Isolation, Purification, and Characterisation of a Phage Tail-Like Bacteriocin from the Insect Pathogenic Bacterium Brevibacillus laterosporus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
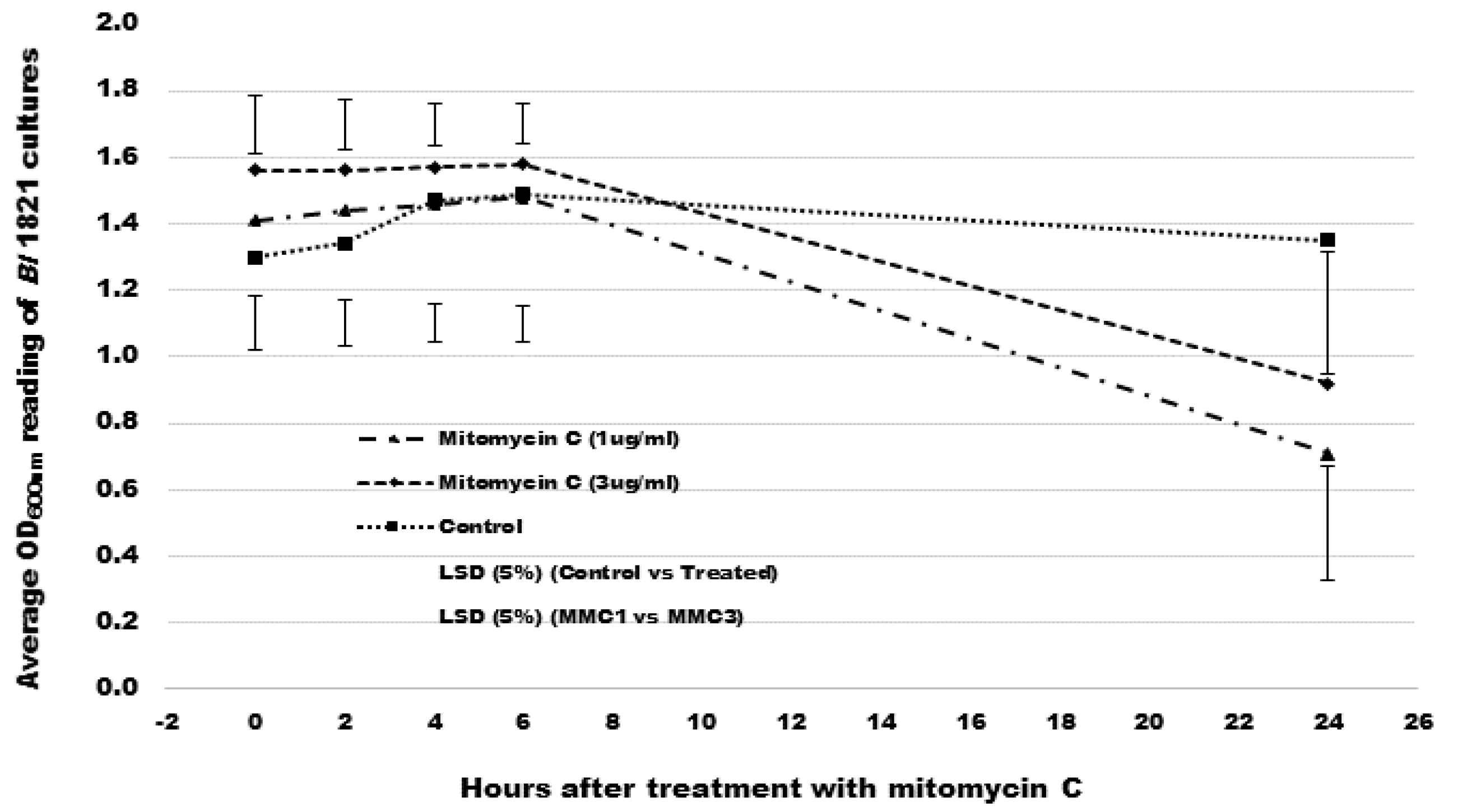
2.2. Mitomycin C Induction of Putative Antibacterial Protein
2.3. Soft-Agar Overlay Method with Polyethylene Glycol (PEG) Precipitation
2.4. Antimicrobial Spectrum of PEG 8000 Precipitated Culture Filtrates
2.5. Purification of Putative Antibacterial Protein
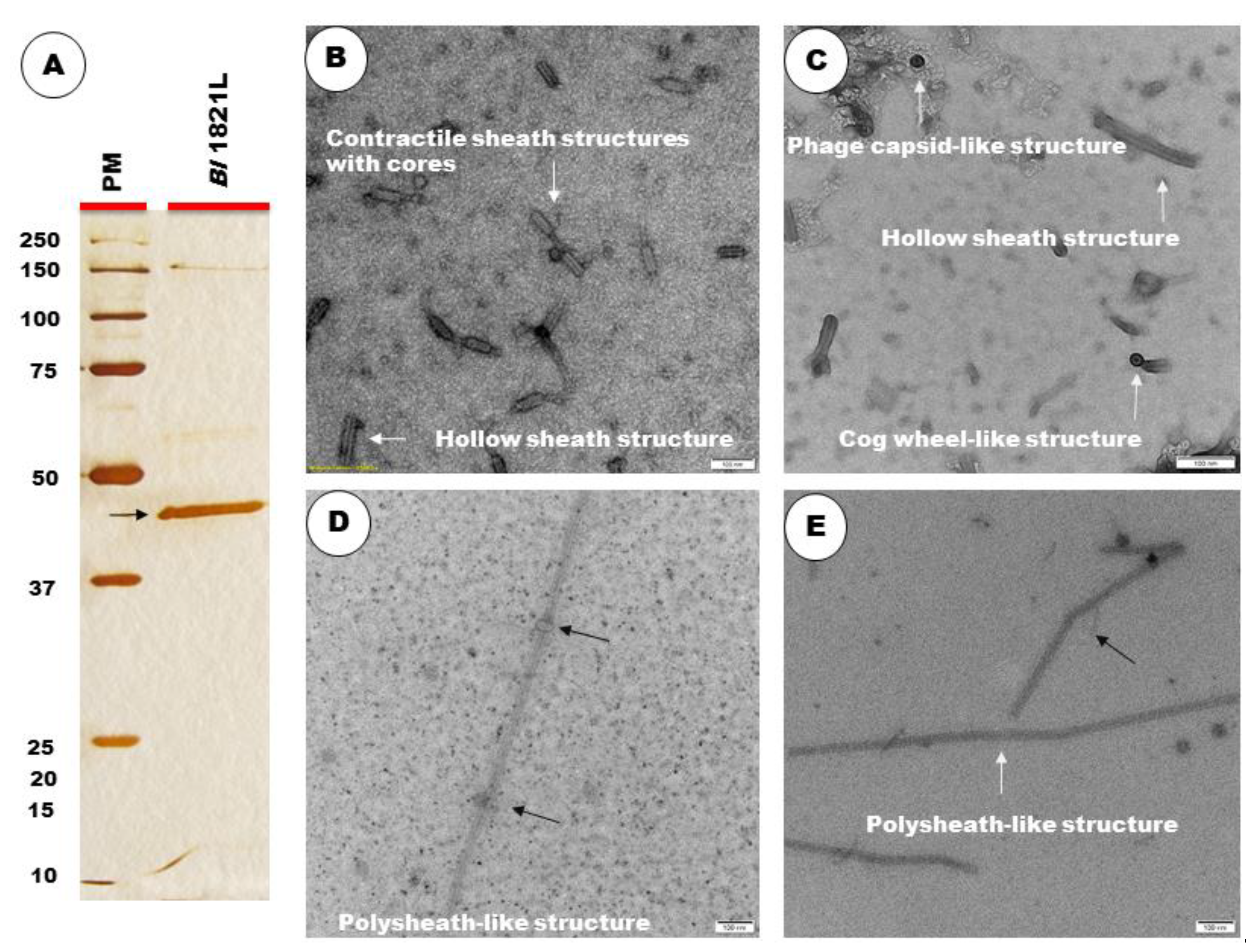
2.6. TEM Examination of Crude and Purified Lysates
2.7. SDS-PAGE Analysis
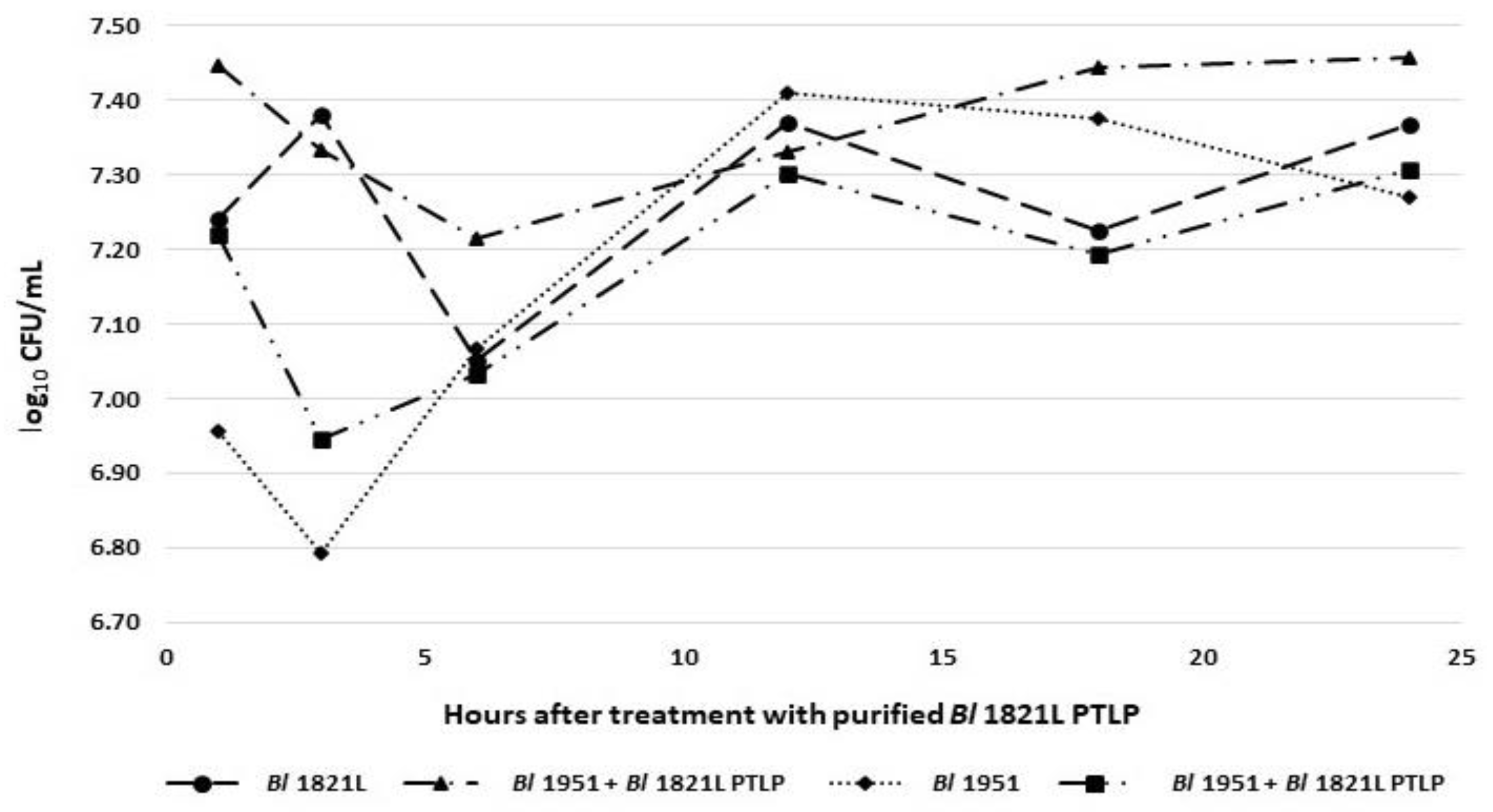
2.8. Bactericidal Activity Assay
2.9. N-Terminal Sequencing and Bioinformatic Analysis
3. Results
3.1. Mitomycin C Induction of Putative Antibacterial Proteins
3.2. Antimicrobial Activity of PEG 8000 Precipitated Culture Filtrates
3.3. TEM and SDS-PAGE Analysis
3.4. Identification of a ~48 kD Putative Antibacterial Protein in Bl 1821L Genome
3.5. Bactericidal Activity of Identified XkdK Protein
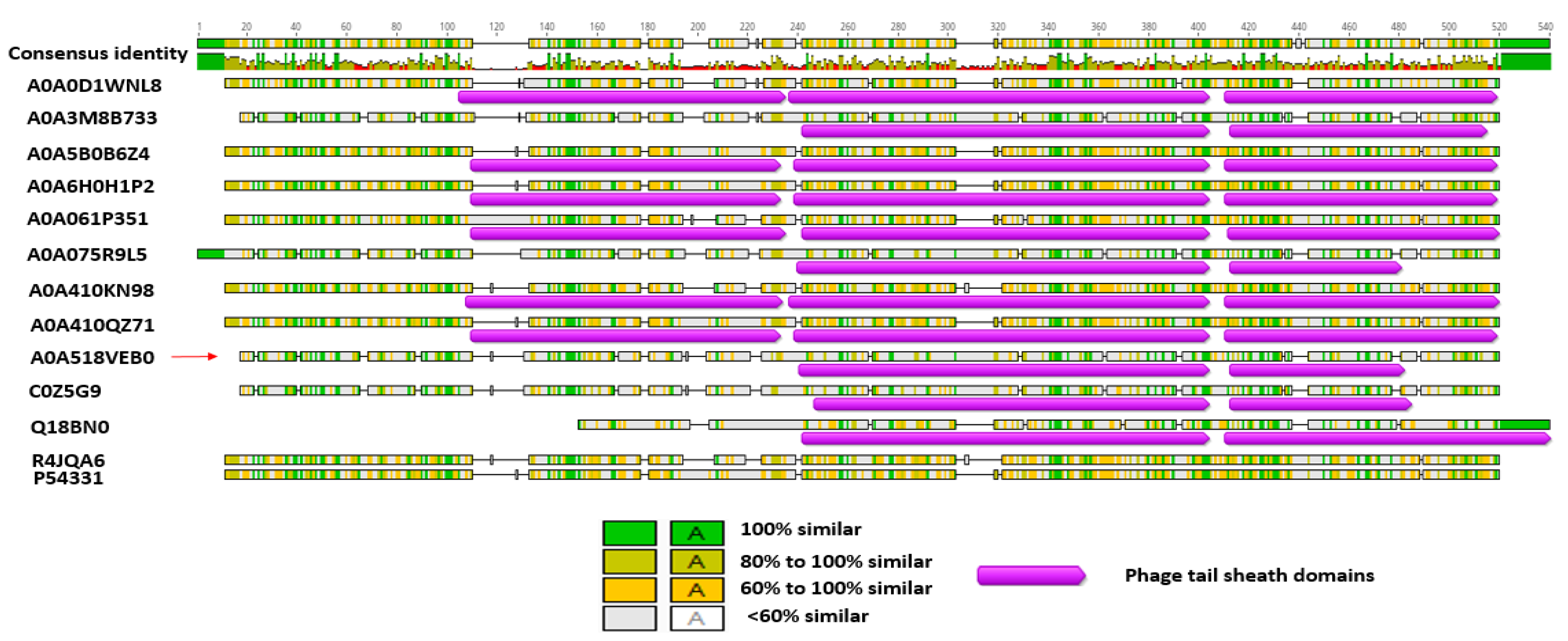
3.6. Bl 1821L and Bl 1951 Phage-Like Element PBSX Protein XkdK Comparison with the Similar Proteins of Other Gram-Positive Bacteria and Bl Phages
3.7. Bioinformatic Analysis of Regions Adjacent to the Bl 1821L and Bl 1951 Putative Antibacterial Protein Encoding Gene
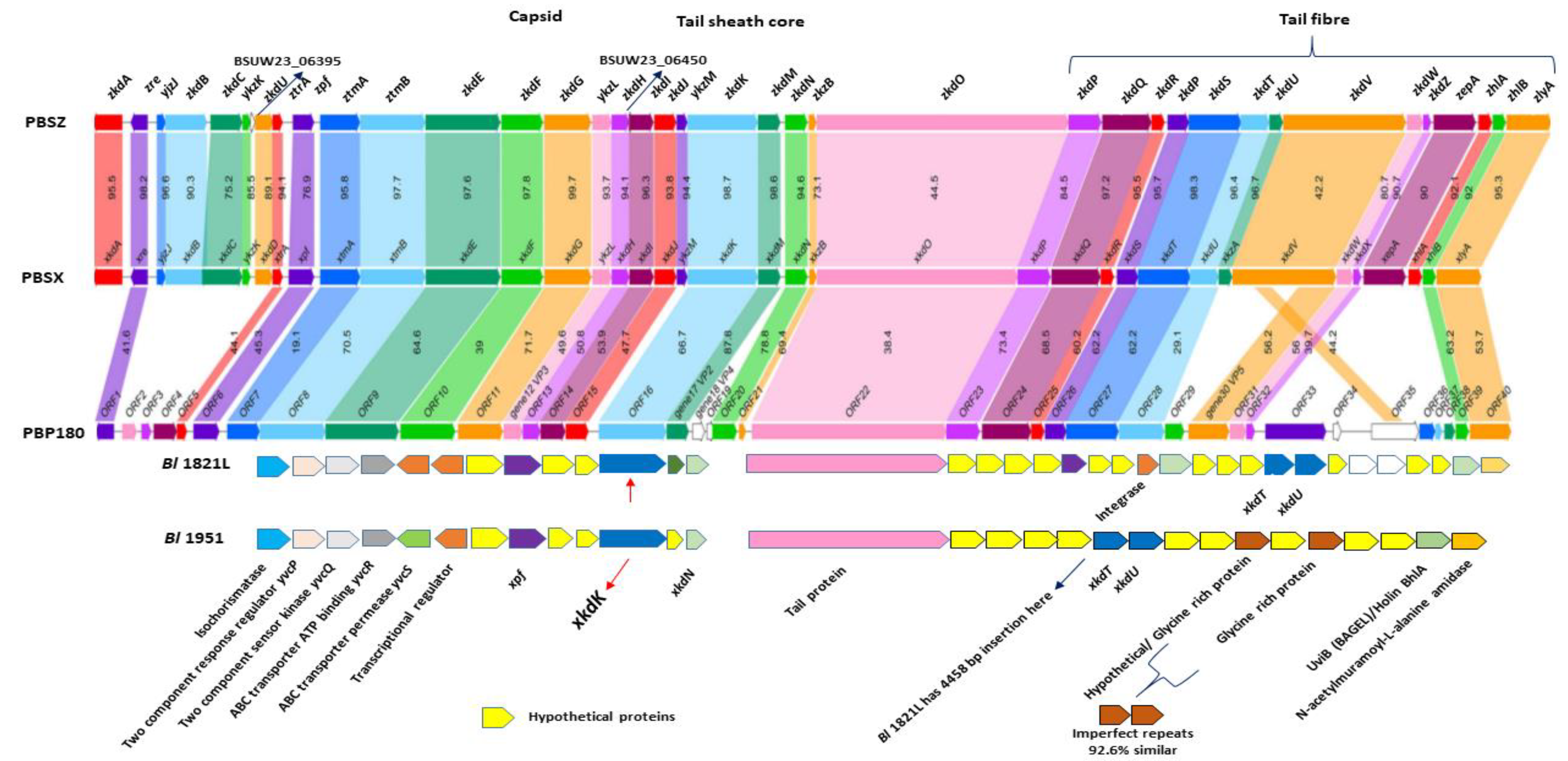
3.8. Genomic Comparison of the Bl 1821L and Bl 1951 PBSX-Like Region with the Defective Prophages PBSZ, PBSX, and PBP180
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shida, O.; Takagi, H.; Kadowaki, K.; Komagata, K. Proposal for two new genera, Brevibacillus gen. nov. and Aneurinibacillus gen. nov. Int. J. Syst. Evol. Microbiol. 1996, 46, 939–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowen, D.; Yin, Y.; Flasinski, S.; Chay, C.; Bean, G.; Milligan, J.; Moar, W.; Pan, A.; Werner, B.; Buckman, K. Cry75Aa (Mpp75Aa) insecticidal proteins for controlling the Western corn rootworm, Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae), isolated from the insect-pathogenic bacterium Brevibacillus laterosporus. Appl. Environ. Microbiol. 2021, 87, e02507–e02520. [Google Scholar] [CrossRef]
- Salama, H.; Foda, M.; El-Bendary, M.; Abdel-Razek, A. Infection of red palm weevil, Rhynchophorus ferrugineus, by spore-forming bacilli indigenous to its natural habitat in Egypt. J. Pest Sci. 2004, 77, 27–31. [Google Scholar] [CrossRef]
- Carramaschi, I.N.; Pereira, L.D.A.; Queiroz, M.M.D.C.; Zahner, V. Preliminary screening of the larvicidal effect of Brevibacillus laterosporus strains against the blowfly Chrysomya megacephala (Fabricius, 1794) (Diptera: Calliphoridae). Rev. Soc. Bras. Med. Trop. 2015, 48, 427–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedini, S.; Muniz, E.R.; Tani, C.; Conti, B.; Ruiu, L. Insecticidal potential of Brevibacillus laterosporus against dipteran pest species in a wide ecological range. J. Invertebr. Pathol. 2020, 177, 107493. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.D.A.; Junqueira, R.M.; Carramaschi, I.N.; Queiroz, M.M.; Zahner, V. Bioactivity under laboratory conditions of Brevibacillus laterosporus towards larvae and adults of Chrysomya putoria (Diptera: Calliphoridae). J. Invertebr. Pathol. 2018, 158, 52–54. [Google Scholar] [CrossRef] [PubMed]
- van Zijll de Jong, E.; Roush, T.L.; Glare, T.R.; Hampton, J.G. Discovery of two Brevibacillus laterosporus isolates from brassica with insecticidal properties against diamondback moth. Biocontrol Sci. Technol. 2016, 26, 426–431. [Google Scholar] [CrossRef]
- Zheng, Z.; Zheng, J.; Zhang, Z.; Peng, D.; Sun, M. Nematicidal spore-forming Bacilli share similar virulence factors and mechanisms. Sci. Rep. 2016, 6, 31341. [Google Scholar] [CrossRef]
- De Oliveira, E.J.; Rabinovitch, L.; Monnerat, R.G.; Passos, L.K.J.; Zahner, V. Molecular characterisation of Brevibacillus laterosporus and its potential use in biological control. Appl. Environ. Microbiol. 2004, 70, 6657–6664. [Google Scholar] [CrossRef] [Green Version]
- Singer, S.; Bair, T.B.; Hammill, T.B.; Berte, A.M.; Correa-Ochoa, M.M.; Stambaugh, A.D. Fermentation and toxin studies of the molluscicidal strains of Bacillus brevis. J. Ind. Microbiol. 1994, 13, 112–119. [Google Scholar] [CrossRef]
- Glare, T.R.; Durrant, A.; Berry, C.; Palma, L.; Ormskirk, M.M.; Cox, M.P. Phylogenetic determinants of toxin gene distribution in genomes of Brevibacillus laterosporus. Genomics 2020, 112, 1042–1053. [Google Scholar] [CrossRef] [PubMed]
- Marche, M.G.; Camiolo, S.; Porceddu, A.; Ruiu, L. Survey of Brevibacillus laterosporus insecticidal protein genes and virulence factors. J. Invertebr. Pathol. 2018, 155, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; CaJacob, C.A.; Feldman, P.; Heck, G.R.; Nooren, I.M.A.; Plaetinck, G.; Maddelein, W.T.; Vaughn, T.T. Methods for Genetic Control of Insect Infestations in Plants and Compositions Thereof. U.S. Patent No. 10,538,783, 21 January 2020. [Google Scholar]
- Boets, A.; Arnaut, G.; Van Rie, J.; Damme, N. Toxins. U.S. Patent No. 7,919,609, 5 April 2011. [Google Scholar]
- Glare, T.R.; Hampton, J.G.; Cox, M.P.; Bienkowski, D.A. Novel Strains of Brevibacillus laterosporus as Biocontrol Agents against Plant Pests, Particularly lepidoptera and Diptera. Google Patent No. WO 2014/045131, 22 October 2018. [Google Scholar]
- Glare, T.R.; Jurat-Fuentes, J.L.; O’callaghan, M. Basic and applied research: Entomopathogenic bacteria. In Microbial Control of Insect and Mite Pests from Theory to Practice; Lacey, L.A., Ed.; Academic Press: Cambridge, MA, USA, 2017. [Google Scholar] [CrossRef]
- Sampson, K.S.; Tomso, D.J.; Guo, R. Pesticidal Genes from Brevibacillus and Methods for Their Use. U.S. Patent No. 9,238,823, 19 January 2016. [Google Scholar]
- Ormskirk, M.M. Brevibacillus laterosporus as a Potential Bio-Control Agent of the Diamondback Moth and Other Insects. Ph.D. Thesis, Lincoln University, Canterbury, New Zealand, 2017. [Google Scholar]
- Touchon, M.; Bernheim, A.; Rocha, E.P.C. Genetic and life-history traits associated with the distribution of prophages in bacteria. Int. Soc. Microbiol. Ecol. J. 2016, 10, 2744–2754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A.A. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 2015, 13, 641–650. [Google Scholar] [CrossRef]
- Bobay, L.-M.; Touchon, M.; Rocha, E.P. Pervasive domestication of defective prophages by bacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 12127–12132. [Google Scholar] [CrossRef] [Green Version]
- Schwemmlein, N.; Pippel, J.; Gazdag, E.M.; Blankenfeldt, W. Crystal structures of R-type bacteriocin sheath and tube proteins CD1363 and CD1364 from Clostridium difficile in the pre-assembled state. Front. Microbiol. 2018, 9, 1750. [Google Scholar] [CrossRef]
- Patz, S.; Becker, Y.; Richert-Pöggeler, K.R.; Berger, B.; Ruppel, S.; Huson, D.H.; Becker, M. Phage tail-like particles are versatile bacterial nanomachines—A mini-review. J. Adv. Res. 2019, 19, 75–84. [Google Scholar] [CrossRef]
- Sarris, P.F.; Ladoukakis, E.D.; Panopoulos, N.J.; Scoulica, E.V. A phage tail-derived element with wide distribution among both prokaryotic domains: A comparative genomic and phylogenetic study. Genome Biol. Evol. 2014, 6, 1739–1747. [Google Scholar] [CrossRef]
- Nagakubo, T. Biological functions and applications of virus-related bacterial nanoparticles: A review. Int. J. Mol. Sci. 2022, 23, 2595. [Google Scholar] [CrossRef]
- Scholl, D. Phage tail–like bacteriocins. Annu. Rev. Virol. 2017, 4, 453–467. [Google Scholar] [CrossRef]
- Michel-Briand, Y.; Baysse, C. The pyocins of Pseudomonas aeruginosa. Biochimie 2002, 84, 499–510. [Google Scholar] [CrossRef]
- Ghequire, M.G.; De Mot, R. The tailocin tale: Peeling off phage tails. Trends Microbiol. 2015, 23, 587–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cascales, E.; Buchanan, S.K.; Duché, D.; Kleanthous, C.; Lloubes, R.; Postle, K.; Riley, M.; Slatin, S.; Cavard, D. Colicin biology. Microbiol. Mol. Biol. Rev. 2007, 71, 158–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, P.; Scholl, D.; Prokhorov, N.S.; Avaylon, J.; Shneider, M.M.; Browning, C.; Buth, S.A.; Plattner, M.; Chakraborty, U.; Ding, K. Action of a minimal contractile bactericidal nanomachine. Nature 2020, 580, 658–662. [Google Scholar] [CrossRef]
- Saha, S.; Ojobor, C.D.; Mackinnon, E.; North, O.I.; Bondy-Denomy, J.; Lam, J.S.; Ensminger, A.W.; Maxwell, K.L.; Davidson, A.R. F-type pyocins are diverse non-contractile phage tail-like weapons for killing Pseudomonas aeruginosa. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dams, D.; Brøndsted, L.; Drulis-Kawa, Z.; Briers, Y. Engineering of receptor-binding proteins in bacteriophages and phage tail-like bacteriocins. Biochem. Soc. Trans. 2019, 47, 449–460. [Google Scholar] [CrossRef]
- Wood, H.E.; Dawson, M.; Devine, K.M.; Mcconnell, D.J. Characterisation of PBSX, a defective prophage of Bacillus subtilis. J. Bacteriol. 1990, 172, 2667–2674. [Google Scholar] [CrossRef] [Green Version]
- Krogh, S.; O’Reilly, M.; Nolan, N.; Devine, K.M. The phage-like element PBSX and part of the skin element, which are resident at different locations on the Bacillus subtilis chromosome, are highly homologous. Microbiology 1996, 142, 2031–2040. [Google Scholar] [CrossRef] [Green Version]
- Yao, G.W.; Duarte, I.; Le, T.T.; Carmody, L.; LiPuma, J.J.; Young, R.; Gonzalez, C.F. A broad-host-range tailocin from Burkholderia cenocepacia. Appl. Environ. Microbiol. 2017, 83, e03414–e03416. [Google Scholar] [CrossRef] [Green Version]
- Jin, T.; Zhang, X.; Zhang, Y.; Hu, Z.; Fu, Z.; Fan, J.; Wu, M.; Wang, Y.; Shen, P.; Chen, X. Biological and genomic analysis of a PBSX-like defective phage induced from Bacillus pumilus AB94180. Arch. Virol. 2014, 159, 739–752. [Google Scholar] [CrossRef]
- Hartford, O.M.; Dowds, B.C. Cloning and characterisation of genes induced by hydrogen peroxide in Bacillus subtilis. Microbiology 1992, 138, 2061–2068. [Google Scholar] [CrossRef] [Green Version]
- Seaman, E.; Tarmy, E.; Marmur, J. Inducible phages of Bacillus subtilis. Biochemistry 1964, 3, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Mauël, C.; Karamata, D. Characterisation of proteins induced by Mitomycin C treatment of Bacillus subtilis. J. Virol. 1984, 49, 806–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thurm, P.; Garro, A.J. Bacteriophage-specific protein synthesis during induction of the defective Bacillus subtilis bacteriophage PBSX. J. Virol. 1975, 16, 179–183. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.; Chakraborty, U.; Gebhart, D.; Govoni, G.R.; Zhou, Z.H.; Scholl, D. F-type bacteriocins of Listeria monocytogenes: A new class of phage tail-like structures reveals broad parallel coevolution between tailed bacteriophages and high molecular-weight bacteriocins. J. Bacteriol. 2016, 198, 2784–2793. [Google Scholar] [CrossRef] [Green Version]
- Nagai, T. A defective bacteriophage produced by Bacillus subtilis MAFF 118147 and a mutant producing no normal particles of the defective bacteriophage. Food Sci. Technol. Res. 2014, 20, 1229–1234. [Google Scholar] [CrossRef] [Green Version]
- Gebhart, D.; Williams, S.R.; Bishop-Lilly, K.A.; Govoni, G.R.; Willner, K.M.; Butani, A.; Sozhamannan, S.; Martin, D.; Fortier, L.C.; Scholl, D. Novel high-molecular-weight, R-type bacteriocins of Clostridium difficile. J. Bacteriol. 2012, 194, 6240–6247. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.; Nishimune, T.; Abe, M.; Kimoto, M.; Hayashi, R. Bacteriocin like killing action of a temperate bacteriophage phiBA1 of Bacillus aneurinolyticus. J. Virol. 1986, 59, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Fernández, A.; Osuna, A.; Vilchez, S. Bacillus pumilus 15.1, a strain active against Ceratitis capitata, contains a novel phage and a phage-related particle with bacteriocin activity. Int. J. Mol. Sci. 2021, 22, 8164. [Google Scholar] [CrossRef]
- Rybakova, D.; Mitra, A.K.; Hurst, M.R.H. Purification and TEM of Afp and Its variants. Bio-Protoc. 2014, 4, e1132. [Google Scholar] [CrossRef]
- Hockett, K.L.; Baltrus, D.A. Use of the soft-agar overlay technique to screen for bacterially produced inhibitory compounds. J. Vis. Exp. JoVE 2017, 119, e55064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirby, W.M.; Yoshihara, G.M.; Sundsted, K.S.; Warren, J.H. Clinical usefulness of a single disc method for antibiotic sensitivity testing. Antibiot. Annu. 1956, 892–897. [Google Scholar]
- Laemmli, U. SDS-PAGE Laemmli method. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Yasumitsu, H.; Ozeki, Y.; Kawsar, S.M.; Fujii, Y.; Sakagami, M.; Matuo, Y.; Toda, T.; Katsuno, H. RAMA stain: A fast, sensitive and less protein-modifying CBB R250 stain. Electrophoresis 2010, 31, 1913–1917. [Google Scholar] [CrossRef]
- Blum, H.; Beier, H.; Gross, H.J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 1987, 8, 93–99. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organisation and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Di Tommaso, P.; Pulido, D.; Nogués, M.V.; Notredame, C.; Boix, E.; Andreu, D. AMPA: An automated web server for prediction of protein antimicrobial regions. Bioinformatics 2012, 28, 130–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, J.A.; Merrill, B.D.; Crockett, J.T.; Esplin, K.P.; Evans, M.R.; Heaton, K.E.; Hilton, J.A.; Hyde, J.R.; McBride, M.S.; Schouten, J.T.; et al. Characterisation of five novel Brevibacillus bacteriophages and genomic comparison of Brevibacillus phages. PLoS ONE 2016, 11, e0156838. [Google Scholar] [CrossRef]
- Bradley, D.E. Ultrastructure of bacteriophage and bacteriocins. Bacteriol. Rev. 1967, 31, 230–314. [Google Scholar] [CrossRef]
- Krogh, S.; Jørgensen, S.T.; Devine, K.M. Lysis genes of the Bacillus subtilis defective prophage PBSX. J. Bacteriol. 1998, 180, 2110–2117. [Google Scholar] [CrossRef] [Green Version]
- Brito, P.H.; Chevreux, B.; Serra, C.R.; Schyns, G.; Henriques, A.O.; Pereira-Leal, J.B. Genetic competence drives genome diversity in Bacillus subtilis. Genome Biol. Evol. 2018, 10, 108–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begot, C.; Desnier, I.; Daudin, J.D.; Labadie, J.C.; Lebert, A. Recommendations for calculating growth parameters by optical density measurements. J. Microbiol. Methods 1996, 25, 225–232. [Google Scholar] [CrossRef]
- Hudson, J.A.; Mott, S.J. Comparison of lag times obtained from optical density and viable count data for a strain of Pseudomonas fragi. J. Food Saf. 1994, 14, 329–339. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S.T. Bacteriophage: Overview. In Schmidt TMBT-E of M; Fourth, E., Ed.; Oxford Academic Press: Oxford, UK, 2019; pp. 441–457. [Google Scholar]
- Young, R. Phage lysis: Three steps, three choices, one outcome. J. Microbiol. 2014, 52, 243–258. [Google Scholar] [CrossRef]
- Young, I.; Wang, I.; Roof, W.D. Phages will out: Strategies of host cell lysis. Trends Microbiol. 2000, 8, 120–128. [Google Scholar] [CrossRef]
- Canchaya, C.; Fournous, G.; Chibani-Chennoufi, S.; Dillmann, M.L.; Brüssow, H. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 2003, 6, 417–424. [Google Scholar] [CrossRef]
- Hockett, K.L.; Renner, T.; Baltrus, D.A. Independent co-option of a tailed bacteriophage into a killing complex in Pseudomonas. ASM J. Mbio 2015, 6, e00452. [Google Scholar] [CrossRef] [Green Version]
- Hayes, F.; Van Melderen, L. Toxins-antitoxins: Diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 386–408. [Google Scholar] [CrossRef]
- Hayes, F.; Kędzierska, B. Regulating toxin-antitoxin expression: Controlled detonation of intracellular molecular timebombs. Toxins 2014, 6, 337–358. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, Y.; Park, J.H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef] [Green Version]
- Steensma, H. Effect of defective phages on the cell membrane of Bacillus subtilis and partial characterisation of a phage protein involved in killing. J. Gen. Virol. 1981, 56, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Pell, L.G.; Cumby, N.; Clark, T.E.; Tuite, A.; Battaile, K.P.; Edwards, A.M.; Chirgadze, N.Y.; Davidson, A.R.; Maxwell, K.L. A conserved spiral structure for highly diverged phage tail assembly chaperones. J. Mol. Biol. 2013, 425, 2436–2449. [Google Scholar] [CrossRef] [PubMed]
- Groth, A.C.; Calos, M.P. Phage integrases: Biology and applications. J. Mol. Biol. 2004, 335, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.; Hupfeld, M.; Klumpp, J.; Loessner, M.J. Molecular basis of bacterial host interactions by gram-positive targeting bacteriophages. Viruses 2018, 10, 397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trojet, S.N.; Caumont-Sarcos, A.; Perrody, E.; Comeau, A.M.; Krisch, H. The Gp38 adhesins of the T4 superfamily: A complex modular determinant of the phage’s host specificity. Genome Biol. Evol. 2011, 3, 674–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.Z.; Fokine, A.; Mahalingam, M.; Zhang, Z.; Garcia-Doval, C.; van Raaij, M.J.; Rossmann, M.G.; Rao, V.B. Molecular anatomy of the receptor binding module of a bacteriophage long tail fibre. PLoS Pathog. 2019, 15, e1008193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holtzman, T.; Globus, R.; Molshanski-Mor, S.; Ben-Shem, A.; Yosef, I.; Qimron, U. A continuous evolution system for contracting the host range of bacteriophage T7. Sci. Rep. 2020, 10, 307. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef]
- Simpson, D.J.; Sacher, J.C.; Szymanski, C.M. Development of an assay for the identification of receptor binding proteins from bacteriophages. Viruses 2016, 8, 17. [Google Scholar] [CrossRef] [Green Version]
- Boeckaerts, D.; Stock, M.; Criel, B.; Gerstmans, H.; De Baets, B.; Briers, Y. Predicting bacteriophage hosts based on sequences of annotated receptor-binding proteins. Sci. Rep. 2021, 11, 1467. [Google Scholar] [CrossRef]
- Blackwell, C.C.; Winstanley, F.P.; Telfer Brunton, W.A. Sensitivity of thermophilic campylobacters to R-type pyocines of Pseudomonas aeruginosa. J. Med. Microbiol. 1982, 15, 247–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, N.J.; John, C.M.; Reinders, L.G.; Gibson, B.W.; Apicella, M.A.; Griffiss, J.M. Structural models for the cell surface lipooligosaccharides of Neisseria gonorrhoeae and Haemophilus influenzae. Biomed. Environ. Mass Spectrom. 1990, 19, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Campagnari, A.A.; Karalus, R.; Apicella, M.; Melaugh, W.; Lesse, A.J.; Gibson, B.W. Use of pyocin to select a Haemophilus ducreyi variant defective in lipooligosaccharide biosynthesis. Infect. Immun. 1994, 62, 2379–2386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, J.C.; Stein, D.C. Cloning, complementation, and characterisation of an rfaE homolog from Neisseria gonorrhoeae. J. Bacteriol. 1996, 178, 4571–4575. [Google Scholar] [CrossRef] [Green Version]
- Morse, S.A.; Vaughan, P.; Johnson, D.; Iglewski, B.H. Inhibition of Neisseria gonorrhoeae by a bacteriocin from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1976, 10, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Filiatrault, M.J.; Munson, R.S., Jr.; Campagnari, A.A. Genetic analysis of a pyocin-resistant lipooligosaccharide (LOS) mutant of Haemophilus ducreyi: Restoration of full-length LOS restores pyocin sensitivity. J. Bacteriol. 2001, 183, 5756–5761. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.R.; Gebhart, D.; Martin, D.W.; Scholl, D. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 2008, 74, 3868–3876. [Google Scholar] [CrossRef] [Green Version]
- Klein, T.A.; Ahmad, S.; Whitney, J.C. Contact-dependent interbacterial antagonism mediated by protein secretion machines. Trends Microbiol. 2020, 28, 387–400. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.P.; Fredrik Inglis, R.; Taddei, F. SYNTHESIS: Evolutionary ecology of microbial wars: Within-host competition and (incidental) virulence. Evol. Appl. 2009, 2, 32–39. [Google Scholar] [CrossRef]
- Foster, K.R.; Bell, T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 2012, 22, 1845–1850. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Bl 1821L as the Host Bacterium | Bl 1951 as the Host Bacterium | ||
|---|---|---|---|
| Dilution Level | Zone of Inhibition Diameter (mm) | Dilution Level | Zone of Inhibition Diameter (mm) |
| FS * | 12.5 | FS | 12.5 |
| 10−1 | – ** | 10−1 | 10.5 |
| 10−2 | – | 10−2 | 13.5 |
| 10−3 | – | 10−3 | 10.5 |
| 10−4 | – | 10−4 | – |
| 10−5 | – | 10−5 | – |
| 10−6 | – | 10−6 | – |
| 10−7 | – | 10−7 | – |
| 10−8 | – | 10−8 | – |
| Control *** | – | Control | – |
| Host Bacterium | Host Bacterium Isolate/Strain | Sensitivity to Induced Bl 1821L CFS |
|---|---|---|
| Bacillus megaterium | 3-2 | – * |
| Bacillus megaterium | S1 | – |
| Bacillus subtilis | EM-13 (Tp5) | – |
| Brevibacillus laterosporus | 1951 | + ** |
| Brevibacillus laterosporus | 1821L | – |
| Brevibacillus laterosporus | Rsp | + |
| Brevibacillus laterosporus | CCEB 342 | – |
| Brevibacillus laterosporus | NRS 590 | + |
| Brevibacillus laterosporus | NCIMB | + |
| Carnobacterium maltaromaticum | 3-1 | + |
| Fictibacillus rigui | EM-14 (FJAT 46895) | – |
| Oceanobacillus sp. | EM-12 (R-31213) | – |
| Oerskovia enterophila | 3-3 | – |
| Paenibacillus sp. | 15.12.1 | – |
| Accession | Protein | Host Bacterium | Amino Acids (%) Similarity to the Identified Accession A0A518VEB0 |
|---|---|---|---|
| A0A075R9L5 | Phage tail sheath | Brevibacillus laterosporus LMG 15441 | 90.3 |
| M8E4N0 | Uncharacterised | Brevibacillus borstelensis AK1 | 69.9 |
| A0A3M8DWU9 | Phage tail | Brevibacillus fluminis | 68.0 |
| C0Z5G9 | Uncharacterised | Brevibacillus brevis (strain 47/JCM 6285/NBRC 100599) | 68.1 |
| A0A6M1UCS4 | Phage tail | Brevibacillus sp. SYP-B805 | 66.6 |
| A0A0X8D434 | Phage tail sheath | Aneurinibacillus sp. XH2 | 49.1 |
| A0A2Z2KS77 | Phage tail sheath | Paenibacillus donghaensis | 48.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babar, T.K.; Glare, T.R.; Hampton, J.G.; Hurst, M.R.H.; Narciso, J.O. Isolation, Purification, and Characterisation of a Phage Tail-Like Bacteriocin from the Insect Pathogenic Bacterium Brevibacillus laterosporus. Biomolecules 2022, 12, 1154. https://doi.org/10.3390/biom12081154
Babar TK, Glare TR, Hampton JG, Hurst MRH, Narciso JO. Isolation, Purification, and Characterisation of a Phage Tail-Like Bacteriocin from the Insect Pathogenic Bacterium Brevibacillus laterosporus. Biomolecules. 2022; 12(8):1154. https://doi.org/10.3390/biom12081154
Chicago/Turabian StyleBabar, Tauseef K., Travis R. Glare, John G. Hampton, Mark R. H. Hurst, and Josefina O. Narciso. 2022. "Isolation, Purification, and Characterisation of a Phage Tail-Like Bacteriocin from the Insect Pathogenic Bacterium Brevibacillus laterosporus" Biomolecules 12, no. 8: 1154. https://doi.org/10.3390/biom12081154
APA StyleBabar, T. K., Glare, T. R., Hampton, J. G., Hurst, M. R. H., & Narciso, J. O. (2022). Isolation, Purification, and Characterisation of a Phage Tail-Like Bacteriocin from the Insect Pathogenic Bacterium Brevibacillus laterosporus. Biomolecules, 12(8), 1154. https://doi.org/10.3390/biom12081154









