The HPV16E7 Affibody as a Novel Potential Therapeutic Agent for Treating Cervical Cancer Is Likely Internalized through Dynamin and Caveolin-1 Dependent Endocytosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Preparation of ZHPV16E7384
2.3. Preparation of FITC-Labelled ZHPV16E7384
2.4. Indirect Immunofluorescence Assay
2.5. Free ZHPV16E7384 Blocking Assay
2.6. Analysis of ZHPV16E7384 Amino Acid Sequence
2.7. Analysis of the Effect of Fetal Bovine Serum on ZHPV16E7384 Internalization
2.8. HPV16E7 Antibody Blocking Assay
2.9. Western Blot
2.10. Endocytosis Inhibition Tests
2.11. Cell Viability Assay
2.12. Colony Formation Assay
2.13. EdU Proliferation Assay
2.14. Flow Cytometry
2.15. Statistical Analysis
3. Results
3.1. ZHPV16E7384 Was Successfully Prepared
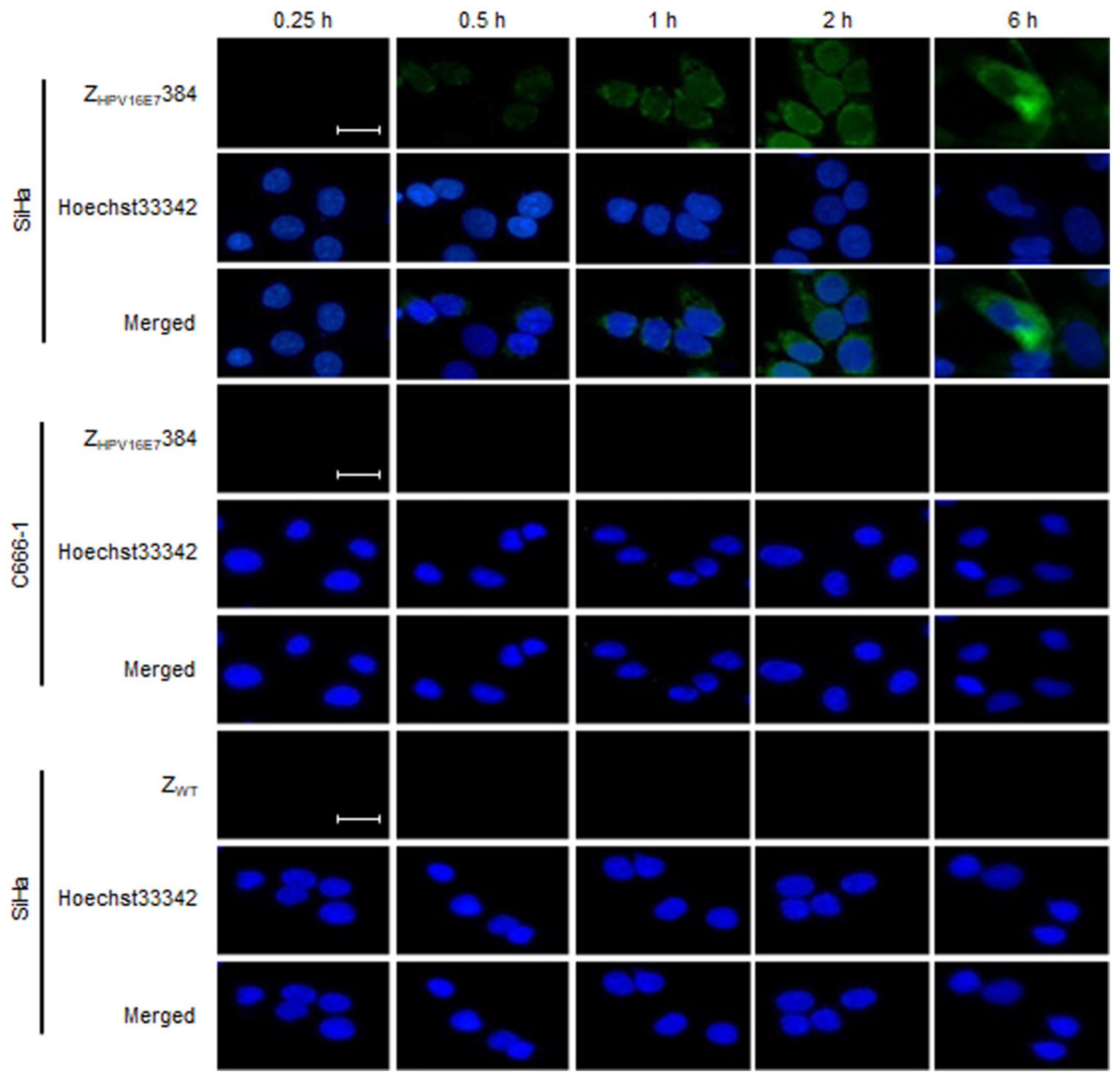
3.2. ZHPV16E7384 Specifically Bound to HPV16 Positive Cervical Cancer Cells
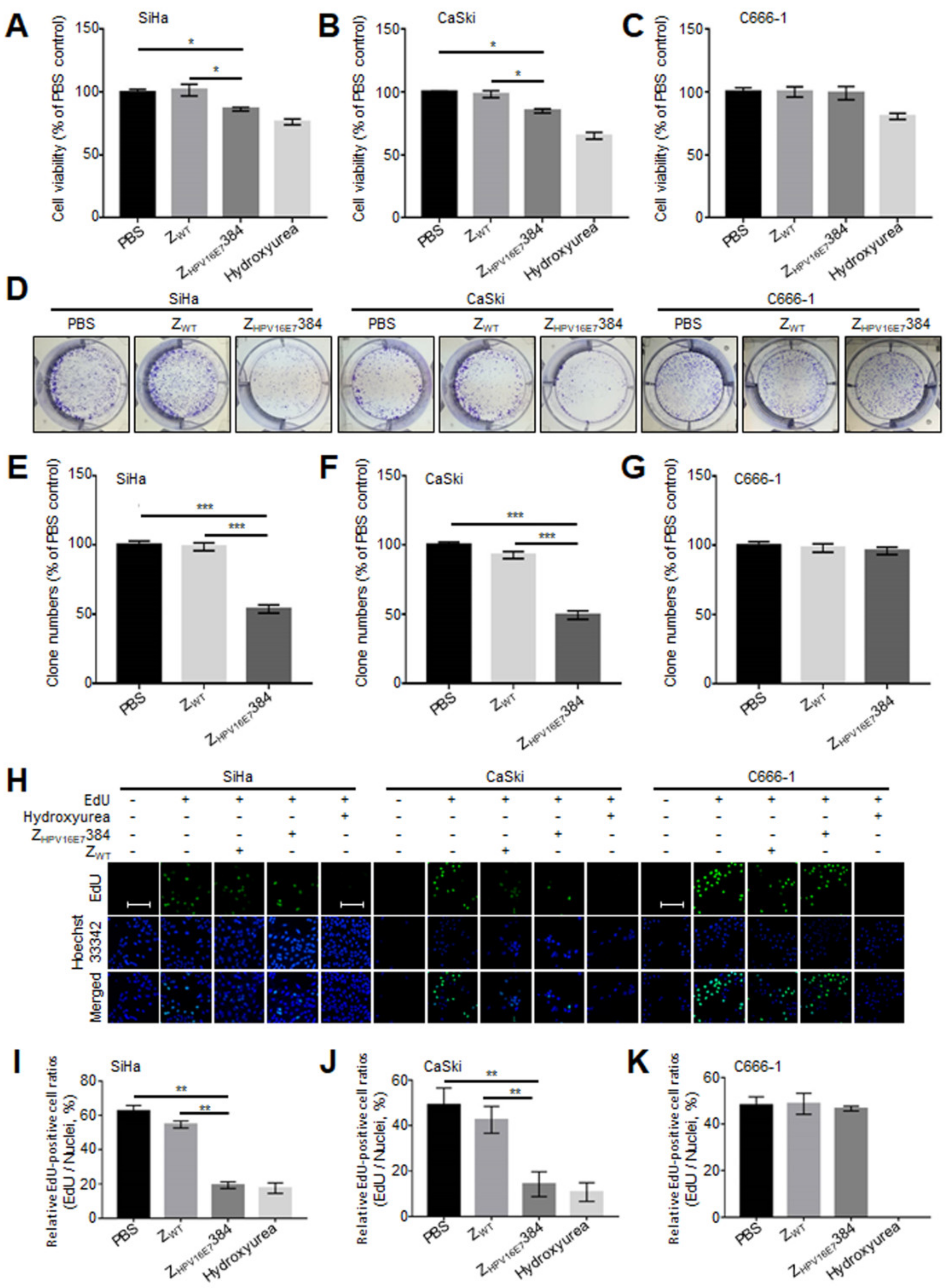
3.3. ZHPV16E7384 Significantly Suppressed the Proliferation of HPV16 Positive Cervical Cancer Cells
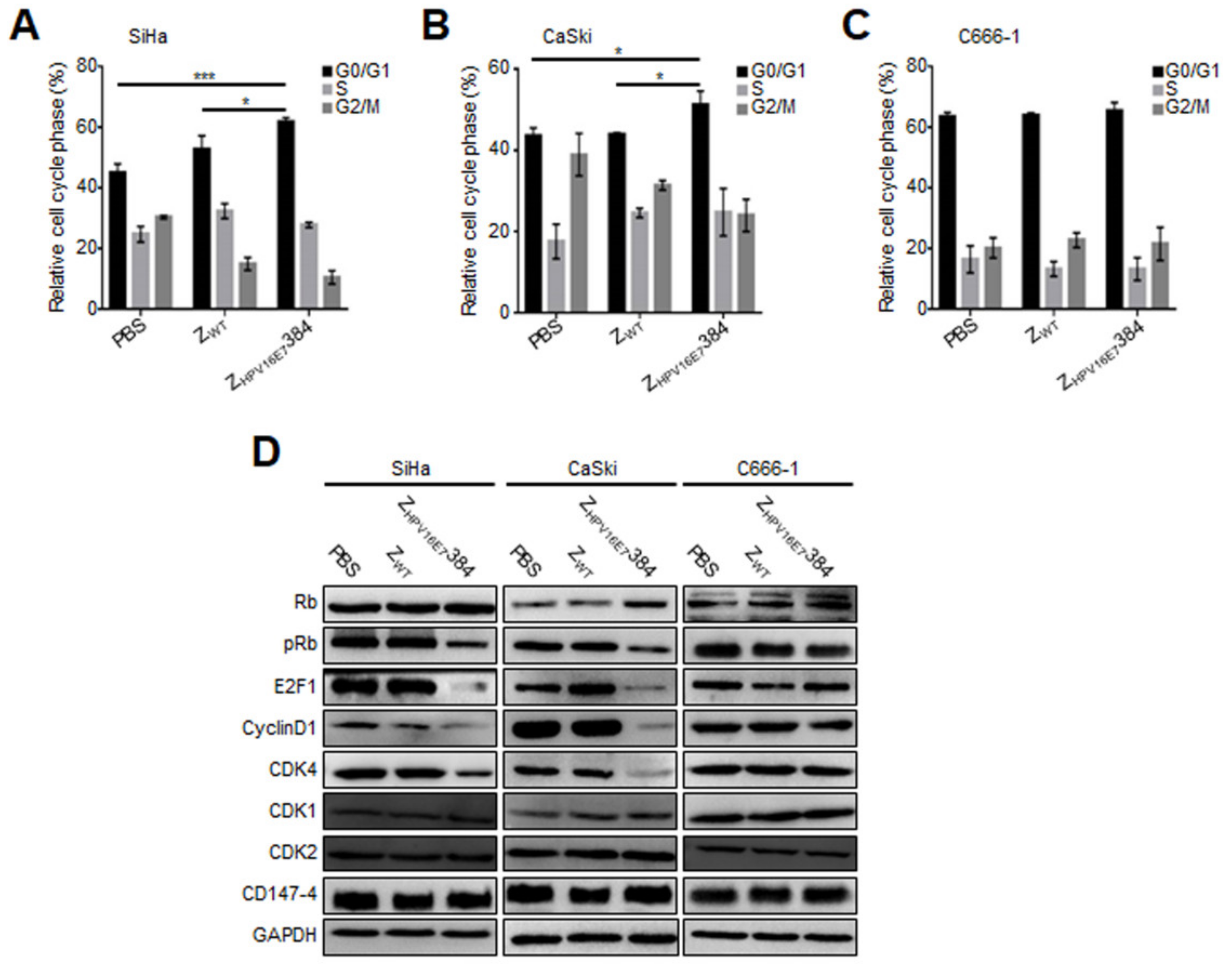
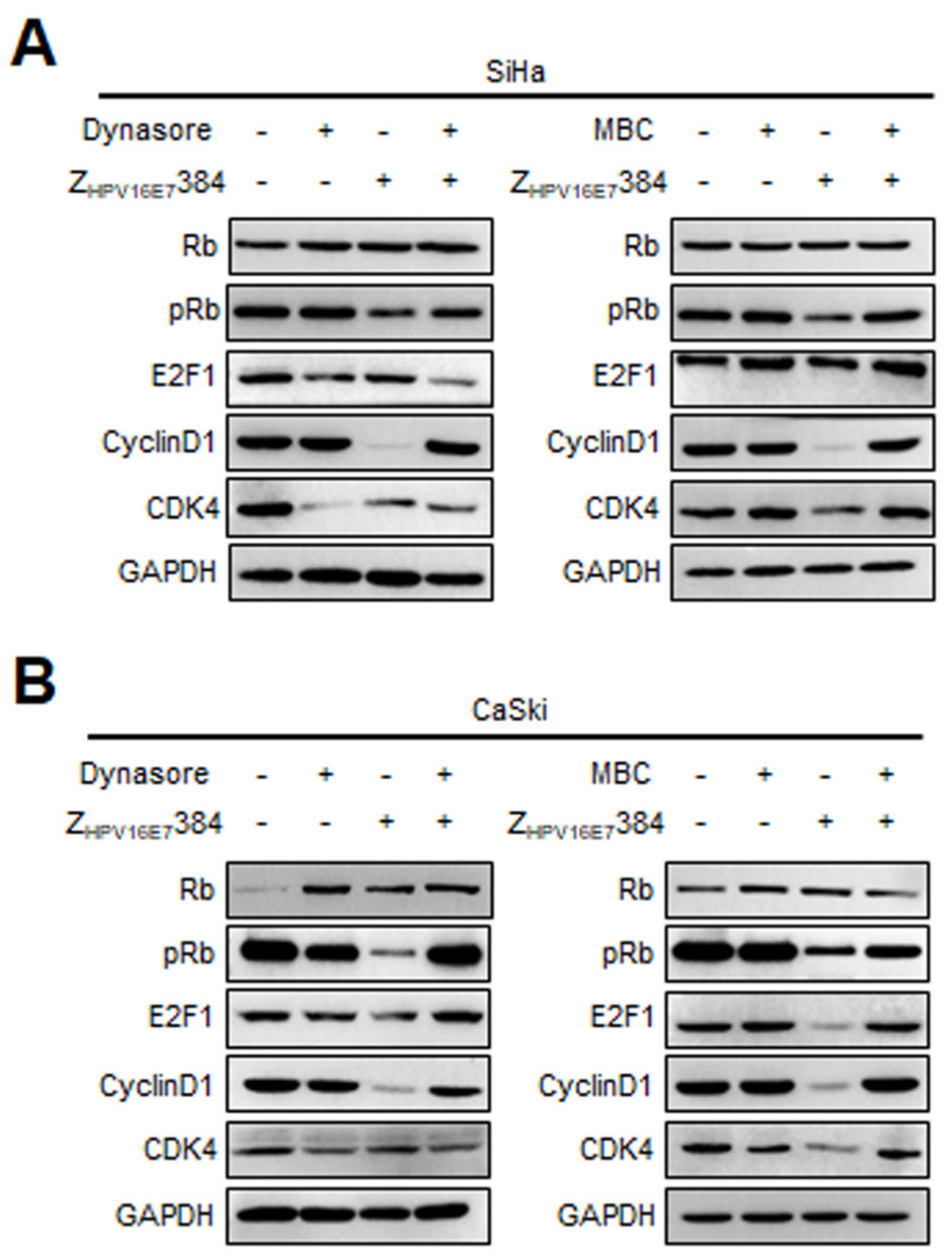
3.4. ZHPV16E7384 Induced G1/S Cell Cycle Arrest through Inhibition of the E7/Rb/E2F1/Cyclin D1/CDK4 Pathway
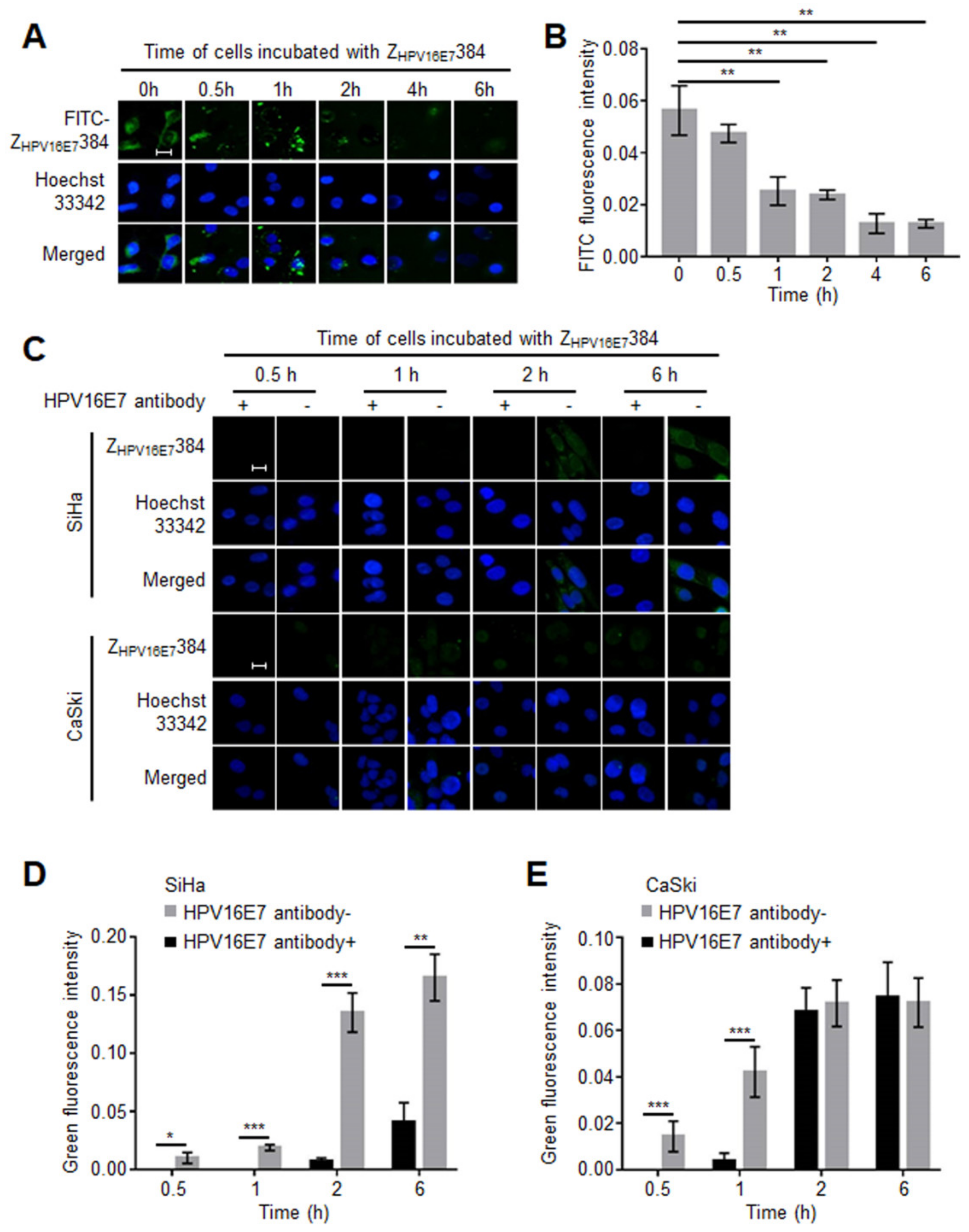
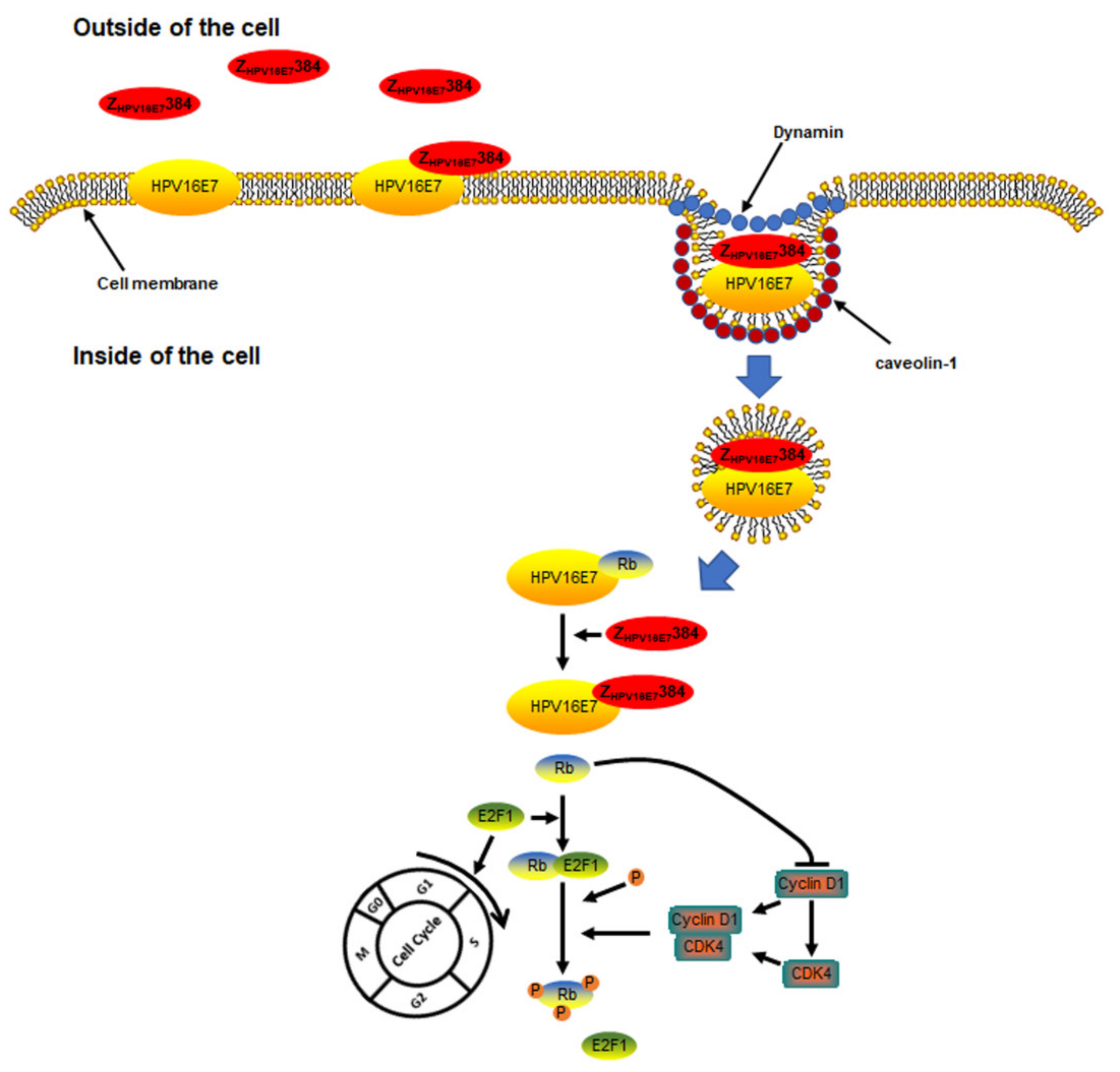
3.5. The HPV16E7 Protein Played an Important Role during the ZHPV16E7384 Internalization
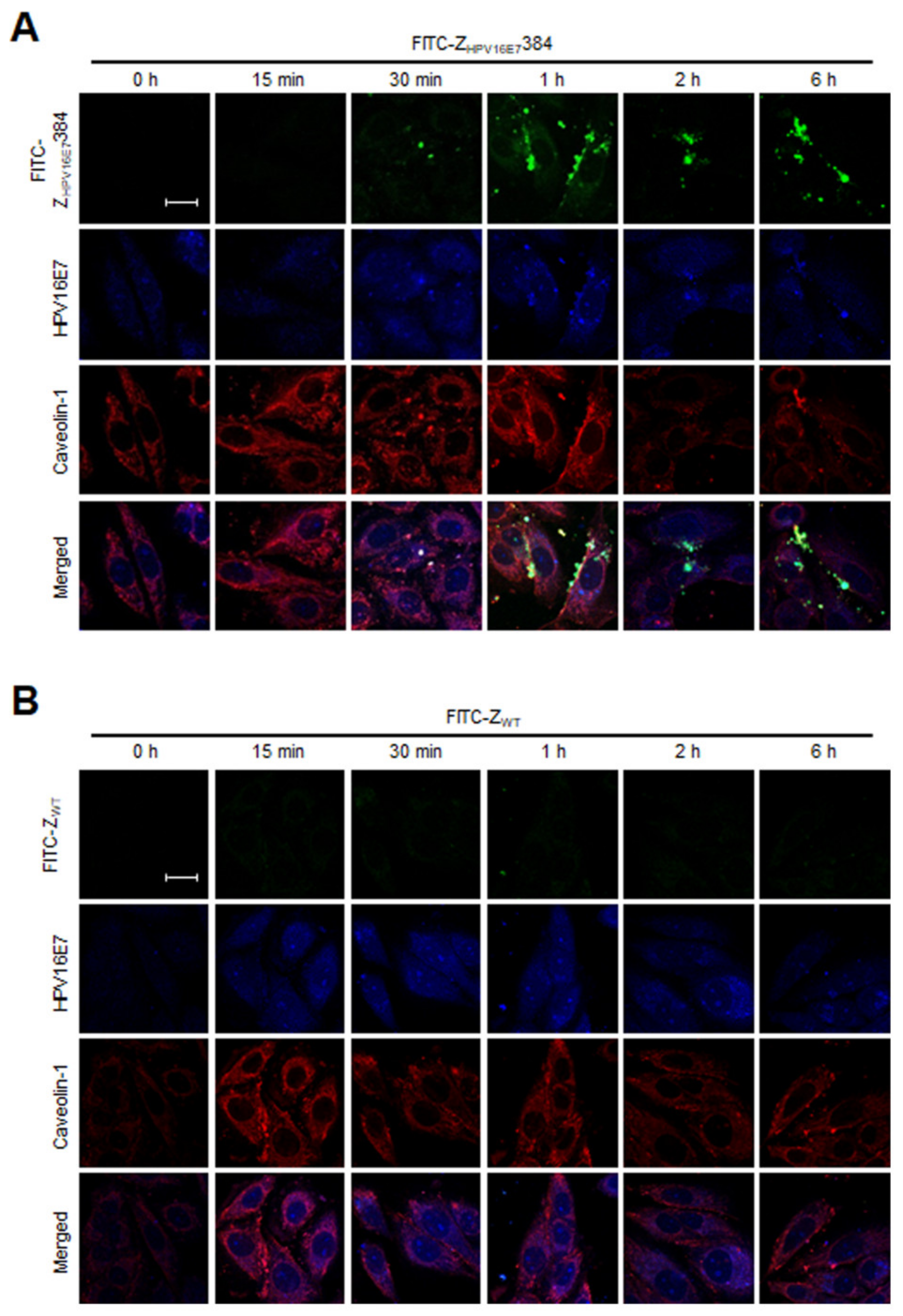
3.6. Dynamin and Caveolin-1 Were Indispensable for the ZHPV16E7384 Internalization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nord, K.; Nilsson, J.; Nilsson, B.; Uhlen, M.; Nygren, P.A. A combinatorial library of an alpha-helical bacterial receptor domain. Protein Eng. 1995, 8, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Nygren, P.A. Alternative binding proteins: Affibody binding proteins developed from a small three-helix bundle scaffold. FEBS J. 2008, 275, 2668–2676. [Google Scholar] [CrossRef] [PubMed]
- De, A.; Kuppusamy, G.; Karri, V. Affibody molecules for molecular imaging and targeted drug delivery in the management of breast cancer. Int. J. Biol. Macromol. 2018, 107, 906–919. [Google Scholar] [CrossRef] [PubMed]
- Ståhl, S.; Gräslund, T.; Eriksson Karlström, A.; Frejd, F.Y.; Nygren, P.; Löfblom, J. Affibody Molecules in Biotechnological and Medical Applications. Trends Biotechnol. 2017, 35, 691–712. [Google Scholar] [CrossRef]
- Akbari, V.; Chou, C.P.; Abedi, D. New insights into affinity proteins for HER2-targeted therapy: Beyond trastuzumab. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188448. [Google Scholar] [CrossRef]
- Xue, X.; Wang, B.; Du, W.; Zhang, C.; Song, Y.; Cai, Y.; Cen, D.; Wang, L.; Xiong, Y.; Jiang, P.; et al. Generation of affibody molecules specific for HPV16 E7 recognition. Oncotarget 2016, 7, 73995–74005. [Google Scholar] [CrossRef]
- Jiang, P.; Wang, L.; Hou, B.; Zhu, J.; Zhou, M.; Jiang, J.; Wang, L.; Chen, S.; Zhu, S.; Chen, J.; et al. A novel HPV16 E7-affitoxin for targeted therapy of HPV16-induced human cervical cancer. Theranostics 2018, 8, 3544–3558. [Google Scholar] [CrossRef]
- Zhu, J.; Kamara, S.; Cen, D.; Tang, W.; Gu, M.; Ci, X.; Chen, J.; Wang, L.; Zhu, S.; Jiang, P.; et al. Generation of novel affibody molecules targeting the EBV LMP2A N-terminal domain with inhibiting effects on the proliferation of nasopharyngeal carcinoma cells. Cell Death Dis. 2020, 11, 213. [Google Scholar] [CrossRef]
- Kamara, S.; Guo, Y.; Mao, S.; Ye, X.; Li, Q.; Zheng, M.; Zhu, J.; Zhang, J.; Du, W.; Chen, J.; et al. Novel EBV LMP1 C-terminal domain binding affibody molecules as potential agents for in vivo molecular imaging diagnosis of nasopharyngeal carcinoma. Appl. Microbiol. Biotechnol. 2021, 105, 7283–7293. [Google Scholar] [CrossRef]
- Li, M.; Shi, W.; Yang, J.; Wang, Q.; Dong, H.; Chen, J.; Zhang, L.; Zhu, S. Generation of a novel affibody molecule targeting Chlamydia trachomatis MOMP. Appl. Microbiol. Biotechnol. 2021, 105, 1477–1487. [Google Scholar] [CrossRef]
- Zhu, J.; Kamara, S.; Wang, Q.; Guo, Y.; Li, Q.; Wang, L.; Chen, J.; Du, Q.; Du, W.; Chen, S.; et al. Novel Affibody Molecules Targeting the HPV16 E6 Oncoprotein Inhibited the Proliferation of Cervical Cancer Cells. Front. Cell Dev. Biol. 2021, 9, 677867. [Google Scholar] [CrossRef]
- Crosbie, E.J.; Einstein, M.H.; Franceschi, S.; Kitchener, H.C. Human papillomavirus and cervical cancer. Lancet 2013, 382, 889–899. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Durst, M.; Gissmann, L.; Ikenberg, H.; zur Hausen, H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl. Acad. Sci. USA 1983, 80, 3812–3815. [Google Scholar] [CrossRef]
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef]
- Garbuglia, A.R.; Lapa, D.; Sias, C.; Capobianchi, M.R.; Del Porto, P. The Use of Both Therapeutic and Prophylactic Vaccines in the Therapy of Papillomavirus Disease. Front. Immunol. 2020, 11, 188. [Google Scholar] [CrossRef]
- Cheung, S.T.; Huang, D.P.; Hui, A.B.; Lo, K.W.; Ko, C.W.; Tsang, Y.S.; Wong, N.; Whitney, B.M.; Lee, J.C. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer 1999, 83, 121–126. [Google Scholar] [CrossRef]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef]
- Cesur, Ö.; Nicol, C.; Groves, H.; Mankouri, J.; Blair, G.E.; Stonehouse, N.J. The Subcellular Localisation of the Human Papillomavirus (HPV) 16 E7 Protein in Cervical Cancer Cells and Its Perturbation by RNA Aptamers. Viruses 2015, 7, 3443–3461. [Google Scholar] [CrossRef]
- Lofblom, J.; Feldwisch, J.; Tolmachev, V.; Carlsson, J.; Stahl, S.; Frejd, F.Y. Affibody molecules: Engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 2010, 584, 2670–2680. [Google Scholar] [CrossRef]
- Nord, K.; Gunneriusson, E.; Ringdahl, J.; Stahl, S.; Uhlen, M.; Nygren, P.A. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat. Biotechnol. 1997, 15, 772–777. [Google Scholar] [CrossRef]
- Ostrowska, K.M.; Garcia, A.; Meade, A.D.; Malkin, A.; Okewumi, I.; O’Leary, J.J.; Martin, C.; Byrne, H.J.; Lyng, F.M. Correlation of p16(INK4A) expression and HPV copy number with cellular FTIR spectroscopic signatures of cervical cancer cells. Analyst 2011, 136, 1365–1373. [Google Scholar] [CrossRef]
- Lin, Z.; Zhao, Y.; Li, Q.; Ci, X.; Ye, X.; Chen, G.; Tu, Q.; Feng, W.; Jiang, P.; Zhu, S.; et al. Sustained Expression of HPV16 E7 Oncoprotein Promotes p-AKT (Ser473)/p-Src (Tyr527) Signalling to Drive Precancerous Lesions to Invasive Cervical Cancer. Carcinogenesis 2022, 4, 479–493. [Google Scholar] [CrossRef]
- Nigg, E.A. Cyclin-dependent protein kinases: Key regulators of the eukaryotic cell cycle. Bioessays 1995, 17, 471–480. [Google Scholar] [CrossRef]
- Araki, K.; Nakajima, Y.; Eto, K.; Ikeda, M.A. Distinct recruitment of E2F family members to specific E2F-binding sites mediates activation and repression of the E2F1 promoter. Oncogene 2003, 22, 7632–7641. [Google Scholar] [CrossRef]
- Sherr, C.J.; Roberts, J.M. Living with or without cyclins and cyclin-dependent kinases. Genes. Dev. 2004, 18, 2699–2711. [Google Scholar] [CrossRef]
- Guo, C.P.; Liu, K.W.; Luo, H.B.; Chen, H.B.; Zheng, Y.; Sun, S.N.; Zhang, Q.; Huang, L. Potent anti-tumor effect generated by a novel human papillomavirus (HPV) antagonist peptide reactivating the pRb/E2F pathway. PLoS ONE 2011, 6, e17734. [Google Scholar] [CrossRef]
- Ohtsubo, M.; Roberts, J.M. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science 1993, 259, 1908–1912. [Google Scholar] [CrossRef]
- Brown, J.R.; Nigh, E.; Lee, R.J.; Ye, H.; Thompson, M.A.; Saudou, F.; Pestell, R.G.; Greenberg, M.E. Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol. 1998, 18, 5609–5619. [Google Scholar] [CrossRef] [PubMed]
- Hitomi, M.; Stacey, D.W. Cyclin D1 production in cycling cells depends on ras in a cell-cycle-specific manner. Curr. Biol. 1999, 9, 1075–1084. [Google Scholar] [CrossRef]
- Dick, F.A.; Rubin, S.M. Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 2013, 14, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.R.; Liban, T.J.; Restrepo, T.; Lee, H.W.; Rubin, S.M. Multiple mechanisms for E2F binding inhibition by phosphorylation of the retinoblastoma protein C-terminal domain. J. Mol. Biol. 2014, 426, 245–255. [Google Scholar] [CrossRef]
- Narasimha, A.M.; Kaulich, M.; Shapiro, G.S.; Choi, Y.J.; Sicinski, P.; Dowdy, S.F. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. eLife 2014, 3, e02872. [Google Scholar] [CrossRef]
- Rashid, N.N.; Rothan, H.A.; Yusoff, M.S. The association of mammalian DREAM complex and HPV16 E7 proteins. Am. J. Cancer Res. 2015, 5, 3525–3533. [Google Scholar]
- Wu, Y.; Zhou, X.; Zheng, P.S. Involvement of CD147 isoform-4 in the proliferation of SiHa cells: A possible molecular mechanism of cervical cancer. Oncol. Rep. 2011, 26, 717–724. [Google Scholar]
- Asghar, U.; Witkiewicz, A.K.; Turner, N.C.; Knudsen, E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 2015, 14, 130–146. [Google Scholar] [CrossRef]
- Yuan, K.; Wang, X.; Dong, H.; Min, W.; Hao, H.; Yang, P. Selective inhibition of CDK4/6: A safe and effective strategy for developing anticancer drugs. Acta Pharm. Sin. B 2021, 11, 30–54. [Google Scholar] [CrossRef]
- Garces, S.; Medeiros, L.J.; Marques-Piubelli, M.L.; Coelho Siqueira, S.A.; Miranda, R.N.; Cuglievan, B.; Sriganeshan, V.; Medina, A.M.; Garces, J.C.; Saluja, K. Cyclin D1 expression in Rosai-Dorfman disease: A near-constant finding that is not invariably associated with mitogen-activated protein kinase/extracellular signal-regulated kinase pathway activation. Hum. Pathol. 2022, 121, 36–45. [Google Scholar] [CrossRef]
- Knapp, A.A.; McManus, P.M.; Bockstall, K.; Moroianu, J. Identification of the nuclear localization and export signals of high risk HPV16 E7 oncoprotein. Virology 2009, 383, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Dreier, K.; Scheiden, R.; Lener, B.; Ehehalt, D.; Pircher, H.; Müller-Holzner, E.; Rostek, U.; Kaiser, A.; Fiedler, M.; Ressler, S.; et al. Subcellular localization of the human papillomavirus 16 E7 oncoprotein in CaSki cells and its detection in cervical adenocarcinoma and adenocarcinoma in situ. Virology 2011, 409, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Doherty, G.J.; McMahon, H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. [Google Scholar] [CrossRef]
- Lajoie, P.; Kojic, L.D.; Nim, S.; Li, L.; Dennis, J.W.; Nabi, I.R. Caveolin-1 regulation of dynamin-dependent, raft-mediated endocytosis of cholera toxin-B sub-unit occurs independently of caveolae. J. Cell Mol. Med. 2009, 13, 3218–3225. [Google Scholar] [CrossRef] [PubMed]
- Damm, E.M.; Pelkmans, L.; Kartenbeck, J.; Mezzacasa, A.; Kurzchalia, T.; Helenius, A. Clathrin-and caveolin-1-independent endocytosis: Entry of simian virus 40 into cells devoid of caveolae. J. Cell Biol. 2005, 168, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Macia, E.; Ehrlich, M.; Massol, R.; Boucrot, E.; Brunner, C.; Kirchhausen, T. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 2006, 10, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Rothberg, K.G.; Ying, Y.S.; Kamen, B.A.; Anderson, R.G. Cholesterol controls the clustering of the glycophospholipid-anchored membrane receptor for 5-methyltetrahydrofolate. J. Cell Biol. 1990, 111, 2931–2938. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tan, S.H.; Ng, S.; Zhou, J.; Yang, N.D.; Koo, G.B.; McMahon, K.-A.; Parton, R.G.; Hill, M.M.; Pozo, M.A.D.; et al. Critical role of CAV1/caveolin-1 in cell stress responses in human breast cancer cells via modulation of lysosomal function and autophagy. Autophagy 2015, 11, 769–784. [Google Scholar] [CrossRef]
- Vercauteren, D.; Vandenbroucke, R.E.; Jones, A.T.; Rejman, J.; Demeester, J.; De Smedt, S.C.; Sanders, N.N.; Braeckmans, K. The use of inhibitors to study endocytic pathways of gene carriers: Optimization and pitfalls. Mol. Ther. 2010, 18, 561–569. [Google Scholar] [CrossRef]
- Dutta, D.; Donaldson, J.G. Search for inhibitors of endocytosis: Intended specificity and unintended consequences. Cell Logist. 2012, 2, 203–208. [Google Scholar] [CrossRef]
- Rennick, J.J.; Johnston, A.P.R.; Parton, R.G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol. 2021, 16, 266–276. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Zhu, H.; Cui, Z.; Li, Y.; Zhuo, J.; Ye, J.; Zhang, Z.; Lian, Z.; Du, Q.; Zhao, K.-N.; et al. The HPV16E7 Affibody as a Novel Potential Therapeutic Agent for Treating Cervical Cancer Is Likely Internalized through Dynamin and Caveolin-1 Dependent Endocytosis. Biomolecules 2022, 12, 1114. https://doi.org/10.3390/biom12081114
Zhang Q, Zhu H, Cui Z, Li Y, Zhuo J, Ye J, Zhang Z, Lian Z, Du Q, Zhao K-N, et al. The HPV16E7 Affibody as a Novel Potential Therapeutic Agent for Treating Cervical Cancer Is Likely Internalized through Dynamin and Caveolin-1 Dependent Endocytosis. Biomolecules. 2022; 12(8):1114. https://doi.org/10.3390/biom12081114
Chicago/Turabian StyleZhang, Qingyuan, Hua Zhu, Zhouying Cui, Yuxiao Li, Jiaying Zhuo, Jingwei Ye, Zhihui Zhang, Zheng Lian, Qianqian Du, Kong-Nan Zhao, and et al. 2022. "The HPV16E7 Affibody as a Novel Potential Therapeutic Agent for Treating Cervical Cancer Is Likely Internalized through Dynamin and Caveolin-1 Dependent Endocytosis" Biomolecules 12, no. 8: 1114. https://doi.org/10.3390/biom12081114





