Aptamers as Theragnostic Tools in Prostate Cancer
Abstract
:1. Introduction
2. Aptamers
| Features | Description | Advantage | Reference |
|---|---|---|---|
| Size | 6–30 kDa 20–100 nt | Aptamers can be better distributed in tissues Less immunogenic than antibodies | [19] |
| Aptamer–antigen interaction | Highly specific | Aptamers can distinguish between molecules with only one methyl group of difference | [20] |
| Chemical modification | Easy chemical transformation | Aptamers can be modified and conjugated to a variety of molecules such as fluorophores, nanoparticles, drugs, or siRNAs | [21] |
| Thermal stability | High resistance to denaturalization | Aptamers can be refolded to their specific 3D conformations after incubation at 65–95 °C | [21] |
| Cost of production | Low cost of production | Aptamers can be produced at a high scale by chemical synthesis | [22] |
3. Aptamer Design
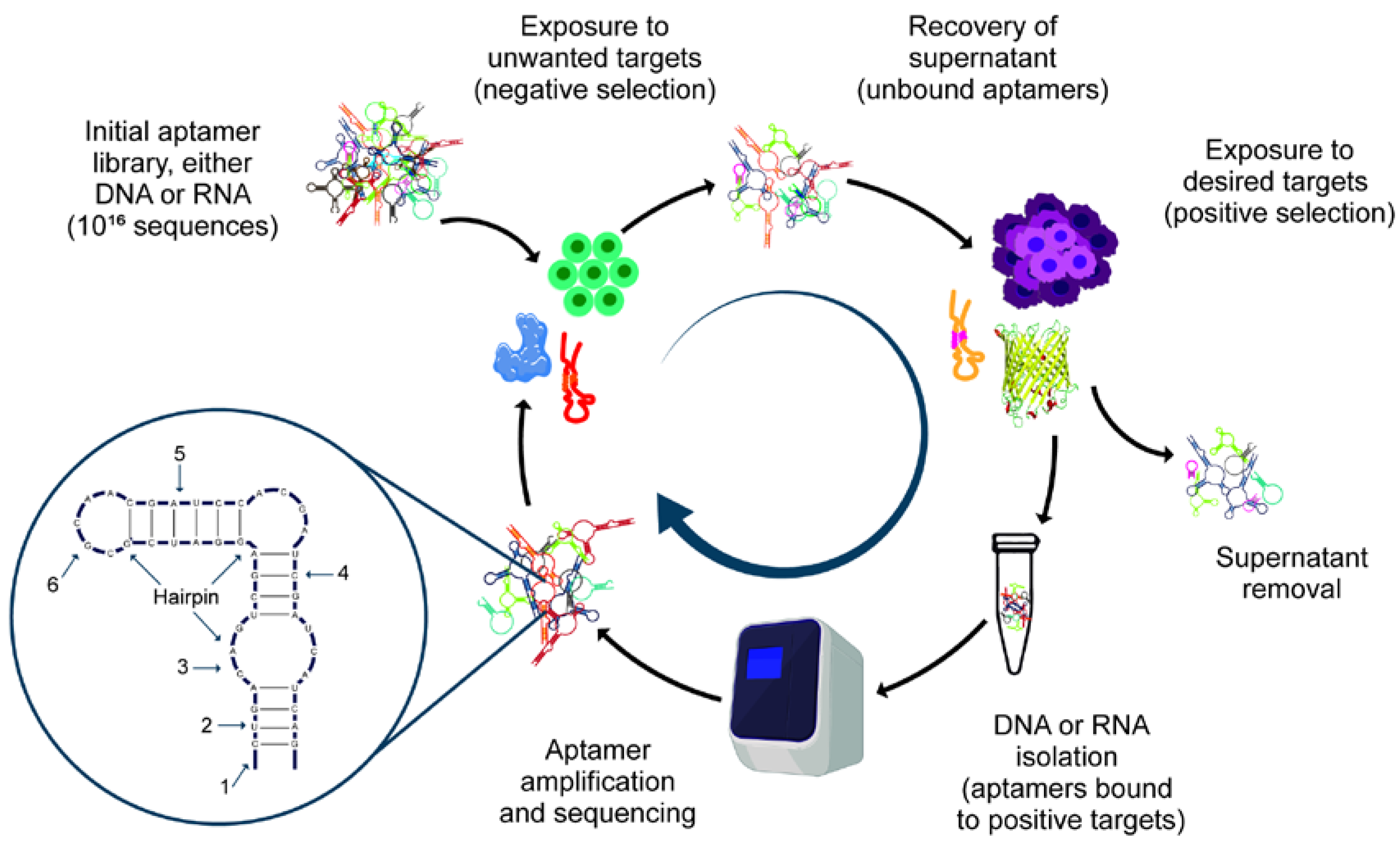
3.1. SELEX
3.2. Variants of SELEX
3.2.1. In Silico SELEX
3.2.2. Negative SELEX
3.2.3. Counter SELEX
3.2.4. Photo-SELEX
3.2.5. Capillary Electrophoresis-SELEX
3.2.6. Microfluidic SELEX
3.2.7. Magnetic Beads SELEX
3.2.8. Toggle-SELEX
3.2.9. Cell-SELEX
3.2.10. In Vivo SELEX
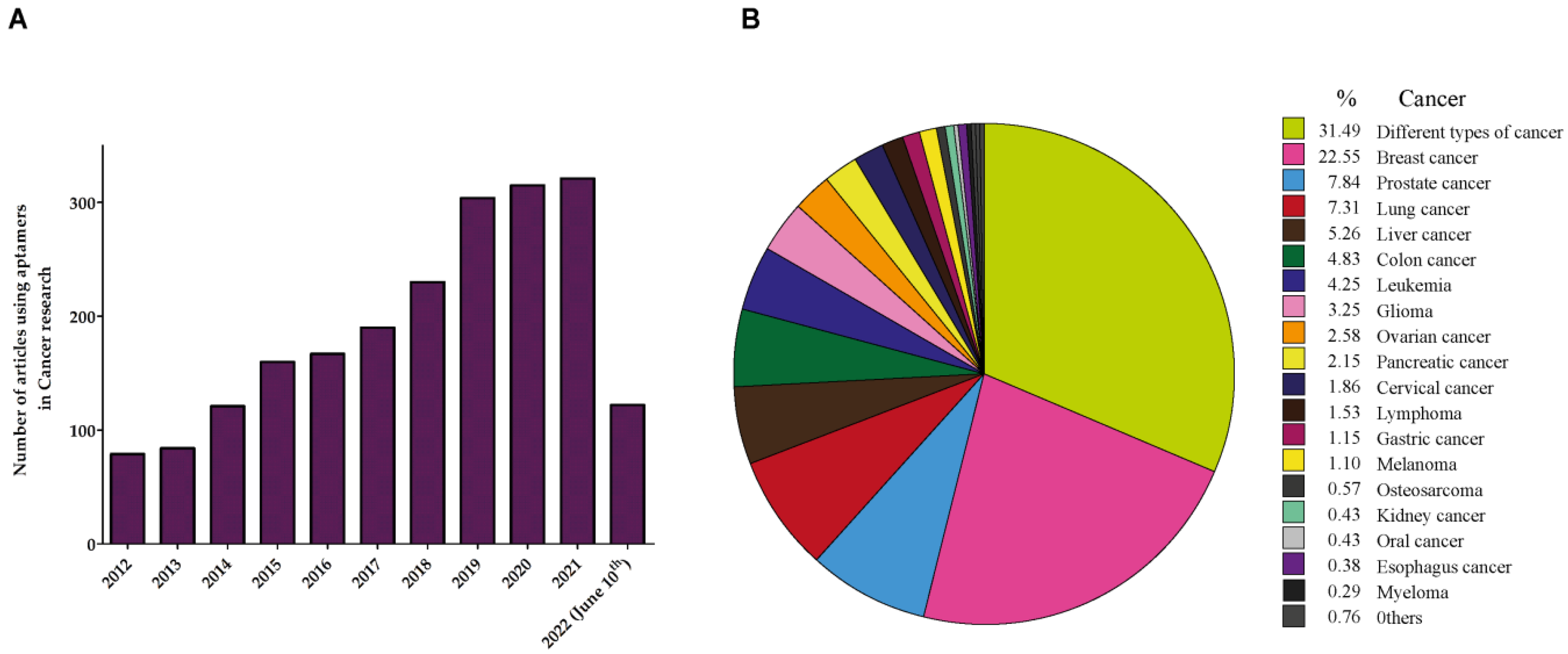
4. Aptamers in Cancer
5. Aptamers in Prostate Cancer (PCa)
6. Aptamers against Prostate-Specific Membrane Antigen (PSMA)
6.1. PSMA Aptamers in PCa Diagnosis
6.2. PSMA Aptamers in PCa Therapy
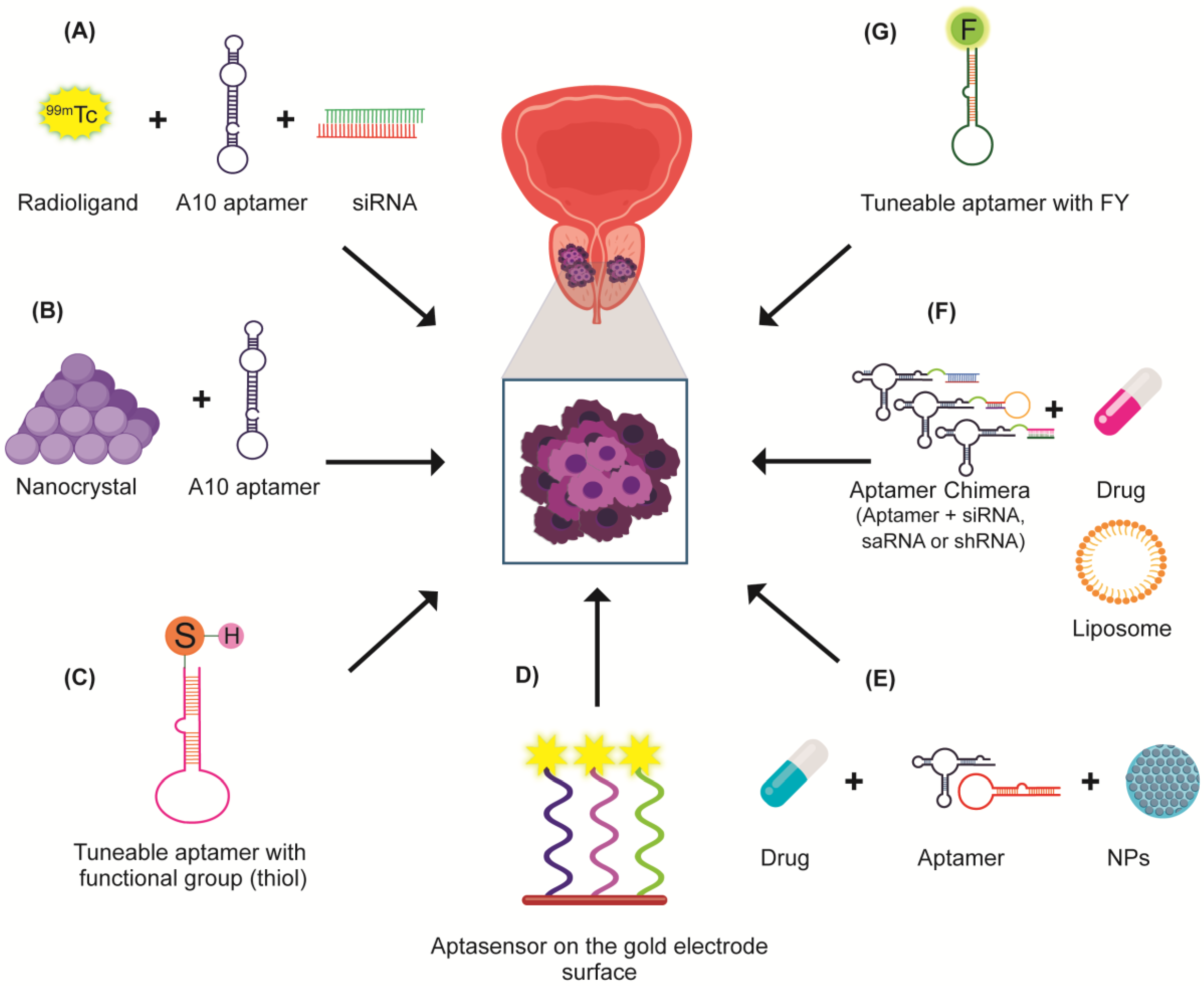
6.3. Chimeras of PSMA
6.4. PSMA Aptamers as Vehicles in PCa
7. Other Aptamers in PCa
| Target | Application | Main Findings | Type of Aptamer | Sensitivity/Efficacy | Disadvantage/Limitation | References |
|---|---|---|---|---|---|---|
| PCA3 | Diagnosis | Aptasensor for the sensitive detection of PCA3 | Thioled hairpin DNA aptamer | Linear detection range at 0–150 ng/ml | The establishment of the PCA3 expression depending on the type of prostate cancer cells is needed | [144] |
| EN2 | Diagnosis | Sensitive and specific enzyme-linked oligonucleotide assay (ELONA) for rapid and sensitive detection of EN2 in urine | ssDNA aptamer | EN2-specific (Kd = 8.26 nM) with a limit of detection of 0.34 nM in buffer and 2.69 nM | The capacity for distinguish between their two targets: bladder and prostate cancers | [145] |
| PSA | Diagnosis | Selective and specific detection of PSA by amperometric electrochemical measurements | A short, single-stranded DNA (ssDNA) pseudoknot forming two stem-loop structural aptamers | Detection range from 10 pg/mL to 500 ng/mL (low detection limit 1.24 pg/mL) | Only control serum samples were used with increasing rPSA concentrations | [146] |
| Glycosylated PSA and total PSA | Diagnosis | An impedimetric aptamer-based sensor to the dual recognition of PSA | DNA aptamers with binary recognition to the peptide region and the innermost sugar residues | A range between 0.26 and 62.5 ng/mL (PSAG-1) | The work was evaluated using serum samples from men with elevated PSA levels | [147] |
| Nucleolin | Diagnostic | Highly selective and specific detection of Nucleolin in peripheral blood mononuclear of PCa patients through ELISA assays | DNA aptamer adopting a G-quadruplex structure (AS1411-N5) | High affinity with Kd = 138.1 ± 5.5 nM | Stability of G4 parallel conformation in the presence of other cations | [148] |
| Neu5Gc | Diagnosis | A sensitive and rapid aptamer-nanoparticle immunochromatographic strip for the visual detection of Neu5Gc was developed | DNA aptamer | The visual limit of detection (LOD) for semi-quantitative detection was 30 ng/mL | Higher LOD than traditional antibody-based ELISA | [149] |
| PSA | Diagnosis | Detection of label-free, potentiometric detection of PSA with silicon nanowire ion-sensitive field-effect transistor (Si NW-ISFET) arrays | DNA aptamer site-specifically immobilized on Si NW-ISFETs | Concentration-dependent measurements were in a wide range of 1 pg/mL to 1 μg/mL | Does not cover the necessary resolution in the most critical concentration range of ~4 ng/mL | [150] |
| PSA | Diagnosis | Detection of PSA based on the affinities of the probe aptamer toward Cu-MOG | DNA aptamer-functionalized Cu-MOG | The linear range was from 0.5 to 8 ng/mL, with a detection limit of 0.33 ng/mL | The optimization of several factors as Cu-MOG concentration, time incubation, and Cu-MOG and PA integration is needed | [151] |
| PSA and VEGF | Diagnosis | Dual biosensor to detect PSA and VEGF released by cancer cells | Thiolated aptamers on gold-covered surface using methylene blue (MB) as redox label | The linear detection ranges (0.08–100 ng/mL for PSA and 0.15 ng–100 ng/mL for VEGF) | Establish the patterns of released proteins by different types of cells to correlate them with cancer aggressiveness | [152] |
| PSA | Diagnosis | The design of a 2D NS-based PSA aptamer sensor system | DNA aptamer functionalized (MoO3, MoS2, and MoSe2) of two-dimensional nanosheets | The detection limit of PSA was achieved to be 13 pM for MoO3 NSs, whereas the MoS2 and MoSe2 systems exhibited detection limits of 72 and 157 pM | Confocal microscopy assay needed for the in vitro imaging | [153] |
| ATP, Bcl-2 | Treatment | Antiproliferative effect using targeted treatment through antiapoptotic Bcl-2 silencing | Duplex DNA–siRNA chimera | Proliferation and inhibition by inducing apoptosis | Demonstrate the complete lack of toxicity in other normal cells with high production of ATP | [154] |
| LNCaP cells | Diagnosis and treatment | Targeted drug delivery to treat prostate cancer cells | Doxorubicin loaded DNA aptamer linked myristate chitosan nanogel | Binding affinity above 70% | Demonstrate the effect in tumors with higher diversity of lineage cells | [155] |
| Vesicle proteins | Profiling study | Improvement in the low abundancy protein analysis in vesicles from plasma and urine samples | SOMAscan | Identification of ~1000 proteins with ~400 proteins present in comparable quantities between plasma and urine vesicles | Standardization of the procedure for obtaining ultrapure vesicles from high proteinous fluids | [156] |
| PCA3 | Diagnosis | Design of novel nucleic acid antibody-like prostate cancer | RNA aptamer | Moderated staining in PCa samples and strong staining in 78% of the cases of BPH | RNA aptamer stability and issues and possible issues in the interaction RNA–RNA | [157] |
8. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Rhea, L.P.; Mendez-Marti, S.; Kim, D.; Aragon-Ching, J.B. Role of immunotherapy in bladder cancer. Cancer Treat. Res. Commun. 2021, 26, 100296. [Google Scholar] [CrossRef] [PubMed]
- Gulley, J.L.; Madan, R.A.; Tsang, K.Y.; Jochems, C.; Marte, J.L.; Farsaci, B.; Tucker, J.A.; Hodge, J.W.; Liewehr, D.J.; Steinberg, S.M.; et al. Immune impact induced by PROSTVAC (PSA-TRICOM), a therapeutic vaccine for prostate cancer. Cancer Immunol. Res. 2014, 2, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.; Xie, Z.; Grotewold, E. SELEX (Systematic Evolution of Ligands by EXponential Enrichment), as a powerful tool for deciphering the protein-DNA interaction space. Methods Mol. Biol. 2011, 754, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef] [Green Version]
- Neves, M.A.D.; Slavkovic, S.; Churcher, Z.R.; Johnson, P.E. Salt-mediated two-site ligand binding by the cocaine-binding aptamer. Nucleic Acids Res. 2017, 45, 1041–1048. [Google Scholar] [CrossRef] [Green Version]
- Deng, B.; Lin, Y.; Wang, C.; Li, F.; Wang, Z.; Zhang, H.; Li, X.F.; Le, X.C. Aptamer binding assays for proteins: The thrombin example—A review. Anal. Chim. Acta 2014, 837, 1–15. [Google Scholar] [CrossRef]
- Jing, Y.; Cai, M.; Xu, H.; Zhou, L.; Yan, Q.; Gao, J.; Wang, H. Aptamer-recognized carbohydrates on the cell membrane revealed by super-resolution microscopy. Nanoscale 2018, 10, 7457–7464. [Google Scholar] [CrossRef]
- Iwagawa, T.; Ohuchi, S.P.; Watanabe, S.; Nakamura, Y. Selection of RNA aptamers against mouse embryonic stem cells. Biochimie 2012, 94, 250–257. [Google Scholar] [CrossRef]
- Percze, K.; Szakacs, Z.; Scholz, E.; Andras, J.; Szeitner, Z.; Kieboom, C.H.; Ferwerda, G.; Jonge, M.I.; Gyurcsanyi, R.E.; Meszaros, T. Aptamers for respiratory syncytial virus detection. Sci. Rep. 2017, 7, 42794. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Weeks, K.M. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 2010, 20, 295–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hori, S.I.; Herrera, A.; Rossi, J.J.; Zhou, J. Current Advances in Aptamers for Cancer Diagnosis and Therapy. Cancers 2018, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- Kumar Kulabhusan, P.; Hussain, B.; Yuce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef]
- Lao, Y.H.; Phua, K.K.; Leong, K.W. Aptamer nanomedicine for cancer therapeutics: Barriers and potential for translation. ACS Nano 2015, 9, 2235–2254. [Google Scholar] [CrossRef]
- Lakhin, A.V.; Tarantul, V.Z.; Gening, L.V. Aptamers: Problems, solutions and prospects. Acta Nat. 2013, 5, 34–43. [Google Scholar] [CrossRef] [Green Version]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202, Erratum in Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [Green Version]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide aptamers: New tools for targeted cancer therapy. Mol. Ther. Nucleic Acids 2014, 3, e182. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.; Tang, T.H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015, 64, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Leslie, M.; Kameyama, H.; Volk, D.E.; Tanaka, T. Aptamer Therapeutics in Cancer: Current and Future. Cancers 2018, 10, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinores, S.A. Pegaptanib in the treatment of wet, age-related macular degeneration. Int. J. Nanomed. 2006, 1, 263–268. [Google Scholar]
- Cerchia, L. Aptamers: Promising Tools for Cancer Diagnosis and Therapy. Cancers 2018, 10, 132. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Latchoumanin, O.; Hebbard, L.; Duan, W.; Liddle, C.; George, J.; Qiao, L. Aptamers as targeting ligands and therapeutic molecules for overcoming drug resistance in cancers. Adv. Drug Deliv. Rev. 2018, 134, 107–121. [Google Scholar] [CrossRef]
- Kulbachinskiy, A.V. Methods for selection of aptamers to protein targets. Biochem. Biokhimiia 2007, 72, 1505–1518. [Google Scholar] [CrossRef]
- Nakatsuka, N.; Cao, H.H.; Deshayes, S.; Melkonian, A.L.; Kasko, A.M.; Weiss, P.S.; Andrews, A.M. Aptamer Recognition of Multiplexed Small-Molecule-Functionalized Substrates. ACS Appl. Mater. Interfaces 2018, 10, 23490–23500. [Google Scholar] [CrossRef]
- Mallikaratchy, P. Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens. Molecules 2017, 22, 215. [Google Scholar] [CrossRef]
- Ma, S.; Tang, N.; Tian, J. DNA synthesis, assembly and applications in synthetic biology. Curr. Opin. Chem. Biol. 2012, 16, 260–267. [Google Scholar] [CrossRef] [Green Version]
- Chushak, Y.; Stone, M.O. In silico selection of RNA aptamers. Nucleic Acids Res. 2009, 37, e87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamada, M. In silico approaches to RNA aptamer design. Biochimie 2018, 145, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Li, H.; Zhao, Y.; Dong, S.; Li, W.; Qiang, W.; Xu, D. Aptamer-functionalized silver nanoparticles for scanometric detection of platelet-derived growth factor-BB. Anal. Chim. Acta 2014, 812, 152–160. [Google Scholar] [CrossRef]
- Ishida, R.; Adachi, T.; Yokota, A.; Yoshihara, H.; Aoki, K.; Nakamura, Y.; Hamada, M. RaptRanker: In silico RNA aptamer selection from HT-SELEX experiment based on local sequence and structure information. Nucleic Acids Res. 2020, 48, e82. [Google Scholar] [CrossRef] [PubMed]
- Orava, E.W.; Abdul-Wahid, A.; Huang, E.H.; Mallick, A.I.; Gariepy, J. Blocking the attachment of cancer cells in vivo with DNA aptamers displaying anti-adhesive properties against the carcinoembryonic antigen. Mol. Oncol. 2013, 7, 799–811. [Google Scholar] [CrossRef]
- Wondergem, J.A.J.; Schiessel, H.; Tompitak, M. Performing SELEX experiments in silico. J. Chem. Phys. 2017, 147, 174101. [Google Scholar] [CrossRef] [Green Version]
- Ellington, A.D.; Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992, 355, 850–852. [Google Scholar] [CrossRef]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef] [Green Version]
- Jenison, R.D.; Gill, S.C.; Pardi, A.; Polisky, B. High-resolution molecular discrimination by RNA. Science 1994, 263, 1425–1429. [Google Scholar] [CrossRef] [Green Version]
- Golden, M.C.; Collins, B.D.; Willis, M.C.; Koch, T.H. Diagnostic potential of PhotoSELEX-evolved ssDNA aptamers. J. Biotechnol. 2000, 81, 167–178. [Google Scholar] [CrossRef]
- Jensen, K.B.; Atkinson, B.L.; Willis, M.C.; Koch, T.H.; Gold, L. Using in vitro selection to direct the covalent attachment of human immunodeficiency virus type 1 Rev protein to high-affinity RNA ligands. Proc. Natl. Acad. Sci. USA 1995, 92, 12220–12224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendonsa, S.D.; Bowser, M.T. In vitro selection of high-affinity DNA ligands for human IgE using capillary electrophoresis. Anal. Chem. 2004, 76, 5387–5392. [Google Scholar] [CrossRef]
- Mosing, R.K.; Mendonsa, S.D.; Bowser, M.T. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal. Chem. 2005, 77, 6107–6112. [Google Scholar] [CrossRef] [PubMed]
- Hybarger, G.; Bynum, J.; Williams, R.F.; Valdes, J.J.; Chambers, J.P. A microfluidic SELEX prototype. Anal. Bioanal. Chem. 2006, 384, 191–198. [Google Scholar] [CrossRef]
- Olsen, T.R.; Tapia-Alveal, C.; Yang, K.A.; Zhang, X.; Pereira, L.J.; Farmakidis, N.; Pei, R.; Stojanovic, M.N.; Lin, Q. Integrated Microfluidic Selex Using Free Solution Electrokinetics. J. Electrochem. Soc. 2017, 164, B3122–B3129. [Google Scholar] [CrossRef]
- Lou, X.; Qian, J.; Xiao, Y.; Viel, L.; Gerdon, A.E.; Lagally, E.T.; Atzberger, P.; Tarasow, T.M.; Heeger, A.J.; Soh, H.T. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. USA 2009, 106, 2989–2994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.M.; Ahn, J.Y.; Jo, M.; Lee, D.K.; Lis, J.T.; Craighead, H.G.; Kim, S. Selection and elution of aptamers using nanoporous sol-gel arrays with integrated microheaters. Lab Chip 2009, 9, 1206–1212. [Google Scholar] [CrossRef]
- Nie, H.; Chen, Y.; Lu, C.; Liu, Z. Efficient selection of glycoprotein-binding DNA aptamers via boronate affinity monolithic capillary. Anal. Chem. 2013, 85, 8277–8283. [Google Scholar] [CrossRef]
- Liu, X.; Li, H.; Jia, W.; Chen, Z.; Xu, D. Selection of aptamers based on a protein microarray integrated with a microfluidic chip. Lab Chip 2016, 17, 178–185. [Google Scholar] [CrossRef]
- Bruno, J.G. In vitro selection of DNA to chloroaromatics using magnetic microbead-based affinity separation and fluorescence detection. Biochem. Biophys. Res. Commun. 1997, 234, 117–120. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 2005, 383, 83–91. [Google Scholar] [CrossRef] [PubMed]
- White, R.; Rusconi, C.; Scardino, E.; Wolberg, A.; Lawson, J.; Hoffman, M.; Sullenger, B. Generation of species cross-reactive aptamers using “toggle” SELEX. Mol. Ther. J. Am. Soc. Gene Ther. 2001, 4, 567–573. [Google Scholar] [CrossRef]
- Kong, H.Y.; Byun, J. Nucleic Acid aptamers: New methods for selection, stabilization, and application in biomedical science. Biomol. Ther. 2013, 21, 423–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Yu, Y.; Jiang, F.; Zhou, J.; Li, Y.; Liang, C.; Dang, L.; Lu, A.; Zhang, G. Development of Cell-SELEX Technology and Its Application in Cancer Diagnosis and Therapy. Int. J. Mol. Sci. 2016, 17, 2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, K.N.; Jensen, K.B.; Julin, C.M.; Weil, M.; Gold, L. High affinity ligands from in vitro selection: Complex targets. Proc. Natl. Acad. Sci. USA 1998, 95, 2902–2907. [Google Scholar] [CrossRef] [Green Version]
- Rong, Y.; Chen, H.; Zhou, X.F.; Yin, C.Q.; Wang, B.C.; Peng, C.W.; Liu, S.P.; Wang, F.B. Identification of an aptamer through whole cell-SELEX for targeting high metastatic liver cancers. Oncotarget 2016, 7, 8282–8294. [Google Scholar] [CrossRef] [Green Version]
- Ohuchi, S. Cell-SELEX Technology. BioResearch Open Access 2012, 1, 265–272. [Google Scholar] [CrossRef]
- Hamula, C.L.; Le, X.C.; Li, X.F. DNA aptamers binding to multiple prevalent M-types of Streptococcus pyogenes. Anal. Chem. 2011, 83, 3640–3647. [Google Scholar] [CrossRef]
- Berezovski, M.V.; Lechmann, M.; Musheev, M.U.; Mak, T.W.; Krylov, S.N. Aptamer-facilitated biomarker discovery (AptaBiD). J. Am. Chem. Soc. 2008, 130, 9137–9143. [Google Scholar] [CrossRef]
- Chen, Z.; Xu, X. Roles of nucleolin. Focus on cancer and anti-cancer therapy. Saudi Med. J. 2016, 37, 1312–1318. [Google Scholar] [CrossRef]
- Mi, J.; Liu, Y.; Rabbani, Z.N.; Yang, Z.; Urban, J.H.; Sullenger, B.A.; Clary, B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010, 6, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Chen, Y.H.; Lennox, K.A.; Behlke, M.A.; Davidson, B.L. In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol. Ther. Nucleic Acids 2013, 2, e67. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Zheng, C.; Zhou, S.F.; Qiao, S.; Tran, P.H.; Pu, C.; Li, Y.; Kong, L.; Kouzani, A.Z.; Lin, J.; et al. Superior Performance of Aptamer in Tumor Penetration over Antibody: Implication of Aptamer-Based Theranostics in Solid Tumors. Theranostics 2015, 5, 1083–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, G.; Chen, X. Aptamer-based targeted therapy. Adv. Drug Deliv. Rev. 2018, 134, 65–78. [Google Scholar] [CrossRef]
- Tan, W.; Wang, H.; Chen, Y.; Zhang, X.; Zhu, H.; Yang, C.; Yang, R.; Liu, C. Molecular aptamers for drug delivery. Trends Biotechnol. 2011, 29, 634–640. [Google Scholar] [CrossRef] [Green Version]
- Pastor, F.; Berraondo, P.; Etxeberria, I.; Frederick, J.; Sahin, U.; Gilboa, E.; Melero, I. An RNA toolbox for cancer immunotherapy. Nat. Rev. Drug Discov. 2018, 17, 751–767. [Google Scholar] [CrossRef]
- Soundararajan, S.; Wang, L.; Sridharan, V.; Chen, W.; Courtenay-Luck, N.; Jones, D.; Spicer, E.K.; Fernandes, D.J. Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol. Pharmacol. 2009, 76, 984–991. [Google Scholar] [CrossRef] [Green Version]
- Kozani, P.S.; Kozani, P.S.; Malik, M.T. AS1411-functionalized delivery nanosystems for targeted cancer therapy. Explor. Med. 2021, 2, 146–166. [Google Scholar] [CrossRef]
- Berger, C.M.; Gaume, X.; Bouvet, P. The roles of nucleolin subcellular localization in cancer. Biochimie 2015, 113, 78–85. [Google Scholar] [CrossRef]
- Soundararajan, S.; Chen, W.; Spicer, E.K.; Courtenay-Luck, N.; Fernandes, D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008, 68, 2358–2365. [Google Scholar] [CrossRef] [Green Version]
- Hoellenriegel, J.; Zboralski, D.; Maasch, C.; Rosin, N.Y.; Wierda, W.G.; Keating, M.J.; Kruschinski, A.; Burger, J.A. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood 2014, 123, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Nachreiner, I.; Hussain, A.F.; Wullner, U.; Machuy, N.; Meyer, T.F.; Fischer, R.; Gattenlohner, S.; Meinhold-Heerlein, I.; Barth, S.; Tur, M.K. Elimination of HER3-expressing breast cancer cells using aptamer-siRNA chimeras. Exp. Ther. Med. 2019, 18, 2401–2412. [Google Scholar] [CrossRef] [PubMed]
- Bagalkot, V.; Farokhzad, O.C.; Langer, R.; Jon, S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. 2006, 45, 8149–8152. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Pei, S.N.; Qi, J.; Zeng, Z.; Iyer, S.P.; Lin, P.; Tung, C.H.; Zu, Y. Oligonucleotide aptamer-drug conjugates for targeted therapy of acute myeloid leukemia. Biomaterials 2015, 67, 42–51. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.; Lee, S.H.; Hwang, Y.; Yoo, H.; Jung, H.; Kim, S.H.; Mok, H. Multivalent Aptamer-RNA Conjugates for Simple and Efficient Delivery of Doxorubicin/siRNA into Multidrug-Resistant Cells. Macromol. Biosci. 2017, 17, 1600343. [Google Scholar] [CrossRef]
- Zhang, J.J.; Cheng, F.F.; Zheng, T.T.; Zhu, J.J. Versatile aptasensor for electrochemical quantification of cell surface glycan and naked-eye tracking glycolytic inhibition in living cells. Biosens. Bioelectron. 2017, 89, 937–945. [Google Scholar] [CrossRef]
- Ushigome, M.; Ubagai, T.; Fukuda, H.; Tsuchiya, N.; Sugimura, T.; Takatsuka, J.; Nakagama, H. Up-regulation of hnRNP A1 gene in sporadic human colorectal cancers. Int. J. Oncol. 2005, 26, 635–640. [Google Scholar] [CrossRef]
- Zerbe, L.K.; Pino, I.; Pio, R.; Cosper, P.F.; Dwyer-Nield, L.D.; Meyer, A.M.; Port, J.D.; Montuenga, L.M.; Malkinson, A.M. Relative amounts of antagonistic splicing factors, hnRNP A1 and ASF/SF2, change during neoplastic lung growth: Implications for pre-mRNA processing. Mol. Carcinog. 2004, 41, 187–196. [Google Scholar] [CrossRef]
- Yan-Sanders, Y.; Hammons, G.J.; Lyn-Cook, B.D. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett. 2002, 183, 215–220. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Liu, F.; Zhou, L.; Shao, N.; Zhao, X. SELEX aptamer used as a probe to detect circulating tumor cells in peripheral blood of pancreatic cancer patients. PLoS ONE 2015, 10, e0121920. [Google Scholar] [CrossRef] [Green Version]
- Tsai, S.C.; Hung, L.Y.; Lee, G.B. An integrated microfluidic system for the isolation and detection of ovarian circulating tumor cells using cell selection and enrichment methods. Biomicrofluidics 2017, 11, 034122. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [Green Version]
- Stokes, M.E.; Ishak, J.; Proskorovsky, I.; Black, L.K.; Huang, Y. Lifetime economic burden of prostate cancer. BMC Health Serv. Res. 2011, 11, 349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catalona, W.J.; Smith, D.S.; Wolfert, R.L.; Wang, T.J.; Rittenhouse, H.G.; Ratliff, T.L.; Nadler, R.B. Evaluation of percentage of free serum prostate-specific antigen to improve specificity of prostate cancer screening. JAMA 1995, 274, 1214–1220. [Google Scholar] [CrossRef] [PubMed]
- Abrahamsson, P.A.; Lilja, H.; Oesterling, J.E. Molecular forms of serum prostate-specific antigen. The clinical value of percent free prostate-specific antigen. Urol. Clin. N. Am. 1997, 24, 353–365. [Google Scholar] [CrossRef]
- Kim, E.H.; Andriole, G.L. Prostate-specific antigen-based screening: Controversy and guidelines. BMC Med. 2015, 13, 61. [Google Scholar] [CrossRef] [Green Version]
- Partin, A.W.; Carter, H.B.; Chan, D.W.; Epstein, J.I.; Oesterling, J.E.; Rock, R.C.; Weber, J.P.; Walsh, P.C. Prostate specific antigen in the staging of localized prostate cancer: Influence of tumor differentiation, tumor volume and benign hyperplasia. J. Urol. 1990, 143, 747–752. [Google Scholar] [CrossRef]
- Carter, H.B.; Pearson, J.D.; Metter, E.J.; Brant, L.J.; Chan, D.W.; Andres, R.; Fozard, J.L.; Walsh, P.C. Longitudinal evaluation of prostate-specific antigen levels in men with and without prostate disease. Jama 1992, 267, 2215–2220. [Google Scholar] [CrossRef] [Green Version]
- Thompson, I.M.; Pauler, D.K.; Goodman, P.J.; Tangen, C.M.; Lucia, M.S.; Parnes, H.L.; Minasian, L.M.; Ford, L.G.; Lippman, S.M.; Crawford, E.D.; et al. Prevalence of prostate cancer among men with a prostate-specific antigen level <or =4.0 ng per milliliter. N. Engl. J. Med. 2004, 350, 2239–2246. [Google Scholar] [CrossRef] [Green Version]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef]
- Robert, G.; Jannink, S.; Smit, F.; Aalders, T.; Hessels, D.; Cremers, R.; Mulders, P.F.; Schalken, J.A. Rational basis for the combination of PCA3 and TMPRSS2:ERG gene fusion for prostate cancer diagnosis. Prostate 2013, 73, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Kohaar, I.; Petrovics, G.; Srivastava, S. A Rich Array of Prostate Cancer Molecular Biomarkers: Opportunities and Challenges. Int. J. Mol. Sci. 2019, 20, 1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, P.A.; Arora, V.K.; Sawyers, C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 2015, 15, 701–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, J.S.; Sheehan, C.E.; Fisher, H.A.; Kaufman, R.P., Jr.; Kaur, P.; Gray, K.; Webb, I.; Gray, G.S.; Mosher, R.; Kallakury, B.V. Correlation of primary tumor prostate-specific membrane antigen expression with disease recurrence in prostate cancer. Clin. Cancer Res. 2003, 9, 6357–6362. [Google Scholar]
- O’Keefe, D.S.; Su, S.L.; Bacich, D.J.; Horiguchi, Y.; Luo, Y.; Powell, C.T.; Zandvliet, D.; Russell, P.J.; Molloy, P.L.; Nowak, N.J.; et al. Mapping, genomic organization and promoter analysis of the human prostate-specific membrane antigen gene. Biochim. Biophys. Acta 1998, 1443, 113–127. [Google Scholar] [CrossRef]
- Chang, S.S.; Reuter, V.E.; Heston, W.D.; Gaudin, P.B. Comparison of anti-prostate-specific membrane antigen antibodies and other immunomarkers in metastatic prostate carcinoma. Urology 2001, 57, 1179–1183. [Google Scholar] [CrossRef]
- Beckett, M.L.; Cazares, L.H.; Vlahou, A.; Schellhammer, P.F.; Wright, G.L., Jr. Prostate-specific membrane antigen levels in sera from healthy men and patients with benign prostate hyperplasia or prostate cancer. Clin. Cancer Res. 1999, 5, 4034–4040. [Google Scholar]
- Salgaller, M.L.; Lodge, P.A.; McLean, J.G.; Tjoa, B.A.; Loftus, D.J.; Ragde, H.; Kenny, G.M.; Rogers, M.; Boynton, A.L.; Murphy, G.P. Report of immune monitoring of prostate cancer patients undergoing T-cell therapy using dendritic cells pulsed with HLA-A2-specific peptides from prostate-specific membrane antigen (PSMA). Prostate 1998, 35, 144–151. [Google Scholar] [CrossRef]
- Bagalkot, V.; Zhang, L.; Levy-Nissenbaum, E.; Jon, S.; Kantoff, P.W.; Langer, R.; Farokhzad, O.C. Quantum dot-aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on bi-fluorescence resonance energy transfer. Nano Lett. 2007, 7, 3065–3070. [Google Scholar] [CrossRef]
- Min, K.; Song, K.M.; Cho, M.; Chun, Y.S.; Shim, Y.B.; Ku, J.K.; Ban, C. Simultaneous electrochemical detection of both PSMA (+) and PSMA (-) prostate cancer cells using an RNA/peptide dual-aptamer probe. Chem. Commun. 2010, 46, 5566–5568. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Guo, Y.; Wang, L.; Xiong, X.; Zhu, L.; Fang, K. Diagnosis of prostate cancer using anti-PSMA aptamer A10-3.2-oriented lipid nanobubbles. Int. J. Nanomed. 2016, 11, 3939–3950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pai, S.S.; Ellington, A.D. Using RNA aptamers and the proximity ligation assay for the detection of cell surface antigens. Methods Mol. Biol. 2009, 504, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Farzin, L.; Shamsipur, M. Recent advances in design of electrochemical affinity biosensors for low level detection of cancer protein biomarkers using nanomaterial-assisted signal enhancement strategies. J. Pharm. Biomed. Anal. 2018, 147, 185–210. [Google Scholar] [CrossRef] [PubMed]
- Spruessel, A.; Steimann, G.; Jung, M.; Lee, S.A.; Carr, T.; Fentz, A.K.; Spangenberg, J.; Zornig, C.; Juhl, H.H.; David, K.A. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. BioTechniques 2004, 36, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Knuuttila, M.; Yatkin, E.; Kallio, J.; Savolainen, S.; Laajala, T.D.; Aittokallio, T.; Oksala, R.; Hakkinen, M.; Keski-Rahkonen, P.; Auriola, S.; et al. Castration induces up-regulation of intratumoral androgen biosynthesis and androgen receptor expression in an orthotopic VCaP human prostate cancer xenograft model. Am. J. Pathol. 2014, 184, 2163–2173. [Google Scholar] [CrossRef]
- Mader, N.; Groener, D.; Tselis, N.; Banek, S.; Nagarajah, J.; Grunwald, F.; Sabet, A. Outcome of (177)Lu-PSMA-617 Radioligand Therapy in Chemo-Refractory Patients with Metastatic Castration-Resistant Early-Onset Prostate Cancer. Cancers 2021, 13, 4193. [Google Scholar] [CrossRef]
- Dhar, S.; Kolishetti, N.; Lippard, S.J.; Farokhzad, O.C. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 1850–1855. [Google Scholar] [CrossRef] [Green Version]
- Farokhzad, O.C.; Cheng, J.; Teply, B.A.; Sherifi, I.; Jon, S.; Kantoff, P.W.; Richie, J.P.; Langer, R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 6315–6320. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Tai, Z.; Gu, F.; Hu, C.; Zhu, Q.; Gao, S. Aptamer-mediated delivery of docetaxel to prostate cancer through polymeric nanoparticles for enhancement of antitumor efficacy. Eur. J. Pharm. Biopharm. 2016, 107, 130–141. [Google Scholar] [CrossRef]
- Baek, S.E.; Lee, K.H.; Park, Y.S.; Oh, D.K.; Oh, S.; Kim, K.S.; Kim, D.E. RNA aptamer-conjugated liposome as an efficient anticancer drug delivery vehicle targeting cancer cells in vivo. J. Control. Release 2014, 196, 234–242. [Google Scholar] [CrossRef]
- Xu, W.; Siddiqui, I.A.; Nihal, M.; Pilla, S.; Rosenthal, K.; Mukhtar, H.; Gong, S. Aptamer-conjugated and doxorubicin-loaded unimolecular micelles for targeted therapy of prostate cancer. Biomaterials 2013, 34, 5244–5253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandekar, A.; Zhu, C.; Jindal, R.; Bruchertseifer, F.; Morgenstern, A.; Sofou, S. Anti-prostate-specific membrane antigen liposomes loaded with 225Ac for potential targeted antivascular alpha-particle therapy of cancer. J. Nucl. Med. 2014, 55, 107–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Jeong, Y.Y.; Jon, S. A drug-loaded aptamer-gold nanoparticle bioconjugate for combined CT imaging and therapy of prostate cancer. ACS Nano 2010, 4, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Zhang, Y.; Zennami, K.; Castanares, M.; Mukherjee, A.; Raval, R.R.; Zhou, H.; DeWeese, T.L.; Lupold, S.E. Systemic Administration and Targeted Radiosensitization via Chemically Synthetic Aptamer-siRNA Chimeras in Human Tumor Xenografts. Mol. Cancer Ther. 2015, 14, 2797–2804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, P.; Cao, S.; Xiao, S.; Zhang, X.; Ke, S.; Ke, F.; Yu, X.; Wang, L.; Wang, S.; Luo, Y.; et al. Enhanced growth inhibition of prostate cancer in vitro and in vivo by a recombinant adenovirus-mediated dual-aptamer modified drug delivery system. Cancer Lett. 2016, 383, 230–242. [Google Scholar] [CrossRef]
- Wullner, U.; Neef, I.; Eller, A.; Kleines, M.; Tur, M.K.; Barth, S. Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr. Cancer Drug Targets 2008, 8, 554–565. [Google Scholar] [CrossRef]
- Dassie, J.P.; Liu, X.Y.; Thomas, G.S.; Whitaker, R.M.; Thiel, K.W.; Stockdale, K.R.; Meyerholz, D.K.; McCaffrey, A.P.; McNamara, J.O., II; Giangrande, P.H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009, 27, 839–849. [Google Scholar] [CrossRef] [Green Version]
- Binzel, D.W.; Shu, Y.; Li, H.; Sun, M.; Zhang, Q.; Shu, D.; Guo, B.; Guo, P. Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol. Ther. J. Am. Soc. Gene Ther. 2016, 24, 1267–1277. [Google Scholar] [CrossRef] [Green Version]
- Jie, F.; Xiao-Lin, W.; Yu-Jie, L.; Jing, Z.; Fang, W.; Shen-Si, X.; Xin, G.; Qing-Qing, W.; Hai-Feng, S. In vivo analysis of treatment of prostate cancer with prostate surface membrane antigen aptamer-cationic liposome-double siRNA complex. Chin. J. Biol. 2016, 29, 151–156. [Google Scholar]
- McNamara, J.O., II; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006, 24, 1005–1015. [Google Scholar] [CrossRef]
- Lupold, S.E.; Hicke, B.J.; Lin, Y.; Coffey, D.S. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002, 62, 4029–4033. [Google Scholar] [PubMed]
- Mathieu, R.; Lucca, I.; Vartolomei, M.D.; Mbeutcha, A.; Klatte, T.; Seitz, C.; Karakiewicz, P.I.; Fajkovic, H.; Sun, M.; Lotan, Y.; et al. Role of survivin expression in predicting biochemical recurrence after radical prostatectomy: A multi-institutional study. BJU Int. 2017, 119, 234–238. [Google Scholar] [CrossRef]
- Leach, J.C.; Wang, A.; Ye, K.; Jin, S. A RNA-DNA Hybrid Aptamer for Nanoparticle-Based Prostate Tumor Targeted Drug Delivery. Int. J. Mol. Sci. 2016, 17, 380. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Li, C. Suppression of Prostate Cancer Metastasis by DPYSL3-Targeted saRNA. Adv. Exp. Med. Biol. 2017, 983, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.; Liu, J.; Ma, Y.; Su, M.; Zhang, H.; Hao, X. A specific aptamer-cell penetrating peptides complex delivered siRNA efficiently and suppressed prostate tumor growth in vivo. Cancer Biol. Ther. 2016, 17, 498–506. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Xu, P.; Luan, S.; Wang, X.; Gao, Y.; Zhao, C.; Fu, P. Molecular imaging and treatment of PSMA-positive prostate cancer with (99m)Tc radiolabeled aptamer-siRNA chimeras. Nucl. Med. Biol. 2022, 104–105, 28–37. [Google Scholar] [CrossRef]
- Dhar, S.; Gu, F.X.; Langer, R.; Farokhzad, O.C.; Lippard, S.J. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc. Natl. Acad. Sci. USA 2008, 105, 17356–17361. [Google Scholar] [CrossRef] [Green Version]
- Farokhzad, O.C.; Jon, S.; Khademhosseini, A.; Tran, T.N.; Lavan, D.A.; Langer, R. Nanoparticle-aptamer bioconjugates: A new approach for targeting prostate cancer cells. Cancer Res. 2004, 64, 7668–7672. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Gordetsky, J.B.; Bae, S.; Acosta, E.P.; Lillard, J.W., Jr.; Singh, R. Selective Targeting of the Hedgehog Signaling Pathway by PBM Nanoparticles in Docetaxel-Resistant Prostate Cancer. Cells 2020, 9, 1976. [Google Scholar] [CrossRef]
- Wu, M.; Wang, Y.; Wang, Y.; Zhang, M.; Luo, Y.; Tang, J.; Wang, Z.; Wang, D.; Hao, L.; Wang, Z. Paclitaxel-loaded and A10-3.2 aptamer-targeted poly(lactide-co-glycolic acid) nanobubbles for ultrasound imaging and therapy of prostate cancer. Int. J. Nanomed. 2017, 12, 5313–5330. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Yu, X.; Han, W.; Kong, Y.; Bao, W.; Zhang, J.; Zhang, W.; Gu, Y. Ultrasensitive and Reversible Nanoplatform of Urinary Exosomes for Prostate Cancer Diagnosis. ACS Sens. 2019, 4, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Hu, C.; Xia, Q.; Gong, C.; Gao, S.; Chen, Z. Aptamer-conjugated multi-walled carbon nanotubes as a new targeted ultrasound contrast agent for the diagnosis of prostate cancer. J. Nanopart. Res. Interdiscip. Forum Nanoscale Sci. Technol. 2018, 20, 303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.Y.; Yu, X.; Liu, H.; Wu, D.; She, J.X. Co-targeting EGFR and survivin with a bivalent aptamer-dual siRNA chimera effectively suppresses prostate cancer. Sci. Rep. 2016, 6, 30346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dogra, V.; Chinni, B.; Singh, S.; Schmitthenner, H.; Rao, N.; Krolewski, J.J.; Nastiuk, K.L. Photoacoustic imaging with an acoustic lens detects prostate cancer cells labeled with PSMA-targeting near-infrared dye-conjugates. J. Biomed. Opt. 2016, 21, 66019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Jiang, W.; Hu, Q.; Li, L.C.; Dong, L.; Chen, R.; Zhang, Y.; Tang, Y.; Thrasher, J.B.; Liu, C.B.; et al. Enhancing DPYSL3 gene expression via a promoter-targeted small activating RNA approach suppresses cancer cell motility and metastasis. Oncotarget 2016, 7, 22893–22910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Zhao, Y.; Huang, Y.; Yang, Q.; Zeng, X.; Jiang, W.; Liu, J.; Thrasher, J.B.; Forrest, M.L.; Li, B. Nanomicellar TGX221 blocks xenograft tumor growth of prostate cancer in nude mice. Prostate 2015, 75, 593–602. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Tai, Z.; Zhu, Q.; Fan, W.; Ding, B.; Zhang, W.; Zhang, L.; Yao, C.; Wang, X.; Ding, X.; et al. Study on the prostate cancer-targeting mechanism of aptamer-modified nanoparticles and their potential anticancer effect in vivo. Int. J. Nanomed. 2014, 9, 5431–5440. [Google Scholar] [CrossRef] [Green Version]
- Zabegina, L.; Nazarova, I.; Nikiforova, N.; Slyusarenko, M.; Sidina, E.; Knyazeva, M.; Tsyrlina, E.; Novikov, S.; Reva, S.; Malek, A. A New Approach for Prostate Cancer Diagnosis by miRNA Profiling of Prostate-Derived Plasma Small Extracellular Vesicles. Cells 2021, 10, 2372. [Google Scholar] [CrossRef]
- Duan, M.; Long, Y.; Yang, C.; Wu, X.; Sun, Y.; Li, J.; Hu, X.; Lin, W.; Han, D.; Zhao, Y.; et al. Selection and characterization of DNA aptamer for metastatic prostate cancer recognition and tissue imaging. Oncotarget 2016, 7, 36436–36446. [Google Scholar] [CrossRef]
- Hu, Y.; Duan, J.; Zhan, Q.; Wang, F.; Lu, X.; Yang, X.D. Novel MUC1 aptamer selectively delivers cytotoxic agent to cancer cells in vitro. PLoS ONE 2012, 7, e31970. [Google Scholar] [CrossRef] [Green Version]
- Noaparast, Z.; Hosseinimehr, S.J.; Piramoon, M.; Abedi, S.M. Tumor targeting with a (99m)Tc-labeled AS1411 aptamer in prostate tumor cells. J. Drug Target. 2015, 23, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Campos-Fernandez, E.; Barcelos, L.S.; Souza, A.G.; Goulart, L.R.; Alonso-Goulart, V. Post-SELEX Optimization and Characterization of a Prostate Cancer Cell-Specific Aptamer for Diagnosis. ACS Omega 2020, 5, 3533–3541. [Google Scholar] [CrossRef] [PubMed]
- Hassani, S.; Salek Maghsoudi, A.; Rezaei Akmal, M.; Rahmani, S.R.; Sarihi, P.; Ganjali, M.R.; Norouzi, P.; Abdollahi, M. A Sensitive Aptamer-Based Biosensor for Electrochemical Quantification of PSA as a Specific Diagnostic Marker of Prostate Cancer. J. Pharm. Pharm. Sci. 2020, 23, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Crulhas, B.P.; Basso, C.R.; Castro, G.R.; Pedrosa, V.A. Detection of Prostate Cancer Biomarker PCA3 by using Aptasensors. Curr. Med. Chem. 2022. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Kang, M.; Ban, C. Aptamer-antibody hybrid ELONA that uses hybridization chain reaction to detect a urinary biomarker EN2 for bladder and prostate cancer. Sci. Rep. 2022, 12, 11523. [Google Scholar] [CrossRef]
- Zheng, J.; Zhao, H.; Ning, G.; Sun, W.; Wang, L.; Liang, H.; Xu, H.; He, C.; Zhao, H.; Li, C.P. A novel affinity peptide-antibody sandwich electrochemical biosensor for PSA based on the signal amplification of MnO2-functionalized covalent organic framework. Talanta 2021, 233, 122520. [Google Scholar] [CrossRef]
- Diaz-Fernandez, A.; Miranda-Castro, R.; de-Los-Santos-Alvarez, N.; Lobo-Castanon, M.J.; Estrela, P. Impedimetric aptamer-based glycan PSA score for discrimination of prostate cancer from other prostate diseases. Biosens. Bioelectron. 2021, 175, 112872. [Google Scholar] [CrossRef]
- Miranda, A.; Santos, T.; Carvalho, J.; Alexandre, D.; Jardim, A.; Caneira, C.F.; Vaz, V.; Pereira, B.; Godinho, R.; Brito, D.; et al. Aptamer-based approaches to detect nucleolin in prostate cancer. Talanta 2021, 226, 122037. [Google Scholar] [CrossRef]
- Gong, S.; Ren, H.; Lin, C.; Hu, P.; Tian, R.; Liu, Z.; Li, Y.; Zhou, Y.; Yang, Y.; Lu, S. Immunochromatographic strip biosensor for the rapid detection of N-glycolylneuraminic acid based on aptamer-conjugated nanoparticle. Anal. Biochem. 2018, 561–562, 52–58. [Google Scholar] [CrossRef]
- Rani, D.; Pachauri, V.; Madaboosi, N.; Jolly, P.; Vu, X.T.; Estrela, P.; Chu, V.; Conde, J.P.; Ingebrandt, S. Top-Down Fabricated Silicon Nanowire Arrays for Field-Effect Detection of Prostate-Specific Antigen. ACS Omega 2018, 3, 8471–8482. [Google Scholar] [CrossRef] [Green Version]
- Zhao, T.T.; Peng, Z.W.; Yuan, D.; Zhen, S.J.; Huang, C.Z.; Li, Y.F. Metal-organic gel enhanced fluorescence anisotropy for sensitive detection of prostate specific antigen. Spectrochim. Acta. Part A Mol. Biomol. Spectrosc. 2018, 192, 328–332. [Google Scholar] [CrossRef]
- Crulhas, B.P.; Karpik, A.E.; Delella, F.K.; Castro, G.R.; Pedrosa, V.A. Electrochemical aptamer-based biosensor developed to monitor PSA and VEGF released by prostate cancer cells. Anal. Bioanal. Chem. 2017, 409, 6771–6780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhenadhayalan, N.; Yadav, K.; Sriram, M.I.; Lee, H.L.; Lin, K.C. Ultra-sensitive DNA sensing of a prostate-specific antigen based on 2D nanosheets in live cells. Nanoscale 2017, 9, 12087–12095. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Y.; Chen, J.; Liang, X.; Han, H.; Yang, Y.; Li, Q.; Wang, Y. Inhibition of cell proliferation through an ATP-responsive co-delivery system of doxorubicin and Bcl-2 siRNA. Int. J. Nanomed. 2017, 12, 4721–4732. [Google Scholar] [CrossRef] [Green Version]
- Atabi, F.; Mousavi Gargari, S.L.; Hashemi, M.; Yaghmaei, P. Doxorubicin Loaded DNA Aptamer Linked Myristilated Chitosan Nanogel for Targeted Drug Delivery to Prostate Cancer. Iran. J. Pharm. Res. IJPR 2017, 16, 35–49. [Google Scholar]
- Welton, J.L.; Brennan, P.; Gurney, M.; Webber, J.P.; Spary, L.K.; Carton, D.G.; Falcon-Perez, J.M.; Walton, S.P.; Mason, M.D.; Tabi, Z.; et al. Proteomics analysis of vesicles isolated from plasma and urine of prostate cancer patients using a multiplex, aptamer-based protein array. J. Extracell. Vesicles 2016, 5, 31209. [Google Scholar] [CrossRef] [Green Version]
- Marangoni, K.; Neves, A.F.; Rocha, R.M.; Faria, P.R.; Alves, P.T.; Souza, A.G.; Fujimura, P.T.; Santos, F.A.; Araujo, T.G.; Ward, L.S.; et al. Prostate-specific RNA aptamer: Promising nucleic acid antibody-like cancer detection. Sci. Rep. 2015, 5, 12090. [Google Scholar] [CrossRef] [Green Version]


| Aptamer | Target | Phase | Patient Number | Goal | Clinical Trial Identifier |
|---|---|---|---|---|---|
| N/A | COVID-19 | Recruiting | 200 | COVID-19 test | NCT04974203 |
| ARC1905 | C5 | I | 47 | Macular degeneration | NCT00950638 |
| E10030 | PDGF | II | 449 | Neovascular age-related macular degeneration | NCT01089517 |
| APT-POCT-01 | Antiretrovirals | Completed | 30 | Test the adherence to antiretrovirals | NCT04302896 |
| Sgc8 | PTK7 | Unknown | 70 | Treatment of colorectal cancer | NCT03385148 |
| REG1 | IXa | I | 107 | Anticoagulation system | NCT00113997 |
| EYE001 | VEGF | I | 5 | Reduce retinal thickening and improve vision in patients with Von Hippel–Lindau syndrome (VHL) | NCT00056199 |
| Oxytocin aptamer | Oxytocin | Unknown | 28 | Test novel aptamer-based electrochemical assay for the detection and quantification of salivary oxytocin | NCT03140709 |
| ARC1779 | Not specified | II | 28 | Treatment of Von Willebrand factor-related platelet disorders | NCT00632242 |
| ApToll | TLR4 | I | 46 | Stroke | NCT04742062 |
| NOX-H94 | Hepcidin | II | 33 | Treatment of anemia of chronic disease | NCT02079896 |
| NOX-E36 | CCL2 | I | 72 | Chronic inflammatory diseases | NCT00976729 |
| NOX-A12 | CXCL2 | I | 48 | To fight solid tumors by modulating the tumor microenvironment | NCT00976378 |
| ApToll | COVID-19 | Recruiting | 30 | Block the progression of patients to cytokine storm syndrome (CSS) | NCT05293236 |
| AS1411 | Nucleolin | II | 90 | Treatment of patients with primary refractory or relapsed acute myeloid leukemia | NCT01034410 |
| Tool | Type | Description |
|---|---|---|
| MPBind | Ranking | Ranks aptamers according to a statistical score. It is based on four types of Z score for each sequence motif (k-mer). |
| FASTAptamer | Clustering | Clusters aptamers based on sequence analysis. |
| MEMERIS | Motif | Finds motifs for its secondary structure and predicts the sequence motif in the loop structure. |
| AptaMut | Optimization | Determines whether mutations contribute to increase the affinity of the aptamers relative to the parent sequence. |
| Rtools | Others | Analyzes secondary RNA structures from a simple sequence of nucleotides. |
| COMPAS | All | Performs quality control, aptamer identification, ranking, clustering, and optimization. |
| Application | Designing Method | PSMA-Aptamer Sequence and Modifications | Sensitivity or Results | Disadvantage/Limitation | Biological Target | References |
|---|---|---|---|---|---|---|
| Diagnosis | Chemical synthesis | NH2-GAATTCGCGTTTTCGCTTTTGCGTTTTGGGTCATCTGCTTACGATAGCAATGCT | ~100 particles/μL in urine | Improvement in the number of particles per microliter to overcome current analysis | Urine | [131] |
| Diagnosis | Chemical synthesis | NH2-GGGAGGACGAUGCGGAUCAGCCAUGUUUACGUCACUCCU | Detection concentration in vitro is 10 μg/mL | Needs to demonstrate the distribution in other tissues apart of the pulmonary system | Cell lines and mice xenografts | [132] |
| Diagnosis | Chemical synthesis | Cy5.5-GGGAGGACGAUGCGGAUCAGCCAUGUUUACGUCACUCCU-spacer-NH2-3′ with 2′-fluoro pyrimidines | AUC of ½ peak intensity (dB·s) is 1507.60 ± 269.33 | Characterization of the nanobubbles’ distribution and their elimination is needed | Cell lines and mice xenografts | [101] |
| Treatment | Cell-SELEX | PSMA aptamer-survivin antisense siRNA: GGGAGGACGAUGCGGAUCAGCCAUGUUUACGU CACUCCUAAAAUGUAGAGAUGCGGUGGUCCUU | Inhibition of tumoral growth in mice | RNA stability and siRNA dosage for keeping therapeutic effect without toxicity in other tissues | Cell lines and mice xenografts | [133] |
| Diagnosis and prognosis | Chemical synthesis | Acid aptamer A10-3.2 (no specified sequence or modifications) | Discrimination of prostate cancer cells that express PSMA | Adjustment in parameters as the intensity of light penetration to deep tissue, and TMIA-chromophore abundance due to target density from small tumors | Cell lines | [134] |
| Treatment | SELEX | A10-3.2-saV2-9: TAA TAC GAC TCA CTA TAG GGA GGA CGA TGC GGA TCA GCC ATG TTT ACG TCA CTC CTA gaa aga aca tga atg ctg c ATGAAGCTTG g cag cat tca tgt tct ttc dTdT | Adjunctive therapy to suppress prostate cancer metastasis | Needs to demonstrate the specific gene activation by saRNA | Cell lines and mice xenografts | [135] |
| Treatment | Chemical synthesis | GGGAGGAAUAGCUGACGGGAGGACGAUGCGGAUCAGCCAUGUUUACGUCACUCCUUGUCAAUAAUAAGGGGC | Cytotoxicity in prostate cancer cells | Needs characterization of the in vivo stability for the biotin–DNA linker | In silico modeling and cell lines | [123] |
| Treatment | Chemical synthesis | GGGAGGACGAUGCGGAUCAGCCAUGUUUACGUCACUCCUUGUCAAUCCUCAUCGGC | Enhancement in the potency of external beam radiation therapy for established PSMA-positive tumors | Possibility of aptamer–siRNA chimera-mediated inflammatory reactions in humans | Cell lines, mice xenografts, and tissue sections | [114] |
| Treatment | Chemical synthesis | TGX221 with PSMA aptamer conjugation (no specified sequence or modifications) | Effective anti-cancer agent for prostate cancer | Possible accumulation of the nanomicellar compounds in tissues | Cell lines, mice xenografts and tissue sections | [136] |
| Treatment | Chemical synthesis | GGGAGGACGAUGCGGAUCAGCCAUGUUUACGUCACUCCU-(CH2)6-S-S-(CH2)6-OH-3′ with 2′-fluoro pyrimidines | Efficient delivery of miRNA expression vectors to prostate cancer cells | Needs the determination of toxicity in other tissues in vivo | Cell lines and mice xenografts | [137] |
| Diagnosis and treatment | Chemical synthesis | GGGAGGACGAUGCGGAUCAGCCAUGUUUACGUCACUCCUAA | To diagnose and treat PSMA-positive PCa in vivo | Determination of the possible undesired effects of 99m Tc radiolabeled aptamer | Cell lines and mice xenografts | [126] |
| Diagnosis | Chemical synthesis | DBCO-5′-GAA TTC GCG TTT TCG CTT TTG CGT TTT GGG TCA TCT GCT TAC GAT AGC AAT GCT-3′ | Diagnostic potency (AUC: miR-145, 0.76; miR-221, 0.7; miR-451a, 0.65; and miR-141, 0.64) | Issues with the high relative amount of PSMA(+)SEVs observed in plasma | Blood samples | [138] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-Hernández, C.D.; Rodríguez-Martínez, G.; Cortés-Ramírez, S.A.; Morales-Pacheco, M.; Cruz-Burgos, M.; Losada-García, A.; Reyes-Grajeda, J.P.; González-Ramírez, I.; González-Covarrubias, V.; Camacho-Arroyo, I.; et al. Aptamers as Theragnostic Tools in Prostate Cancer. Biomolecules 2022, 12, 1056. https://doi.org/10.3390/biom12081056
Cruz-Hernández CD, Rodríguez-Martínez G, Cortés-Ramírez SA, Morales-Pacheco M, Cruz-Burgos M, Losada-García A, Reyes-Grajeda JP, González-Ramírez I, González-Covarrubias V, Camacho-Arroyo I, et al. Aptamers as Theragnostic Tools in Prostate Cancer. Biomolecules. 2022; 12(8):1056. https://doi.org/10.3390/biom12081056
Chicago/Turabian StyleCruz-Hernández, Carlos David, Griselda Rodríguez-Martínez, Sergio A. Cortés-Ramírez, Miguel Morales-Pacheco, Marian Cruz-Burgos, Alberto Losada-García, Juan Pablo Reyes-Grajeda, Imelda González-Ramírez, Vanessa González-Covarrubias, Ignacio Camacho-Arroyo, and et al. 2022. "Aptamers as Theragnostic Tools in Prostate Cancer" Biomolecules 12, no. 8: 1056. https://doi.org/10.3390/biom12081056
APA StyleCruz-Hernández, C. D., Rodríguez-Martínez, G., Cortés-Ramírez, S. A., Morales-Pacheco, M., Cruz-Burgos, M., Losada-García, A., Reyes-Grajeda, J. P., González-Ramírez, I., González-Covarrubias, V., Camacho-Arroyo, I., Cerbón, M., & Rodríguez-Dorantes, M. (2022). Aptamers as Theragnostic Tools in Prostate Cancer. Biomolecules, 12(8), 1056. https://doi.org/10.3390/biom12081056






