] |
| Crotonase/enoyl-CoA hydratase family protein | PRK06023 (PRK06023) | - | KAA1296466.1 | [65] |
| Cobalamin binding protein BtuF | cd01144 (BtuF) | CxxU | RUA21600.1 | [65] |
| KCU-star family selenoprotein (or DUF466 protein) | NF033934 (KCU-star) | - | WP_052061029.1 | [67] |
| Thioredoxin-like selenoprotein Sec.1 | pfam13192 (Thioredoxin_3) | CxU | WP_232817751.1 | [68] |
| Thioredoxin-like selenoprotein Sec.2 | pfam13192 (Thioredoxin_3) | UxC | WP_218069652.1 | [68] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Jin, J.; Huang, B.; Ying, H.; He, J.; Jiang, L. Selenium Metabolism and Selenoproteins in Prokaryotes: A Bioinformatics Perspective. Biomolecules 2022, 12, 917. https://doi.org/10.3390/biom12070917
Zhang Y, Jin J, Huang B, Ying H, He J, Jiang L. Selenium Metabolism and Selenoproteins in Prokaryotes: A Bioinformatics Perspective. Biomolecules. 2022; 12(7):917. https://doi.org/10.3390/biom12070917
Chicago/Turabian StyleZhang, Yan, Jiao Jin, Biyan Huang, Huimin Ying, Jie He, and Liang Jiang. 2022. "Selenium Metabolism and Selenoproteins in Prokaryotes: A Bioinformatics Perspective" Biomolecules 12, no. 7: 917. https://doi.org/10.3390/biom12070917
APA StyleZhang, Y., Jin, J., Huang, B., Ying, H., He, J., & Jiang, L. (2022). Selenium Metabolism and Selenoproteins in Prokaryotes: A Bioinformatics Perspective. Biomolecules, 12(7), 917. https://doi.org/10.3390/biom12070917





