Single-Cell FISH Analysis Reveals Distinct Shifts in PKM Isoform Populations during Drug Resistance Acquisition
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Semi-Quantitative Reverse Transcription (RT)-PCR and Real-Time Quantitative PCR (qRT-PCR)
2.4. Cell Viability Measurement
2.5. RNA Interference
2.6. Preparation of FISH Probe for Hybridization
2.7. In Situ Hybridization
2.8. SM-FISH Imaging and Analysis
2.9. SDS-PAGE and Western Blot Analysis
2.10. Subcellular Fractionation
2.11. Quantification of Relative mRNA ratio of PKM Isoform by qRT-PCR
2.12. Statistical Analysis
3. Results and Discussion
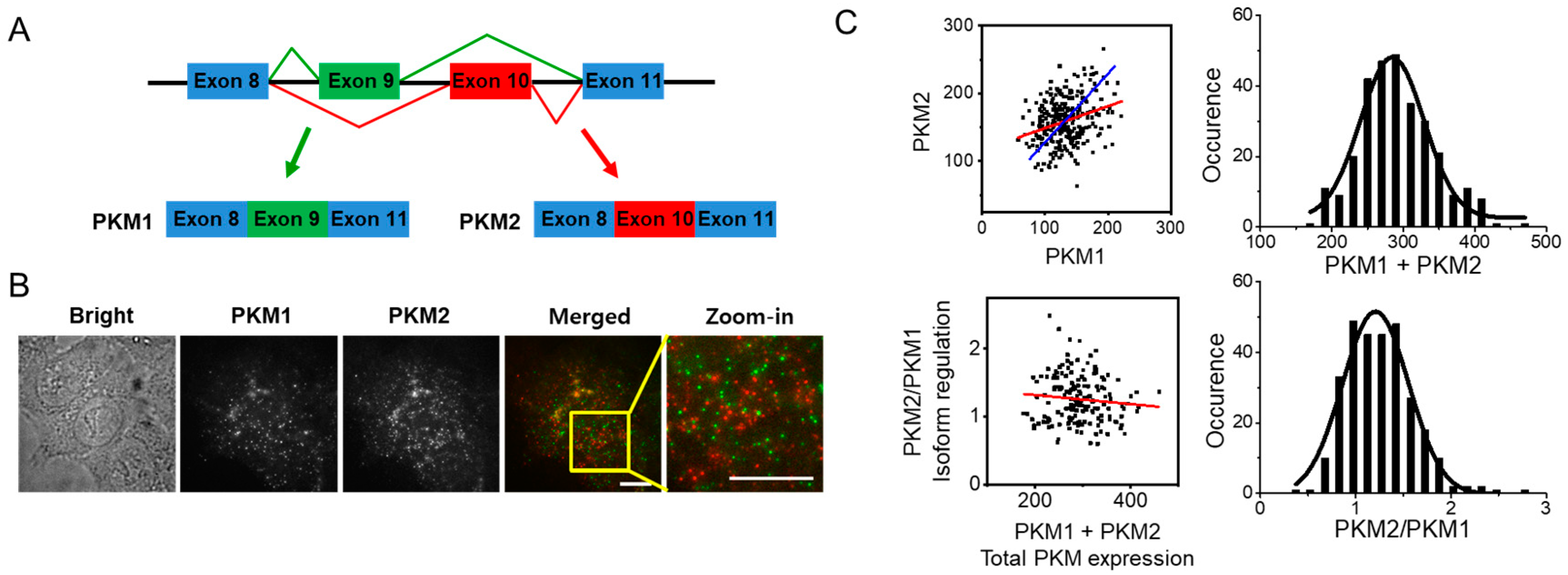
3.1. Two Splicing Isoforms of PKM Can Be Distinctly Quantified at a Single-Cell Level
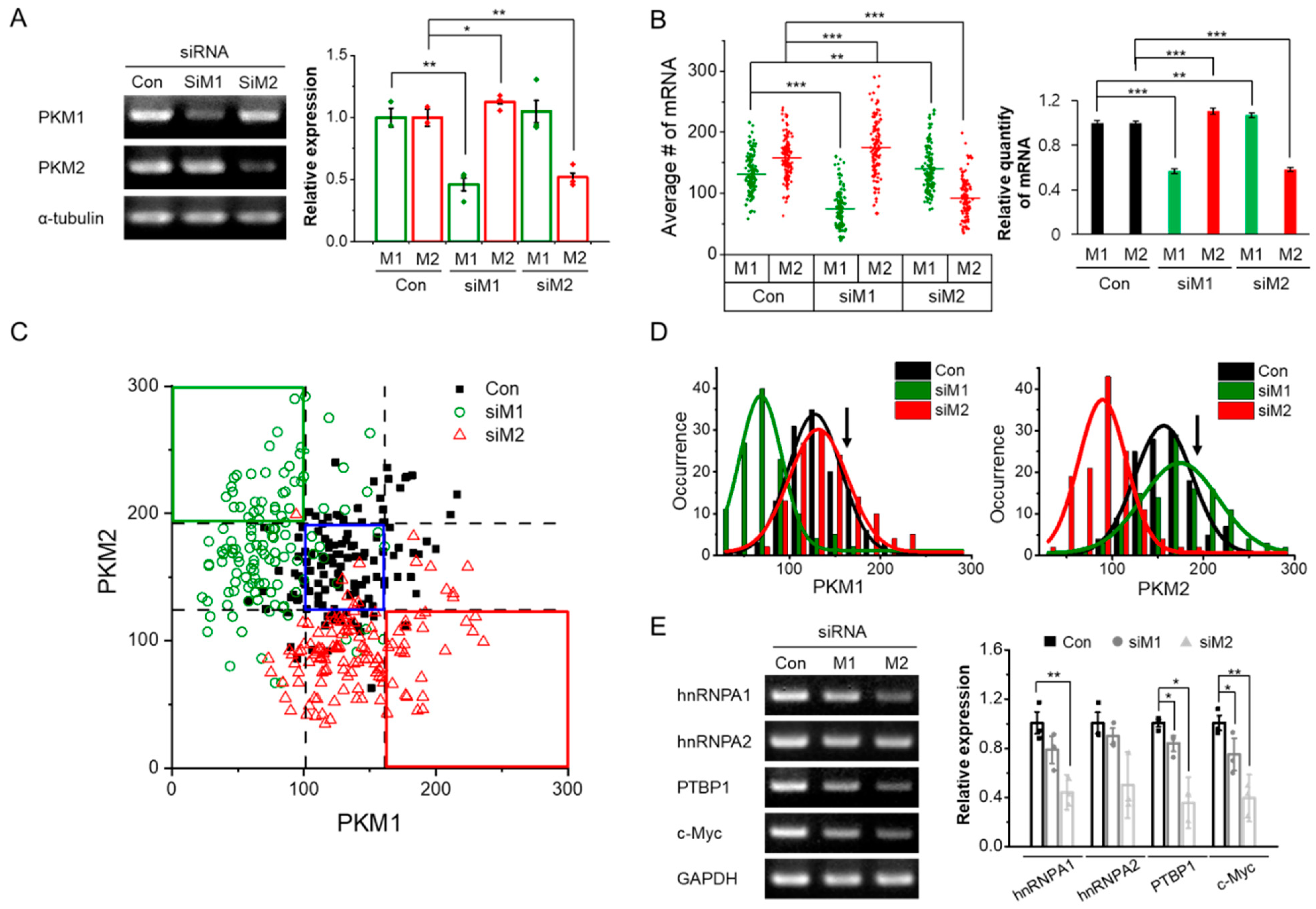
3.2. Depletion of PKM1 Induces a Heterogeneous PKM2 Increase
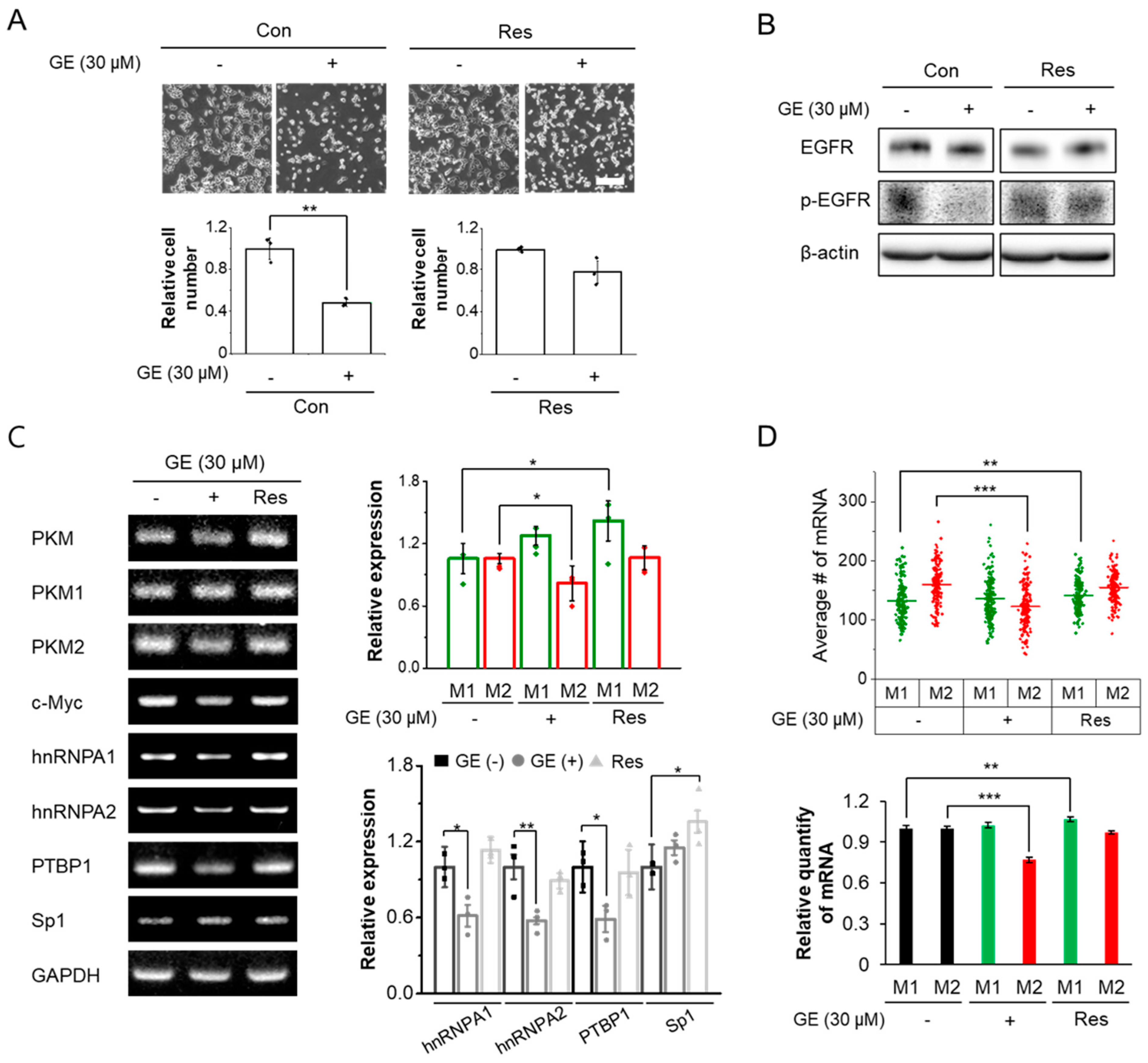
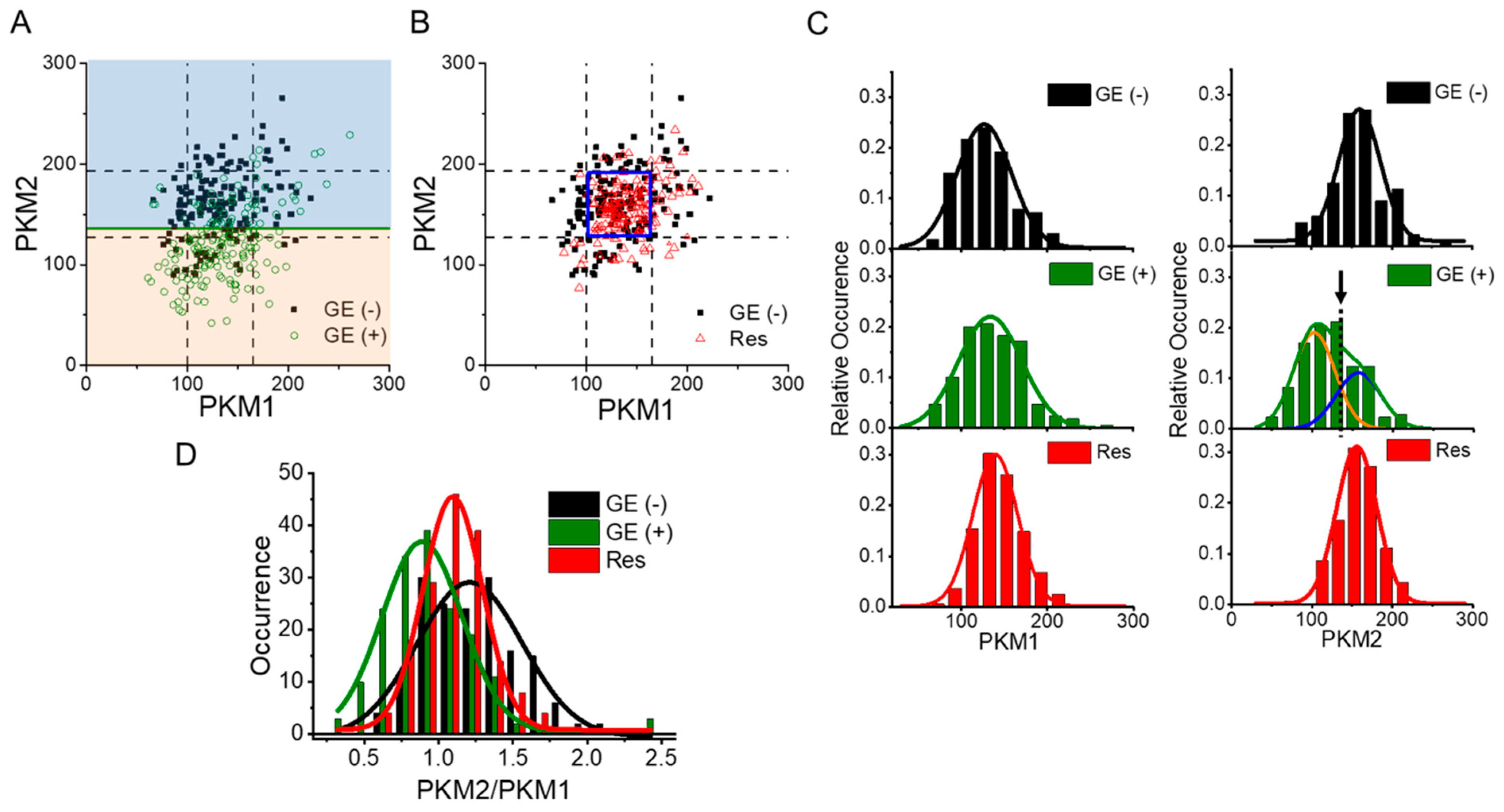
3.3. Population Shifts of PKM Isoforms Can Be Identified during Drug Resistance Acquisition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Konieczkowski, D.J.; Johannessen, C.M.; Garraway, L.A. A Convergence-Based Framework for Cancer Drug Resistance. Cancer Cell 2018, 33, 801–815. [Google Scholar] [CrossRef] [PubMed]
- Lawson, D.A.; Kessenbrock, K.; Davis, R.T.; Pervolarakis, N.; Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 2018, 20, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Icard, P.; Shulman, S.; Farhat, D.; Steyaert, J.M.; Alifano, M.; Lincet, H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist. Updat 2018, 38, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zaal, E.A.; Berkers, C.R. The Influence of Metabolism on Drug Response in Cancer. Front. Oncol. 2018, 8, 500. [Google Scholar] [CrossRef]
- Bhattacharya, B.; Mohd Omar, M.F.; Soong, R. The Warburg effect and drug resistance. Br. J. Pharmacol. 2016, 173, 970–979. [Google Scholar] [CrossRef]
- Xiao, Z.; Dai, Z.; Locasale, J.W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 2019, 10, 3763. [Google Scholar] [CrossRef]
- Zhang, Z.; Deng, X.; Liu, Y.; Liu, Y.; Sun, L.; Chen, F. PKM2, function and expression and regulation. Cell Biosci. 2019, 9, 52. [Google Scholar] [CrossRef]
- Hsu, M.C.; Hung, W.C. Pyruvate kinase M2 fuels multiple aspects of cancer cells: From cellular metabolism, transcriptional regulation to extracellular signaling. Mol. Cancer 2018, 17, 35. [Google Scholar] [CrossRef]
- Wiese, E.K.; Hitosugi, T. Tyrosine Kinase Signaling in Cancer Metabolism: PKM2 Paradox in the Warburg Effect. Front. Cell Dev. Biol. 2018, 6, 79. [Google Scholar] [CrossRef]
- Christofk, H.R.; Vander Heiden, M.G.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.D.; Schreiber, S.L.; Cantley, L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Lv, F.; Liu, F.; Guo, X.; Fan, Y.; Gu, F.; Gu, J.; Fu, L. High Expression of Pyruvate Kinase M2 is Associated with Chemosensitivity to Epirubicin and 5-Fluorouracil in Breast Cancer. J. Cancer 2015, 6, 1130–1139. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.C.; Ku, J.-L.; Hong, S.-H.; Shin, Y.-K.; Park, S.Y.; Kim, H.K.; Park, J.-G. Decreased pyruvate kinase M2 activity linked to cisplatin resistance in human gastric carcinoma cell lines. Int. J. Cancer 2003, 108, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Balibrea, E.; Plasencia, C.; Gines, A.; Martinez-Cardus, A.; Musulen, E.; Aguilera, R.; Manzano, J.L.; Neamati, N.; Abad, A. A proteomic approach links decreased pyruvate kinase M2 expression to oxaliplatin resistance in patients with colorectal cancer and in human cell lines. Mol. Cancer 2009, 8, 771–778. [Google Scholar] [CrossRef]
- Gines, A.; Bystrup, S.; Ruiz de Porras, V.; Guardia, C.; Musulen, E.; Martinez-Cardus, A.; Manzano, J.L.; Layos, L.; Abad, A.; Martinez-Balibrea, E. PKM2 Subcellular Localization Is Involved in Oxaliplatin Resistance Acquisition in HT29 Human Colorectal Cancer Cell Lines. PLoS ONE 2015, 10, e0123830. [Google Scholar] [CrossRef]
- Zhu, H.; Wu, J.; Zhang, W.; Luo, H.; Shen, Z.; Cheng, H.; Zhu, X. PKM2 enhances chemosensitivity to cisplatin through interaction with the mTOR pathway in cervical cancer. Sci. Rep. 2016, 6, 30788. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, D.; Chen, X.; He, L.; Li, T.; Xu, X.; Li, M. Nuclear PKM2 contributes to gefitinib resistance via upregulation of STAT3 activation in colorectal cancer. Sci. Rep. 2015, 5, 16082. [Google Scholar] [CrossRef]
- Calabretta, S.; Bielli, P.; Passacantilli, I.; Pilozzi, E.; Fendrich, V.; Capurso, G.; Fave, G.D.; Sette, C. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene 2016, 35, 2031–2039. [Google Scholar] [CrossRef]
- Su, Q.; Luo, S.; Tan, Q.; Deng, J.; Zhou, S.; Peng, M.; Tao, T.; Yang, X. The role of pyruvate kinase M2 in anticancer therapeutic treatments (Review). Oncol. Lett. 2019, 18, 5663. [Google Scholar] [CrossRef]
- Taniguchi, K.; Sakai, M.; Sugito, N.; Kuranaga, Y.; Kumazaki, M.; Shinohara, H.; Ueda, H.; Futamura, M.; Yoshida, K.; Uchiyama, K.; et al. PKM1 is involved in resistance to anti-cancer drugs. Biochem. Biophys. Res Commun. 2016, 473, 174–180. [Google Scholar] [CrossRef]
- Okazaki, M.; Fushida, S.; Tsukada, T.; Kinoshita, J.; Oyama, K.; Miyashita, T.; Ninomiya, I.; Harada, S.; Ohta, T. The effect of HIF-1α and PKM1 expression on acquisition of chemoresistance. Cancer Manag. Res. 2018, 10, 1865–1874. [Google Scholar] [CrossRef] [PubMed]
- Bluemlein, K.; Grüning, N.-M.; Feichtinger, R.G.; Lehrach, H.; Kofler, B.; Ralser, M. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget 2011, 2, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.; Ding, M.; Wang, B.; Lu, Z.; Zhao, Q.; Shaw, K.; Yung, W.K.; Weinstein, J.N.; Tan, M.; Yao, J. Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers. Oncotarget 2014, 5, 8202–8210. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, C.; Kim, W.; Arif, M.; Gao, C.; Hober, A.; Kotol, D.; Strandberg, L.; Forsström, B.; Sivertsson, Å.; et al. Discovery of functional alternatively spliced PKM transcripts in human cancers. Cancers 2021, 13, 348. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, J.R.; Hao, J.; Wang, G.; Chen, K.H.; Babcock, H.P.; Zhuang, X. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl. Acad. Sci. USA 2016, 113, 11046–11051. [Google Scholar] [CrossRef]
- Urbanek, M.O.; Krzyzosiak, W.J. Discriminating RNA variants with single-molecule allele-specific FISH. Mutat. Res. 2017, 773, 230–241. [Google Scholar] [CrossRef]
- Shaffer, S.M.; Dunagin, M.C.; Torborg, S.R.; Torre, E.A.; Emert, B.; Krepler, C.; Beqiri, M.; Sproesser, K.; Brafford, P.A.; Xiao, M.; et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 2017, 546, 431–435. [Google Scholar] [CrossRef]
- Cortes-Cros, M.; Hemmerlin, C.; Ferretti, S.; Zhang, J.; Gounarides, J.S.; Yin, H.; Muller, A.; Haberkorn, A.; Chene, P.; Sellers, W.R.; et al. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proc. Natl. Acad. Sci. USA 2013, 110, 489–494. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, X.; Mo, L.; Liu, Y.; He, F.; Zhang, F.; Huang, K.H.; Wu, X.R. Role of isoenzyme M2 of pyruvate kinase in urothelial tumorigenesis. Oncotarget 2016, 7, 23947–23960. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, K.R.; Ahn, D.R.; Lee, J.E.; Yang, E.G.; Kim, S.Y. Reversible Regulation of Enzyme Activity by pH-Responsive Encapsulation in DNA Nanocages. ACS Nano 2017, 11, 9352–9359. [Google Scholar] [CrossRef]
- Mueller, F.; Senecal, A.; Tantale, K.; Marie-Nelly, H.; Ly, N.; Collin, O.; Basyuk, E.; Bertrand, E.; Darzacq, X.; Zimmer, C. FISH-quant: Automatic counting of transcripts in 3D FISH images. Nat. Methods 2013, 10, 277–278. [Google Scholar] [CrossRef] [PubMed]
- Westoby, J.; Artemov, P.; Hemberg, M.; Ferguson-Smith, A. Obstacles to detecting isoforms using full-length scRNA-seq data. Genome Biol. 2020, 21, 74. [Google Scholar] [CrossRef] [PubMed]
- Waks, Z.; Klein, A.M.; Silver, P.A. Cell-to-cell variability of alternative RNA splicing. Mol. Syst. Biol. 2011, 7, 506. [Google Scholar] [CrossRef]
- Sinnamon, J.R.; Czaplinski, K. RNA detection in situ with FISH-STICs. RNA 2014, 20, 260–266. [Google Scholar] [CrossRef]
- Feng, Y.-H.; Tsao, C.-J.; Wu, C.-L.; Chang, J.-G.; Lu, P.-J.; Yeh, K.-T.; Shieh, G.-S.; Shiau, A.-L.; Lee, J.-C. Sprouty2 protein enhances the response to gefitinib through epidermal growth factor receptor in colon cancer cells. Cancer Sci. 2010, 101, 2033–2038. [Google Scholar] [CrossRef]
- Morgillo, F.; Martinelli, E.; Troiani, T.; Orditura, M.; De Vita, F.; Ciardiello, F. Antitumor Activity of Sorafenib in Human Cancer Cell Lines with Acquired Resistance to EGFR and VEGFR Tyrosine Kinase Inhibitors. PLoS ONE 2011, 6, e28841. [Google Scholar] [CrossRef] [PubMed]
- Eigenbrodt, E.; Basenau, D.; Holthusen, S.; Mazurek, S.; Fischer, G. Quantification of tumor type M2 pyruvate kinase (Tu M2-PK) in human carcinomas. Anticancer Res. 1997, 17, 3153–3156. [Google Scholar] [PubMed]
- Morita, M.; Sato, T.; Nomura, M.; Sakamoto, Y.; Inoue, Y.; Tanaka, R.; Ito, S.; Kurosawa, K.; Yamaguchi, K.; Sugiura, Y.; et al. PKM1 Confers Metabolic Advantages and Promotes Cell-Autonomous Tumor Cell Growth. Cancer Cell 2018, 33, 355–367. [Google Scholar] [CrossRef]
- Israelsen, W.J.; Dayton, T.L.; Davidson, S.M.; Fiske, B.P.; Hosios, A.M.; Bellinger, G.; Li, J.; Yu, Y.; Sasaki, M.; Horner, J.W.; et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 2013, 155, 397–409. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Muralidhar, V.; Hosios, A.M.; Israelsen, W.J.; Gui, D.Y.; Newhouse, L.; Ogrodzinski, M.; Hecht, V.; Xu, K.; Acevedo, P.N.; et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol. Cell 2015, 57, 95–107. [Google Scholar] [CrossRef]
- Dayton, T.L.; Gocheva, V.; Miller, K.M.; Israelsen, W.J.; Bhutkar, A.; Clish, C.B.; Davidson, S.M.; Luengo, A.; Bronson, R.T.; Jacks, T.; et al. Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev. 2016, 30, 1020–1033. [Google Scholar] [CrossRef] [PubMed]
- David, C.J.; Chen, M.; Assanah, M.; Canoll, P.; Manley, J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010, 463, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Clower, C.V.; Chatterjee, D.; Wang, Z.; Cantley, L.C.; Vander Heiden, M.G.; Krainer, A.R. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 1894–1899. [Google Scholar] [CrossRef]
- Prakasam, G.; Iqbal, M.A.; Bamezai, R.N.K.; Mazurek, S. Posttranslational Modifications of Pyruvate Kinase M2: Tweaks that Benefit Cancer. Front. Oncol. 2018, 8, 22. [Google Scholar] [CrossRef]
- Ciardiello, F.; Tortora, G. A novel approach in the treatment of cancer: Targeting the epidermal growth factor receptor. Clin. Cancer Res. 2001, 7, 2958–2970. [Google Scholar] [PubMed]
- Cohen, R.B. Epidermal Growth Factor Receptor as a Therapeutic Target in Colorectal Cancer. Clin. Colorectal Cancer 2003, 2, 246–251. [Google Scholar] [CrossRef]
- De Roock, W.; Jonker, D.J.; Di Nicolantonio, F.; Sartore-Bianchi, A.; Tu, D.; Siena, S.; Lamba, S.; Arena, S.; Frattini, M.; Piessevaux, H.; et al. Association of KRAS p.G13D Mutation With Outcome in Patients With Chemotherapy-Refractory Metastatic Colorectal Cancer Treated With Cetuximab. JAMA 2010, 304, 1812. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Xia, Y.; Cao, Y.; Zheng, Y.; Bu, W.; Zhang, L.; You, M.J.; Koh, M.Y.; Cote, G.; Aldape, K.; et al. EGFR-induced and PKCepsilon monoubiquitylation-dependent NF-kappaB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell 2012, 48, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Xia, Y.; Hawke, D.; Li, X.; Liang, J.; Xing, D.; Aldape, K.; Hunter, T.; Alfred Yung, W.K.; Lu, Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 2012, 150, 685–696. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, H.-X.; Zhou, S.-Y.; Wang, S.-X.; Zheng, K.; Xu, D.-D.; Liu, Y.-T.; Wang, X.-Y.; Wang, X.; Yan, H.-Z.; et al. Sp1 and c-Myc modulate drug resistance of leukemia stem cells by regulating survivin expression through the ERK-MSK MAPK signaling pathway. Mol. Cancer 2015, 14, 56. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, N.; Lai, W.; Yan, B.; Chen, L.; Liu, S.; Liu, S.; Wang, X.; Xiao, D.; Liu, X.; et al. Nuclear EGFR-PKM2 axis induces cancer stem cell-like characteristics in irradiation-resistant cells. Cancer Lett. 2018, 422, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zheng, Y.; Xia, Y.; Ji, H.; Chen, X.; Guo, F.; Lyssiotis, C.A.; Aldape, K.; Cantley, L.C.; Lu, Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 2012, 14, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Ru, P.; Williams, T.; Chakravarti, A.; Guo, D. Tumor Metabolism of Malignant Gliomas. Cancers 2013, 5, 1469–1484. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Park, Y.S.; Kang, M.G.; You, Y.M.; Jung, Y.; Koo, H.; Kim, J.A.; Kim, M.J.; Hong, S.M.; Lee, K.B.; et al. Pyruvate kinase isoenzyme M2 is a therapeutic target of gemcitabine-resistant pancreatic cancer cells. Exp. Cell Res. 2015, 336, 119–129. [Google Scholar] [CrossRef]
- Goldberg, M.S.; Sharp, P.A. Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J. Exp. Med. 2012, 209, 217–224. [Google Scholar] [CrossRef]
- Ma, C.; Zu, X.; Liu, K.; Bode, A.M.; Dong, Z.; Liu, Z.; Kim, D.J. Knockdown of Pyruvate Kinase M Inhibits Cell Growth and Migration by Reducing NF-kB Activity in Triple-Negative Breast Cancer Cells. Mol. Cells 2019, 42, 628–636. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, D.; Jin, F.; Bian, Z.; Li, L.; Liang, H.; Li, M.; Shi, L.; Pan, C.; Zhu, D.; et al. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat. Commun. 2017, 8, 14041. [Google Scholar] [CrossRef]
- Dierolf, J.G.; Watson, A.J.; Betts, D.H. Differential localization patterns of pyruvate kinase isoforms in murine naïve, formative and primed pluripotent states. Exp. Cell Res. 2021, 405, 112714. [Google Scholar] [CrossRef]
- Chaneton, B.; Hillmann, P.; Zheng, L.; Martin, A.C.L.; Maddocks, O.D.K.; Chokkathukalam, A.; Coyle, J.E.; Jankevics, A.; Holding, F.P.; Vousden, K.H.; et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 2012, 491, 458–462. [Google Scholar] [CrossRef]
- Zhao, W.-F.; Wang, H.-B.; Xie, B.; Hu, L.-J.; Xu, L.-H.; Kuang, B.-H.; Li, M.-Z.; Zhang, X. Sp1 and Sp3 are involved in the full transcriptional activity of centromere protein H in human nasopharyngeal carcinoma cells. FEBS J. 2012, 279, 2714–2726. [Google Scholar] [CrossRef]
- Loh, T.; Moon, H.; Cho, S.; Jang, H.; Liu, Y.; Tai, H.; Jung, D.-W.; Williams, D.; Kim, H.-R.; Shin, M.-G.; et al. CD44 alternative splicing and hnRNP A1 expression are associated with the metastasis of breast cancer. Oncol. Rep. 2015, 34, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Koo, O.J.; Park, H.J.; Moon, J.H.; Da Torre, B.R.; Javaregowda, P.K.; Kang, J.T.; Park, S.J.; Saadeldin, I.M.; Choi, J.Y.; et al. Oct4 overexpression facilitates proliferation of porcine fibroblasts and development of cloned embryos. Zygote 2015, 23, 704–711. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, H.; Woo, D.J.; Kim, S.Y.; Yang, E.G. p21-activated kinase 4 regulates HIF-1alpha translation in cancer cells. Biochem. Biophys. Res. Commun. 2017, 486, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-W.; Ryu, H.-C.; Ng, Y.C.; Kim, C.; Wei, J.-D.; Sabaratnam, V.; Kim, J.-H. 12(S)-Hydroxyheptadeca-5Z,8E,10E-trienoic acid suppresses UV-induced IL-6 synthesis in keratinocytes, exerting an anti-inflammatory activity. Exp. Mol. Med. 2012, 44, 378. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.H.; Wi, J.H.; Gwak, H.; Yang, E.G.; Kim, S.Y. Single-Cell FISH Analysis Reveals Distinct Shifts in PKM Isoform Populations during Drug Resistance Acquisition. Biomolecules 2022, 12, 1082. https://doi.org/10.3390/biom12081082
Kim SH, Wi JH, Gwak H, Yang EG, Kim SY. Single-Cell FISH Analysis Reveals Distinct Shifts in PKM Isoform Populations during Drug Resistance Acquisition. Biomolecules. 2022; 12(8):1082. https://doi.org/10.3390/biom12081082
Chicago/Turabian StyleKim, Seong Ho, Ji Hun Wi, HyeRan Gwak, Eun Gyeong Yang, and So Yeon Kim. 2022. "Single-Cell FISH Analysis Reveals Distinct Shifts in PKM Isoform Populations during Drug Resistance Acquisition" Biomolecules 12, no. 8: 1082. https://doi.org/10.3390/biom12081082
APA StyleKim, S. H., Wi, J. H., Gwak, H., Yang, E. G., & Kim, S. Y. (2022). Single-Cell FISH Analysis Reveals Distinct Shifts in PKM Isoform Populations during Drug Resistance Acquisition. Biomolecules, 12(8), 1082. https://doi.org/10.3390/biom12081082





