Benchmarking the Accuracy of AlphaFold 2 in Loop Structure Prediction
Abstract
1. Introduction
2. Methods
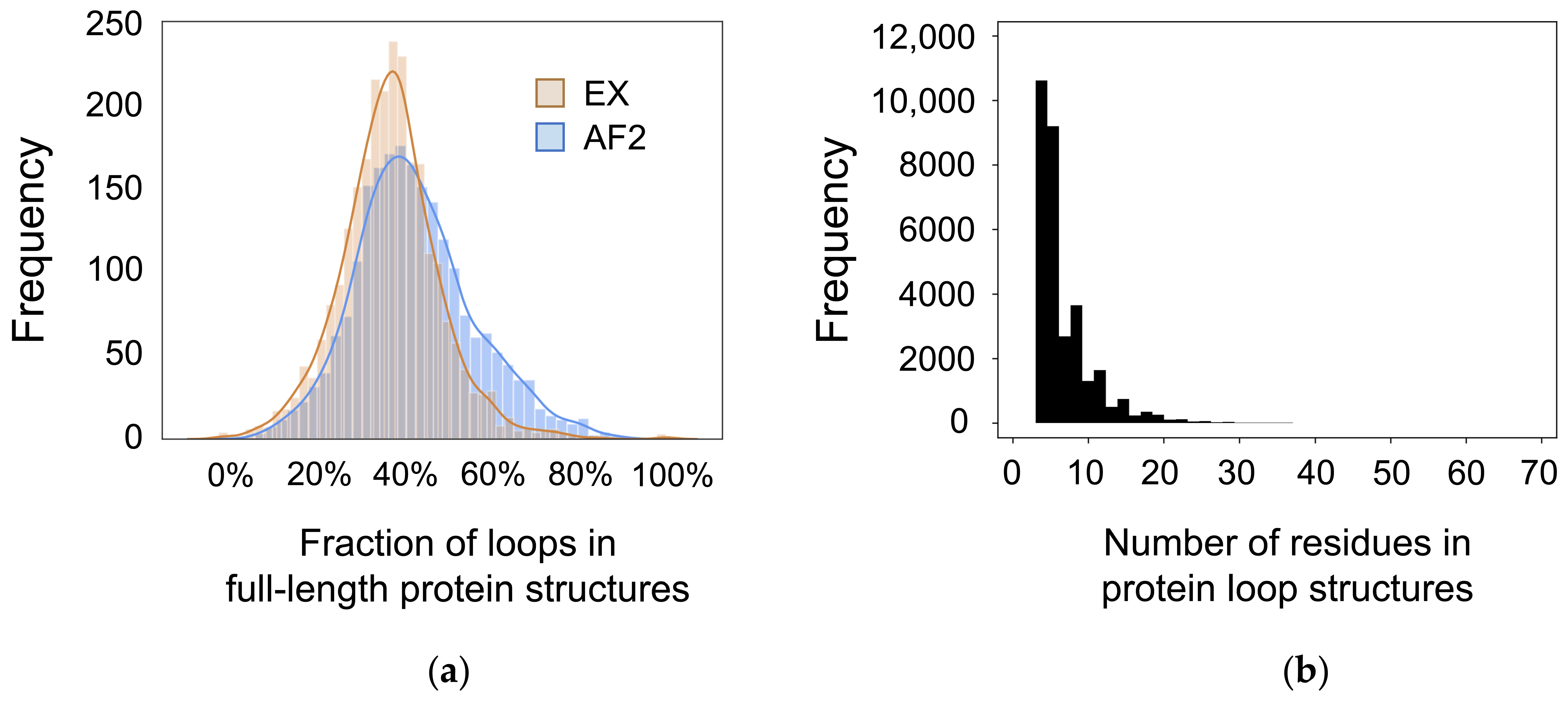
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, M.S.; Srinivasan, N.; Sowdhamini, R.; Blundell, T.L. Knowledge-based protein modeling. Crit. Rev. Biochem. Mol. Biol. 1994, 29, 1–68. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Brandt, B.W.; Heringa, J.; Leunissen, J.A.M. SEQATOMS: A web tool for identifying missing regions in PDB in sequence context. Nucleic Acids Res. 2008, 36, W255–W259. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lins, L.; Thomas, A.; Brasseur, R. Analysis of accessible surface of residues in proteins. Protein Sci. 2003, 12, 1406–1417. [Google Scholar] [CrossRef]
- Papaleo, E.; Saladino, G.; Lambrughi, M.; Lindorff-Larsen, K.; Gervasio, F.L.; Nussinov, R. The Role of Protein Loops and Linkers in Conformational Dynamics and Allostery. Chem. Rev. 2016, 116, 6391–6423. [Google Scholar] [CrossRef]
- Wu, S.; Dean, D. Functional significance of loops in the receptor binding domain ofBacillus thuringiensisCryIIIA δ-endotoxin. J. Mol. Biol. 1996, 255, 628–640. [Google Scholar] [CrossRef]
- Shi, L.; Javitch, J.A. The second extracellular loop of the dopamine D2 receptor lines the binding-site crevice. Proc. Natl. Acad. Sci. USA 2004, 101, 440–445. [Google Scholar] [CrossRef]
- Jones, S.; Thornton, J.M. Prediction of protein-protein interaction sites using patch analysis. J. Mol. Biol. 1997, 272, 133–143. [Google Scholar] [CrossRef]
- Fiser, A.; Bioinformatics, A.S. ModLoop: Automated modeling of loops in protein structures. Bioinformatics 2003, 19, 2500–2501. [Google Scholar] [CrossRef]
- Martí-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sánchez, R.; Melo, F.; Šali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. [Google Scholar] [CrossRef]
- Cohen, B.I.; Presnell, S.R.; Cohen, F.E. Origins of structural diversity within sequentially identical hexapeptides. Protein Sci. 1993, 2, 2134–2145. [Google Scholar] [CrossRef]
- Ring, C.S.; Kneller, D.G.; Langridge, R.; Cohen, F.E. Taxonomy and conformational analysis of loops in proteins. J. Mol. Biol. 1992, 224, 685–699. [Google Scholar] [CrossRef]
- Rufino, S.D.; Donate, L.E.; Canard, L.H.J.; Blundell, T.L. Predicting the conformational class of short and medium size loops connecting regular secondary structures: Application to comparative modelling. J. Mol. Biol. 1997, 267, 352–367. [Google Scholar] [CrossRef]
- Wojcik, J.; Mornon, J.P.; Chomilier, J. New efficient statistical sequence-dependent structure prediction of short to medium-sized protein loops based on an exhaustive loop classification. J. Mol. Biol. 1999, 289, 1469–1490. [Google Scholar] [CrossRef]
- Oliva, B.; Bates, P.A.; Querol, E.; Avilés, F.X.; Sternberg, M.J.E. An automated classification of the structure of protein loops. J. Mol. Biol. 1997, 266, 814–830. [Google Scholar] [CrossRef]
- Tippana, R.; Xiao, W.; Myong, S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014, 42, 8106–8114. [Google Scholar] [CrossRef]
- Panchenko, A.R.; Madej, T. Structural similarity of loops in protein families: Toward the understanding of protein evolution. BMC Evol. Biol. 2005, 5, 10. [Google Scholar] [CrossRef]
- Moult, J.; Fidelis, K.; Kryshtafovych, A.; Schwede, T.; Tramontano, A. Critical assessment of methods of protein structure prediction (CASP)—Round XII. Proteins Struct. Funct. Bioinforma. 2018, 86, 7–15. [Google Scholar] [CrossRef]
- Bonet, J.; Planas-Iglesias, J.; Garcia-Garcia, J.; Marín-López, M.A.; Fernandez-Fuentes, N.; Oliva, B. ArchDB 2014: Structural classification of loops in proteins. Nucleic Acids Res. 2014, 42, D315–D319. [Google Scholar] [CrossRef]
- Fernandez-Fuentes, N.; Fiser, A. Saturating representation of loop conformational fragments in structure databanks. BMC Struct. Biol. 2006, 6, 1–12. [Google Scholar] [CrossRef]
- Marks, C.; Nowak, J.; Klostermann, S.; Georges, G.; Dunbar, J.; Shi, J.; Kelm, S.; Deane, C.M. Sphinx: Merging knowledge-based and ab initio approaches to improve protein loop prediction. Bioinformatics 2017, 33, 1346–1353. [Google Scholar] [CrossRef]
- Stein, A.; Kortemme, T. Improvements to Robotics-Inspired Conformational Sampling in Rosetta. PLoS ONE 2013, 8, e63090. [Google Scholar] [CrossRef]
- Park, H.; Lee, G.R.; Heo, L.; Seok, C. Protein loop modeling using a new hybrid energy function and its application to modeling in inaccurate structural environments. PLoS ONE 2014, 9, e113811. [Google Scholar] [CrossRef]
- Karami, Y.; Guyon, F.; De Vries, S.; Tufféry, P. DaReUS-Loop: Accurate loop modeling using fragments from remote or unrelated proteins. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Fernandez-Fuentes, N.; Zhai, J.; Fiser, A. ArchPRED: A template based loop structure prediction server. Nucleic Acids Res. 2006, 34, W173–W176. [Google Scholar] [CrossRef]
- Choi, Y.; Deane, C.M. FREAD revisited: Accurate loop structure prediction using a database search algorithm. Wiley Online Libr. 2010, 78, 1431–1440. [Google Scholar] [CrossRef]
- Ismer, J.; Rose, A.S.; Tiemann, J.K.S.; Goede, A.; Preissner, R.; Hildebrand, P.W. SL2: An interactive webtool for modeling of missing segments in proteins. Nucleic Acids Res. 2016, 44, W390–W394. [Google Scholar] [CrossRef]
- Messih, M.A.; Lepore, R.; Tramontano, A. LoopIng: A template-based tool for predicting the structure of protein loops. Bioinformatics 2015, 31, 3767–3772. [Google Scholar] [CrossRef]
- Deane, C.M.; Blundell, T.L. CODA: A combined algorithm for predicting the structurally variable regions of protein models. Wiley Online Libr. 2001, 10, 599–612. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Touw, W.G.; Baakman, C.; Black, J.; Te Beek, T.A.H.; Krieger, E.; Joosten, R.P.; Vriend, G. A series of PDB-related databanks for everyday needs. Nucleic Acids Res. 2015, 43, D364–D368. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef] [PubMed]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Proteins Struct. Funct. Bioinforma. 1995, 23, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Binder, J.L.; Berendzen, J.; Stevens, A.O.; He, Y.; Wang, J.; Dokholyan, N.V.; Oprea, T.I. AlphaFold illuminates half of the dark human proteins. Curr. Opin. Struct. Biol. 2022, 74, 102372. [Google Scholar] [CrossRef]
- Ashraf, U.; Tengo, L.; Le Corre, L.; Fournier, G.; Busca, P.; McCarthy, A.A.; Rameix-Welti, M.A.; Gravier-Pelletier, C.; Ruigrok, R.W.H.; Jacob, Y.; et al. Destabilization of the human RED–SMU1 splicing complex as a basis for host-directed antiinfluenza strategy. Proc. Natl. Acad. Sci. USA 2019, 166, 10968–10977. [Google Scholar] [CrossRef]
- Sok, P.; Gógl, G.; Kumar, G.S.; Alexa, A.; Singh, N.; Kirsch, K.; Sebő, A.; Drahos, L.; Gáspári, Z.; Peti, W.; et al. MAP Kinase-Mediated Activation of RSK1 and MK2 Substrate Kinases. Structure 2020, 28, 1101–1113.e5. [Google Scholar] [CrossRef]
- Rittner, A.; Paithankar, K.S.; Himmler, A.; Grininger, M. Type I fatty acid synthase trapped in the octanoyl-bound state. Protein Sci. 2020, 29, 589–605. [Google Scholar] [CrossRef]
- Khanra, N.; Brown, P.M.G.E.; Perozzo, A.M.; Bowie, D.; Meyerson, J.R. Architecture and structural dynamics of the heteromeric gluk2/k5 kainate receptor. Elife 2021, 10, e66097. [Google Scholar] [CrossRef]
- Lu, T.W.; Aoto, P.C.; Weng, J.H.; Nielsen, C.; Cash, J.N.; Hall, J.; Zhang, P.; Simon, S.M.; Cianfrocco, M.A.; Taylor, S.S. Structural analyses of the PKA RIIβ holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma. PLoS Biol. 2020, 18, e3001018. [Google Scholar] [CrossRef]
- Bussiere, D.E.; Xie, L.; Srinivas, H.; Shu, W.; Burke, A.; Be, C.; Zhao, J.; Godbole, A.; King, D.; Karki, R.G.; et al. Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. Nat. Chem. Biol. 2019, 16, 15–23. [Google Scholar] [CrossRef]







| DSSP Secondary Structure Type | Experimental Structures | AlphaFold 2 Structures |
|---|---|---|
| None | 78.64% | 76.66% |
| Turn | 12.92% | 14.51% |
| Bend | 8.44% | 6.55% |
| Parallel beta sheet | 0% * | 0.10% |
| Antiparallel beta sheet | 0% * | 0.16% |
| Alpha helix | 0% * | 0.74% |
| Pi helix | 0% * | 0.03% |
| 3–10 helix | 0% * | 1.25% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stevens, A.O.; He, Y. Benchmarking the Accuracy of AlphaFold 2 in Loop Structure Prediction. Biomolecules 2022, 12, 985. https://doi.org/10.3390/biom12070985
Stevens AO, He Y. Benchmarking the Accuracy of AlphaFold 2 in Loop Structure Prediction. Biomolecules. 2022; 12(7):985. https://doi.org/10.3390/biom12070985
Chicago/Turabian StyleStevens, Amy O., and Yi He. 2022. "Benchmarking the Accuracy of AlphaFold 2 in Loop Structure Prediction" Biomolecules 12, no. 7: 985. https://doi.org/10.3390/biom12070985
APA StyleStevens, A. O., & He, Y. (2022). Benchmarking the Accuracy of AlphaFold 2 in Loop Structure Prediction. Biomolecules, 12(7), 985. https://doi.org/10.3390/biom12070985







