Bioinformatics Analysis of LGR4 in Colon Adenocarcinoma as Potential Diagnostic Biomarker, Therapeutic Target and Promoting Immune Cell Infiltration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Differentially Expressed Genes Screening and Protein Expression
2.3. Protein–Protein Interaction Network Analysis
2.4. Pathway Enrichment Analysis
2.5. Immune Cell Infiltration Analysis
2.6. Gene Set Enrichment Analysis
2.7. Cell Culture
2.8. RT-PCR Analysis
2.9. Statistical Methods
3. Results
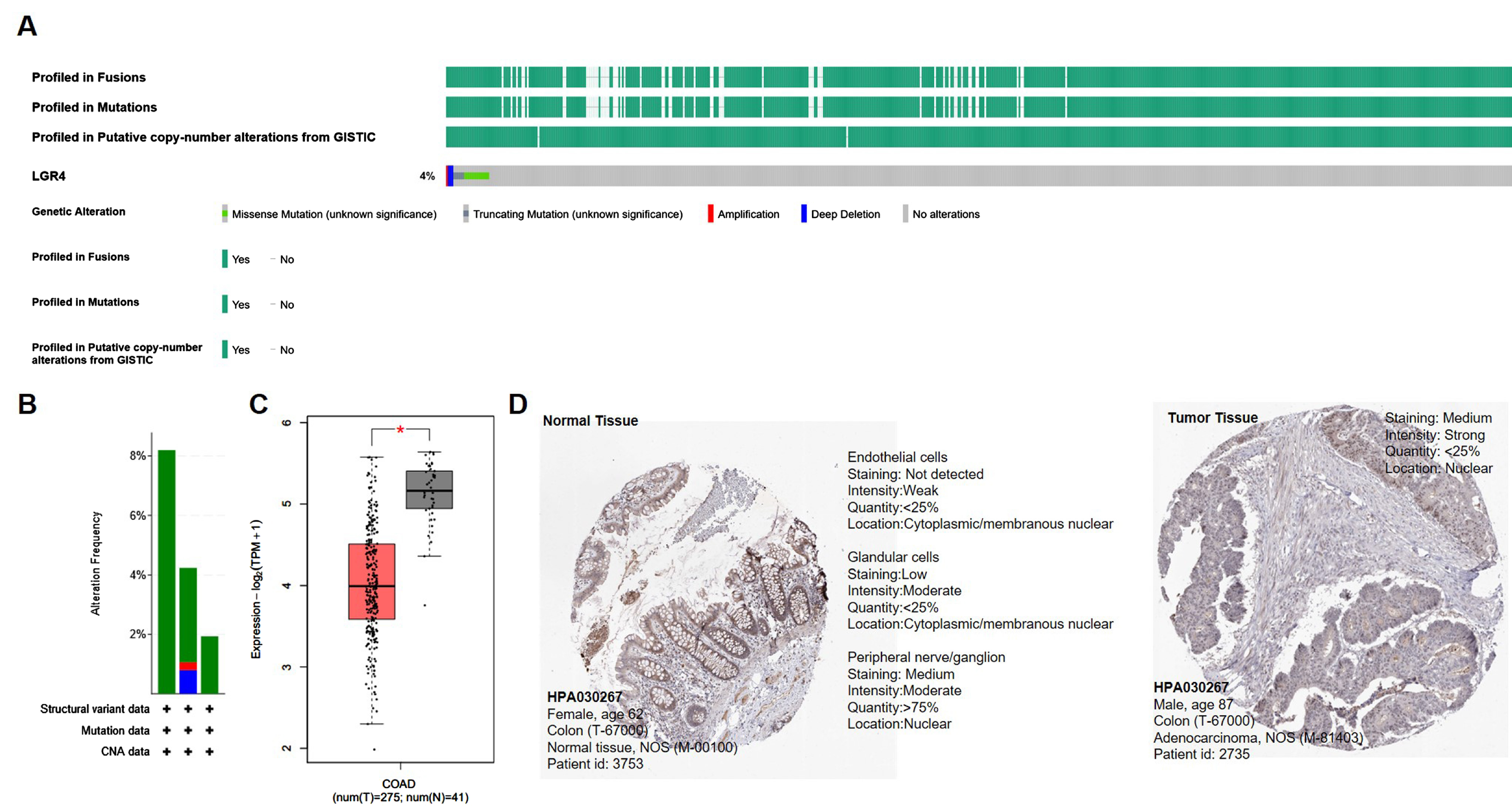
3.1. Clinical Characteristics and LGR4 Expression of COAD Patients
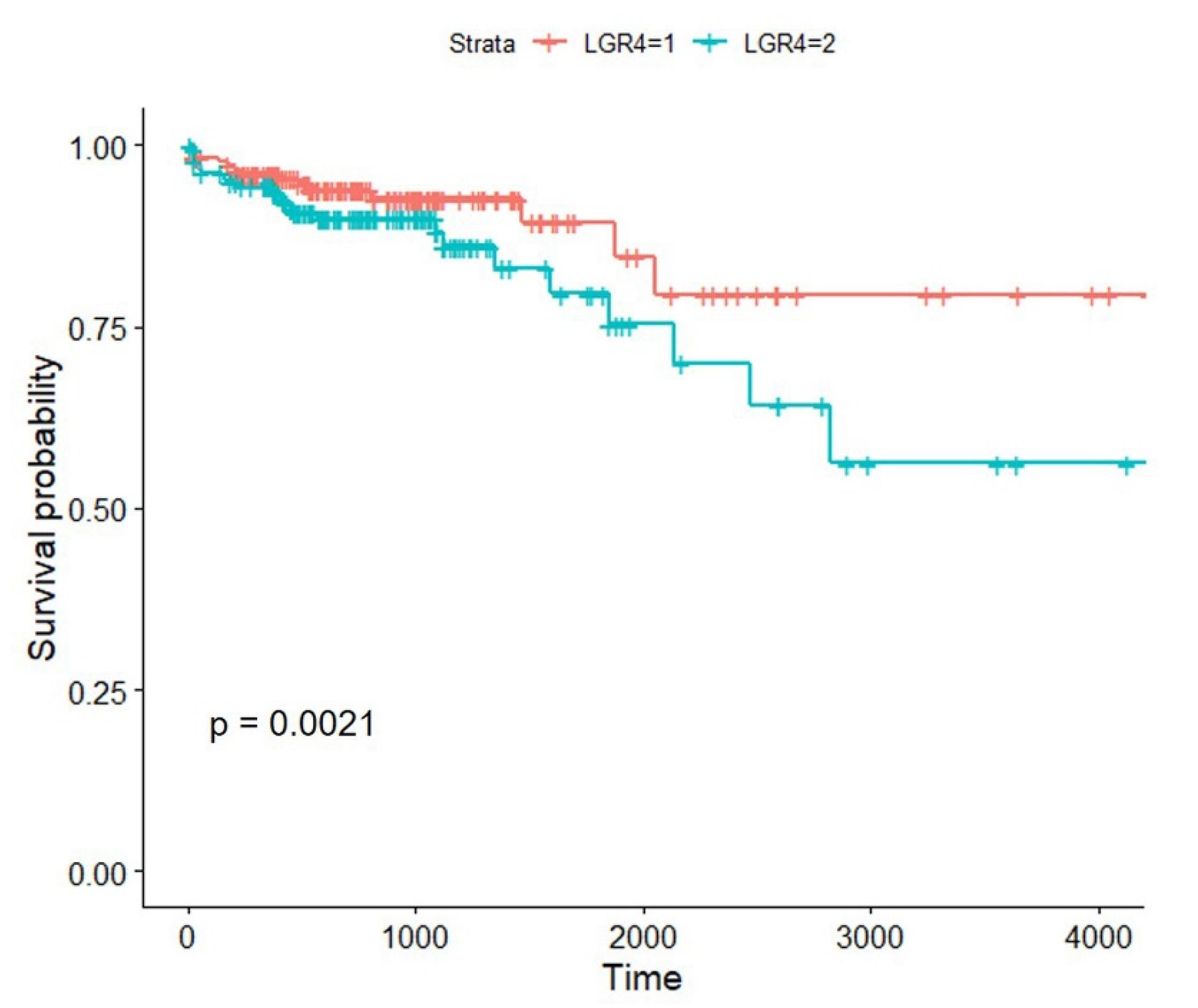
3.2. High Expression of LGR4 in COAD Worsens the Overall Survival of Patients
3.3. Identification of Differentially Expressed Genes
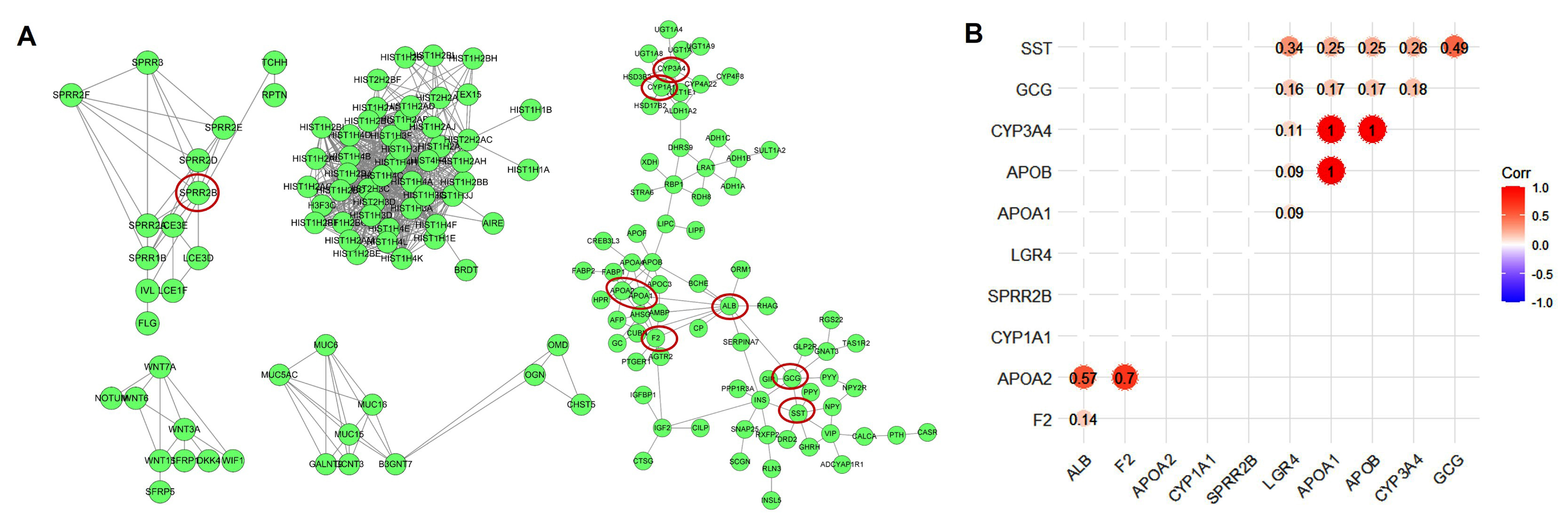
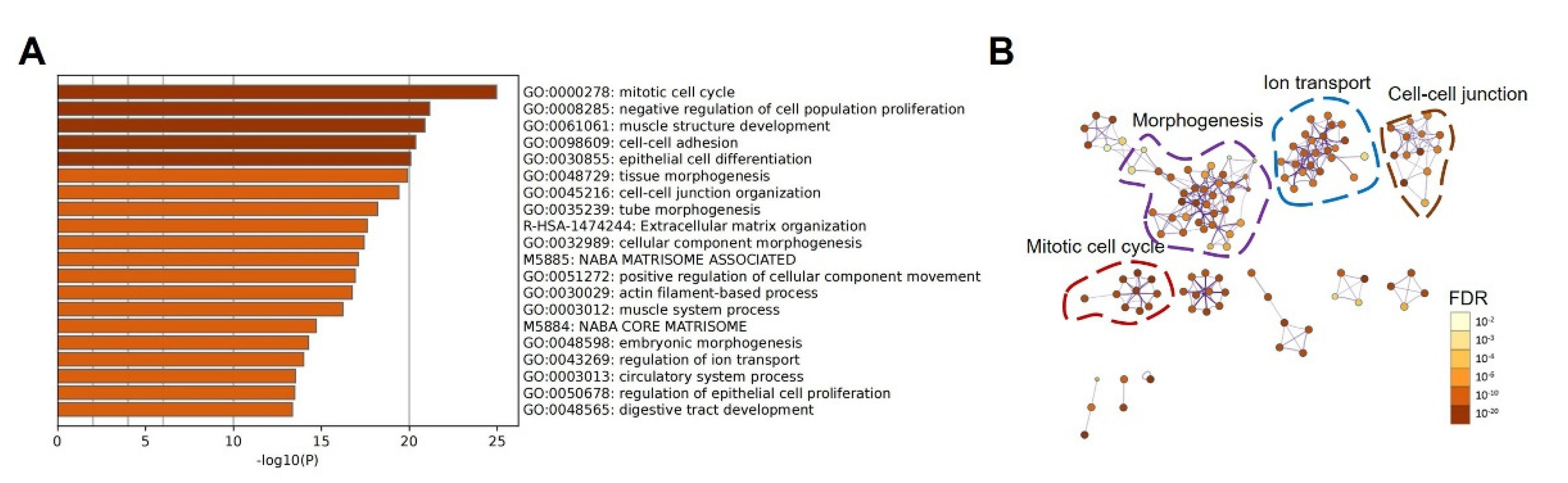
3.4. Protein–Protein Interaction Network Analysis
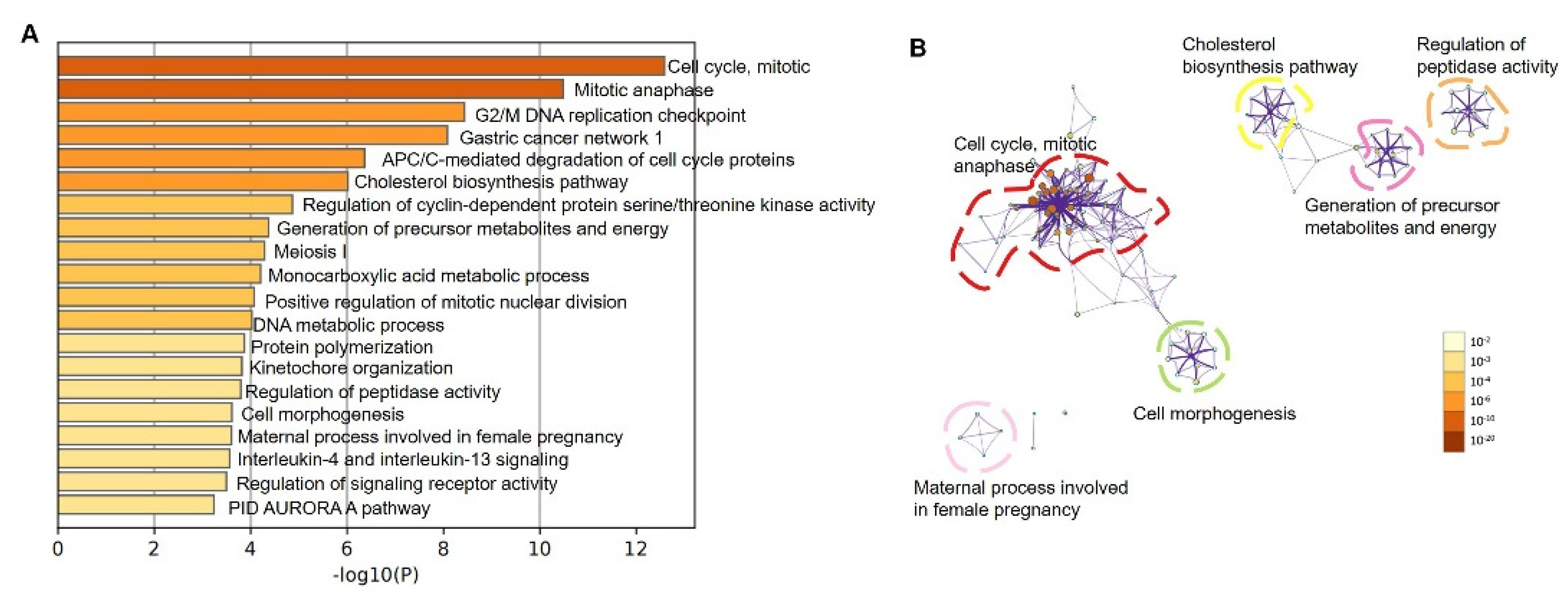
3.5. Modulation of Pathways by Up-Regulated LGR4 in COAD
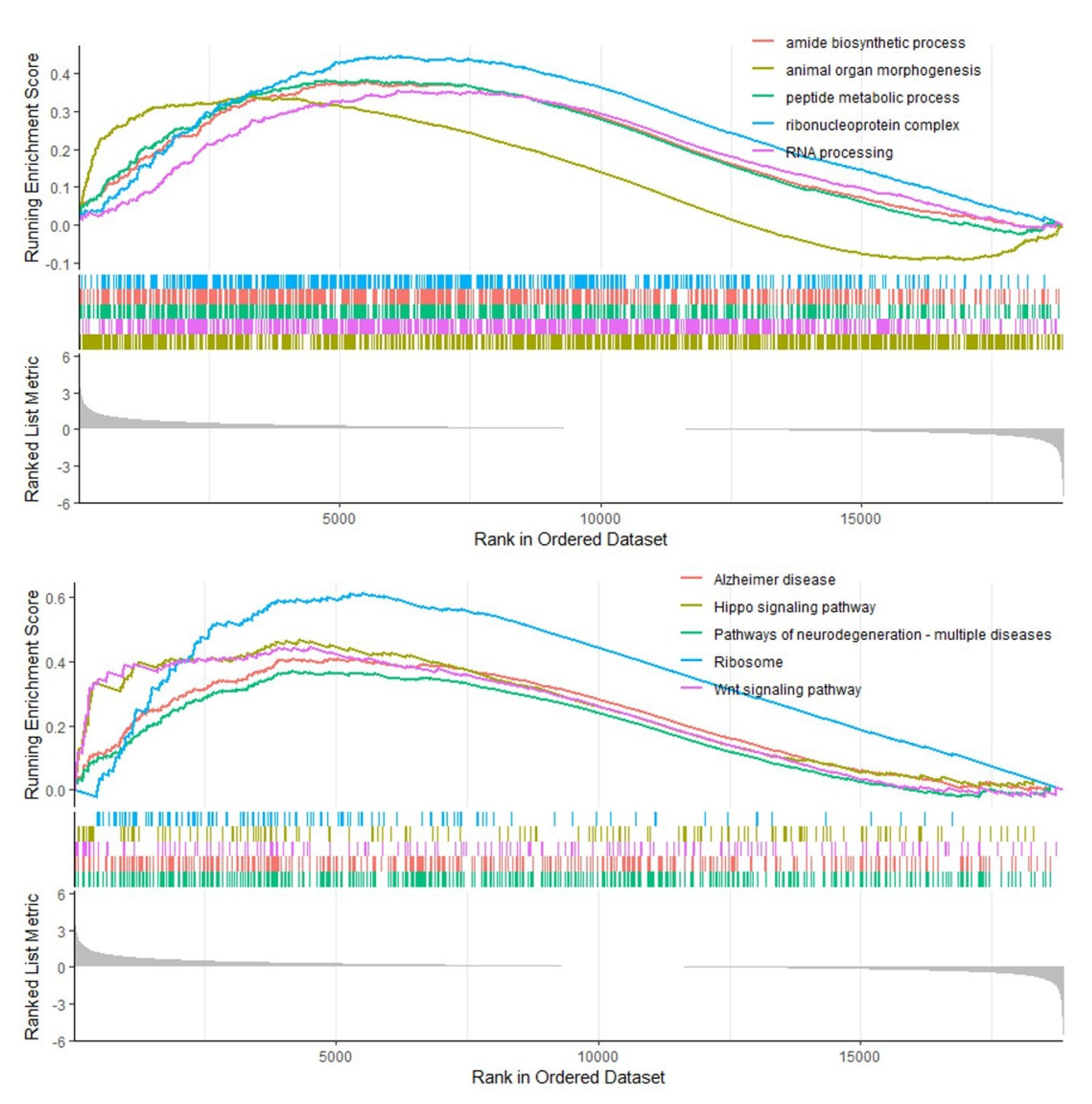
3.6. Gene Set Enrichment Analysis
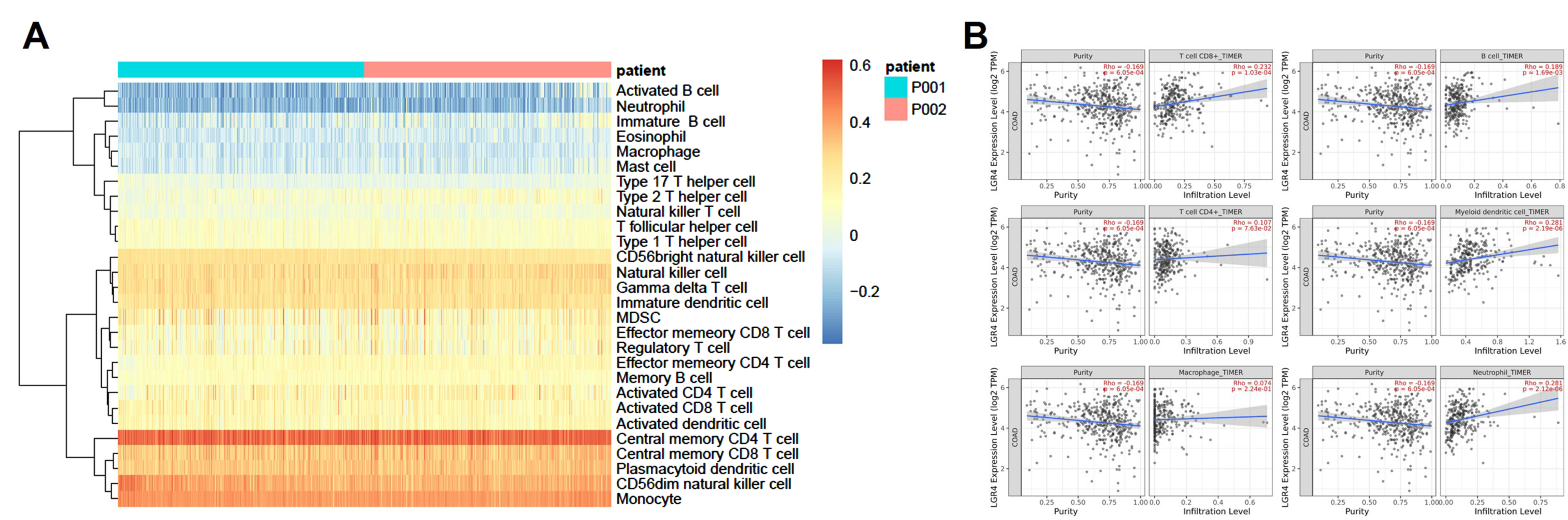
3.7. Immune Cell Infiltration Analysis
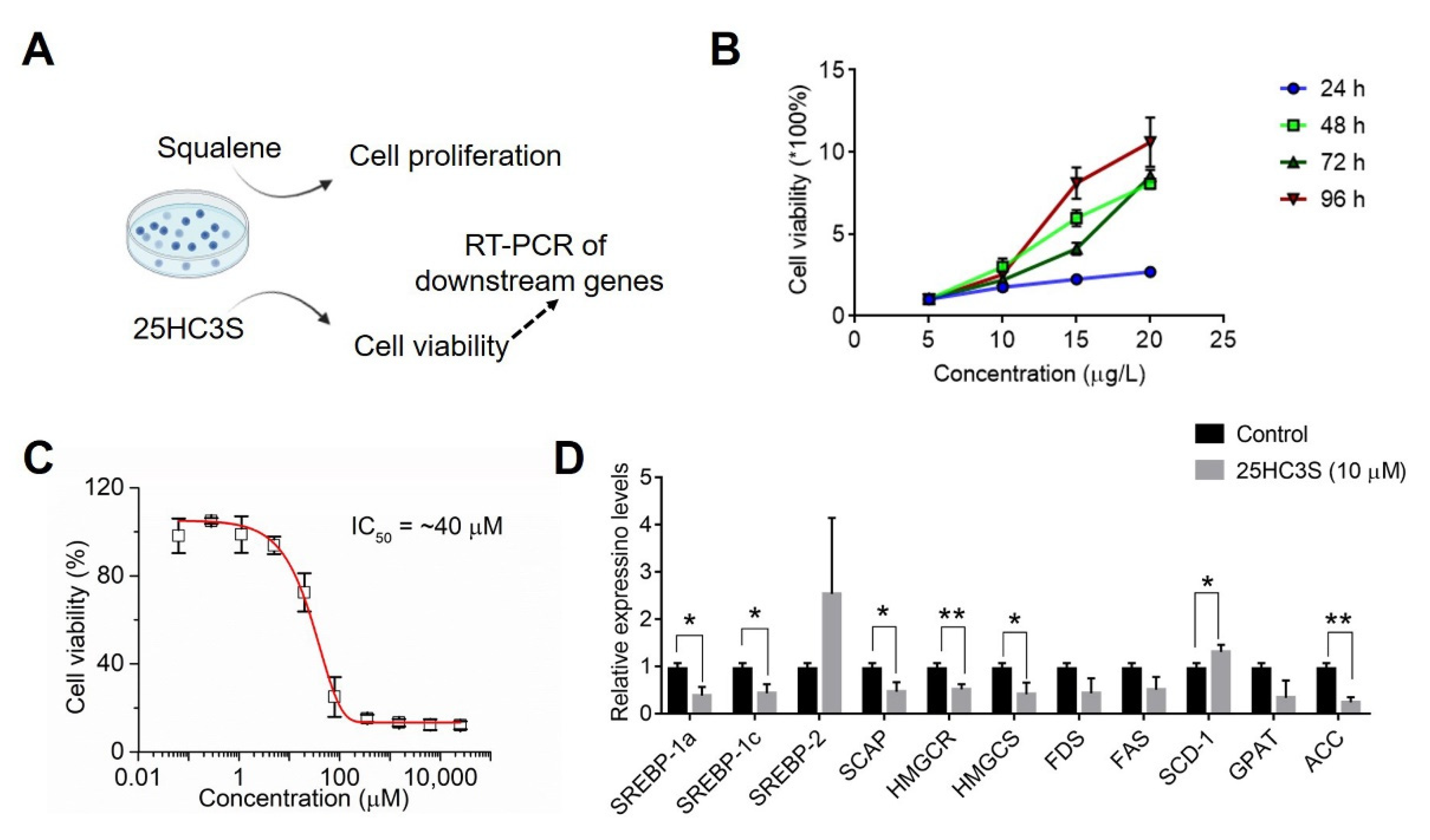
3.8. Analysis of Cholesterol Biosynthesis in COAD
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
- COAD: Colon Adenocarcinoma
- TCGA: The Cancer Genome Atlas
- TIMER: Tumor Immune Estimation Resource
- RANKL: Receptor activator of nuclear factor kappa-B ligand
- DEGs: Differentially expressed genes
- PPI: Protein–protein interaction
- GO: Gene Ontology
- GSEA: Gene Set Enrichment Analysis
- ssGSEA: Single-sample GSEA
- DMEM: Dulbecco’s Modified Eagle Medium
- MTT: 2,5-diphenyl-2H-tetrazolium bromide
- GTEx: The Genotype-Tissue Expression
- KEGG: Kyoto Encyclopedia of Genes and Genomes
- NES: Normalized enrichment score
- ff-MAS: Follicular fluid meiosis-activating sterol
- 25HC3S: 25-hydroxycholesterol-3-sulfate
- HPA: The Human Protein Atlas
- K–M analysis: Kaplan–Meier analysis
- FDR: False Discovery Rate
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.H.; Tseng, P.L.; Tung, H.Y.; Cheng, S.C.; Ching, C.Y.; Chang, S.C.; Wu, S.F. Comparison of risk factors between colon cancer and rectum cancer in a single medical center hospital, Taiwan. Arch. Med. Sci. AMS 2020, 16, 102–111. [Google Scholar] [CrossRef]
- Laudi, N.; Arora, M.; Burns, L.; McGlave, P.; Miller, J.; Bohac, G.; Rogers, T.; Barker, J.; Coad, J.; Weisdorf, D. Efficacy of high-dose therapy and hematopoietic stem cell transplantation for mantle cell lymphoma. Am. J. Hematol. 2006, 81, 519–524. [Google Scholar] [CrossRef]
- Pan, J.H.; Zhou, H.; Cooper, L.; Huang, J.L.; Zhu, S.B.; Zhao, X.X.; Ding, H.; Pan, Y.L.; Rong, L. LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Front. Immunol. 2019, 10, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Jia, S.; Jin, X.; Zhu, S.; Zhang, S. HOXC11 Expression Is Associated with the Progression of Colon Adenocarcinoma and Is a Prognostic Biomarker. DNA Cell Biol. 2021, 40, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jiao, N.; Sun, T.; Ma, Y.; Zhang, X.; Chen, H.; Hong, J.; Zhang, Y. CXCL11 Correlates With Antitumor Immunity and an Improved Prognosis in Colon Cancer. Front. Cell Dev. Biol. 2021, 9, 646252. [Google Scholar] [CrossRef]
- Pang, C.; Wang, H.; Shen, C.; Liang, H. Application Potential of CTHRC1 as a Diagnostic and Prognostic Indicator for Colon Adenocarcinoma. Front. Mol. Biosci. 2022, 9, 849771. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Deng, D.; Wang, X.; Wang, X.; Jiang, S.; Cui, R. LINC00491 as a new molecular marker can promote the proliferation, migration and invasion of colon adenocarcinoma cells. OncoTargets Ther. 2019, 12, 6471–6480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, J.; Luo, J.; Cheng, X.; Jin, C.; Zhou, X.; Qu, J.; Tu, L.; Ai, D.; Li, D.; Wang, J.; et al. Deletion of G protein-coupled receptor 48 leads to ocular anterior segment dysgenesis (ASD) through down-regulation of Pitx2. Proc. Natl. Acad. Sci. USA 2008, 105, 6081–6086. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Zhou, W.; Zhou, X.; Li, D.; Weng, J.; Yi, Z.; Cho, S.G.; Li, C.; Yi, T.; Wu, X.; et al. Regulation of bone formation and remodeling by G-protein-coupled receptor 48. Development 2009, 136, 2747–2756. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Rodriguez, M.; Valdez, J.M.; Zhu, X.; Tan, K.; Li, D.; Siwko, S.; Xin, L.; Liu, M. Lgr4 is a key regulator of prostate development and prostate stem cell differentiation. Stem Cells 2013, 31, 2492–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Yang, H.; Wei, D.; Zhang, X.; Wang, J.; Wu, X.; Chang, J. Mitochondria-targeted nanoparticles in treatment of neurodegenerative diseases. Exploration 2021, 1, 20210115. [Google Scholar] [CrossRef]
- Pan, H.; Cui, H.; Liu, S.; Qian, Y.; Wu, H.; Li, L.; Guan, Y.; Guan, X.; Zhang, L.; Fan, H.Y.; et al. Lgr4 gene regulates corpus luteum maturation through modulation of the WNT-mediated EGFR-ERK signaling pathway. Endocrinology 2014, 155, 3624–3637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siwko, S.; Lai, L.; Weng, J.; Liu, M. Lgr4 in ocular development and glaucoma. J. Ophthalmol. 2013, 2013, 987494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustata, R.C.; Vasile, G.; Fernandez-Vallone, V.; Strollo, S.; Lefort, A.; Libert, F.; Monteyne, D.; Pérez-Morga, D.; Vassart, G.; Garcia, M.I. Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. 2013, 5, 421–432. [Google Scholar] [CrossRef]
- Oyama, K.; Mohri, Y.; Sone, M.; Nawa, A.; Nishimori, K. Conditional knockout of Lgr4 leads to impaired ductal elongation and branching morphogenesis in mouse mammary glands. Sex. Dev. 2011, 5, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, S.; Lou, J.; Mulholland, M.; Zhang, W. LGR4 protects hepatocytes from injury in mouse. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 316, G123–G131. [Google Scholar] [CrossRef] [PubMed]
- Kinzel, B.; Pikiolek, M.; Orsini, V.; Sprunger, J.; Isken, A.; Zietzling, S.; Desplanches, M.; Dubost, V.; Breustedt, D.; Valdez, R.; et al. Functional roles of Lgr4 and Lgr5 in embryonic gut, kidney and skin development in mice. Dev. Biol. 2014, 390, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Ordaz-Ramos, A.; Rosales-Gallegos, V.H.; Melendez-Zajgla, J.; Maldonado, V.; Vazquez-Santillan, K. The Role of LGR4 (GPR48) in Normal and Cancer Processes. Int. J. Mol. Sci. 2021, 22, 4690. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Z.; Pan, Z.; Liu, Y. Advanced bioactive nanomaterials for biomedical applications. Exploration 2021, 1, 20210089. [Google Scholar] [CrossRef]
- Gao, Y.; Kitagawa, K.; Hiramatsu, Y.; Kikuchi, H.; Isobe, T.; Shimada, M.; Uchida, C.; Hattori, T.; Oda, T.; Nakayama, K.; et al. Up-regulation of GPR48 induced by down-regulation of p27Kip1 enhances carcinoma cell invasiveness and metastasis. Cancer Res. 2006, 66, 11623–11631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, G.; He, J.; Liu, J.; Yan, X.; Fan, K. Nanozyme for tumor therapy: Surface modification matters. Exploration 2021, 1, 75–89. [Google Scholar] [CrossRef]
- Liu, J.; Chen, C.; Wei, T.; Gayet, O.; Loncle, C.; Borge, L.; Dusetti, N.; Ma, X.; Marson, D.; Laurini, E. Dendrimeric nanosystem consistently circumvents heterogeneous drug response and resistance in pancreatic cancer. Exploration 2021, 1, 21–34. [Google Scholar] [CrossRef]
- Wu, J.; Xie, N.; Xie, K.; Zeng, J.; Cheng, L.; Lei, Y.; Liu, Y.; Song, L.; Dong, D.; Chen, Y.; et al. GPR48, a poor prognostic factor, promotes tumor metastasis and activates β-catenin/TCF signaling in colorectal cancer. Carcinogenesis 2013, 34, 2861–2869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, L.; Liao, Z.; Luo, Z.; Wu, Y.L.; Herrmann, A.; Huo, S. Ultrasound-controlled drug release and drug activation for cancer therapy. Exploration 2021, 1, 20210023. [Google Scholar] [CrossRef]
- Yi, M.; Nissley, D.V.; McCormick, F.; Stephens, R.M. ssGSEA score-based Ras dependency indexes derived from gene expression data reveal potential Ras addiction mechanisms with possible clinical implications. Sci. Rep. 2020, 10, 10258. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT-qPCR—Publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef]
- Xue, X.; Li, X.; Pan, Z.; Zhao, L.; Ding, Y. Comparison of clinicopathological features and KRAS gene mutation of left-sided and right-sided colon cancers. Int. J. Clin. Exp. Pathol. 2017, 10, 11353–11359. [Google Scholar] [PubMed]
- Zhang, H.; Pan, X.; Wu, Q.; Guo, J.; Wang, C.; Liu, H. Manganese carbonate nanoparticles-mediated mitochondrial dysfunction for enhanced sonodynamic therapy. Exploration 2021, 1, 20210010. [Google Scholar] [CrossRef]
- Ramanathan, V.; Jin, G.; Westphalen, C.B.; Whelan, A.; Dubeykovskiy, A.; Takaishi, S.; Wang, T.C. P53 gene mutation increases progastrin dependent colonic proliferation and colon cancer formation in mice. Cancer Investig. 2012, 30, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Pan, D. Hippo signaling in organ size control. Genes Dev. 2007, 21, 886–897. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5, e138999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsche, M.A.; McDonald, J.G.; Hobbs, H.H.; Cohen, J.C. Flux analysis of cholesterol biosynthesis in vivo reveals multiple tissue and cell-type specific pathways. eLife 2015, 4, e07999. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, Y.; Hu, Q. Recent advances in overcoming barriers to cell-based delivery systems for cancer immunotherapy. Exploration 2022, 2, 20210106. [Google Scholar] [CrossRef]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin. 2022; online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Rajurkar, M.; Parikh, A.R.; Solovyov, A.; You, E.; Kulkarni, A.S.; Chu, C.; Xu, K.H.; Jaicks, C.; Taylor, M.S.; Wu, C. Reverse Transcriptase Inhibition Disrupts Repeat Element Life Cycle in Colorectal Cancer. Cancer Discov. 2022, 12, 1462–1481. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; Coussens, L.M.; Kroemer, G.; Galluzzi, L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019, 30, 36–50. [Google Scholar] [CrossRef]
- Yuan, Y.; Li, H.; Pu, W.; Chen, L.; Guo, D.; Jiang, H.; He, B.; Qin, S.; Wang, K.; Li, N. Cancer metabolism and tumor microenvironment: Fostering each other? Sci. China Life Sci. 2021, 65, 236–279. [Google Scholar] [CrossRef]
- Zheng, Y.; Han, Y.; Sun, Q.; Li, Z. Harnessing anti-tumor and tumor-tropism functions of macrophages via nanotechnology for tumor immunotherapy. Exploration 2022, 2, 20210166. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, H.; Zou, J.; Feng, X.; Feng, D. Bisphenol A induces cholesterol biosynthesis in HepG2 cells via SREBP-2/HMGCR signaling pathway. J. Toxicol. Sci. 2019, 44, 481–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Name | Forward Sequence | Reverse Sequence |
|---|---|---|
| GAPDH | CAATGACCCCTTCATTGACC | TTGATTTTGGAGGGATCTCG |
| HMGR | GTCATTCCAGCCAAGGTTGT | GGGACCACTTGCTTCCATTA |
| HMGCS | CAGCTGCTGTVTTVAATGCTGTTA | AGCTACTGCTCCAACTCGGCATC |
| FAS | TGTGGACATGGTCACGGAC | GGCATCAAACCTAGACAGGTC |
| ACC1 | TCGCTTTGGGGGAAATAAAGTG | ACCACCTACGGATAGACCGC |
| SREBP-1a | GCGCCATGGAGGAGCTGCCCTTCG | GTCACTGTCTTGGTTGTTGATG |
| SREBP-1c | TGCGGACGCAGTCTGGGCAAC-3 | GTCACTGTCTTGGTTGTTGATG |
| SREBP-2 | AACGGTCATTCACCCAGGTC | GGCTGAAGAATAGGAGTTGCC |
| GPAT | ATCTTCAGAACAGCAAAATCGAAA | CAGCGGAAAACTCCAAATCC |
| SCD1 | TTCTTGCGATACACTCTGGTGC | CGGGATTGAATCTTCTTGTCGT |
| FDS | TCCATGGCGGATCTGAAGTCAACT | CATCCAGTCTTTGTCCATGTATCTG |
| SCAP | ACTGGGCATCATCCTCATTG | GGCACTGTCTGGTTCTCTGG |
| Clinical Factor | TCGA Database |
|---|---|
| Age (years, mean ± SD) | 66.92 ± 13.08 |
| Sex (female, male) | 230/285 |
| Stage of COAD | |
| Stage I/IA | 75/1 |
| Stage II/IIA/IIB/IIC | 30/137/10/1 |
| Stage III/IIIA/IIIB/IIIC | 20/8/60/41 |
| Stage IV/IVA/IVB | 46/17/2 |
| NA | 14 |
| Tissue_or_organ_of_origin | |
| Ascending colon | 91 |
| Cecum | 88 |
| Colon, NOS | 104 |
| Descending colon | 17 |
| Hepatic flexure of colon | 12 |
| Rectosigmoid junction | 7 |
| Sigmoid colon | 114 |
| Splenic flexure of colon | 5 |
| Transverse colon | 21 |
| NA | 2 |
| Primary_diagnosis | |
| Adenocarcinoma with mixed subtypes | 1 |
| Adenocarcinoma with neuroendocrine differentiation | 1 |
| Adenocarcinoma, NOS | 388 |
| Adenosquamous carcinoma | 1 |
| Carcinoma, NOS | 1 |
| Mucinous adenocarcinoma/ | 65 |
| Papillary adenocarcinoma, NOS | 2 |
| NA | 2 |
| Death (alive/death/no report) | 357/102/2 |
| Radiation therapy (yes/no/no report) | 379/12/70 |
| Pharmaceutical therapy (yes/no/no report) | 150/241/70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Tian, X.; Du, H.; Liu, X.; Wu, H. Bioinformatics Analysis of LGR4 in Colon Adenocarcinoma as Potential Diagnostic Biomarker, Therapeutic Target and Promoting Immune Cell Infiltration. Biomolecules 2022, 12, 1081. https://doi.org/10.3390/biom12081081
Wu L, Tian X, Du H, Liu X, Wu H. Bioinformatics Analysis of LGR4 in Colon Adenocarcinoma as Potential Diagnostic Biomarker, Therapeutic Target and Promoting Immune Cell Infiltration. Biomolecules. 2022; 12(8):1081. https://doi.org/10.3390/biom12081081
Chicago/Turabian StyleWu, Lijuan, Xiaoxiao Tian, Hao Du, Xiaomin Liu, and Haigang Wu. 2022. "Bioinformatics Analysis of LGR4 in Colon Adenocarcinoma as Potential Diagnostic Biomarker, Therapeutic Target and Promoting Immune Cell Infiltration" Biomolecules 12, no. 8: 1081. https://doi.org/10.3390/biom12081081






