Illuminating Intrinsically Disordered Proteins with Integrative Structural Biology
Abstract
1. Introduction
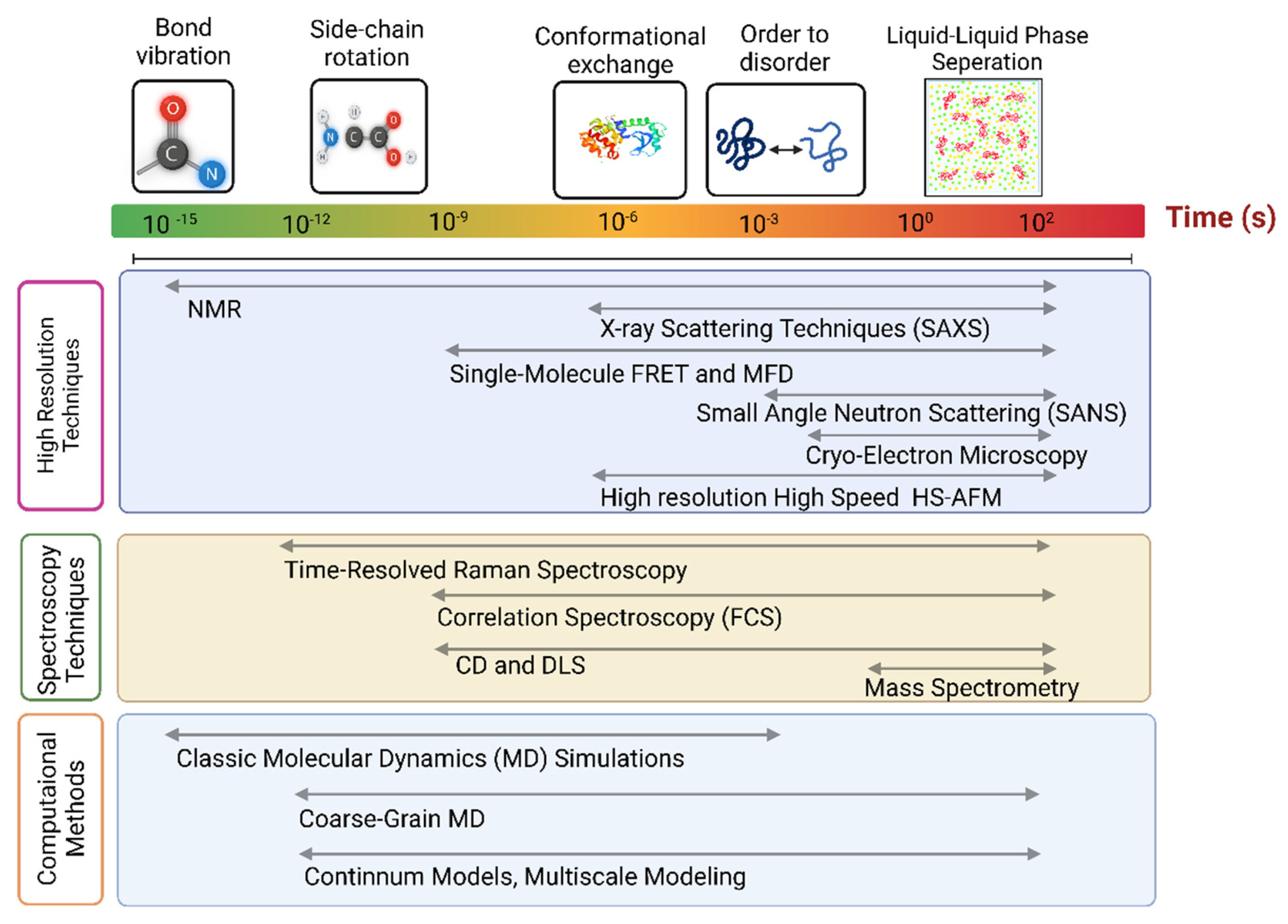
2. Summary of Methods
2.1. Nuclear Magnetic Resonance
2.2. Scattering Methods
2.3. Label-Based Approaches
2.4. Single-Molecule Approaches
2.5. Atomic Force Microscopy
2.6. Cryo-EM and X-ray Crystallography
2.7. Solvent Accessibility Methods
2.7.1. Hydrogen-Deuterium-Exchange
2.7.2. Crosslinking Mass Spectrometry
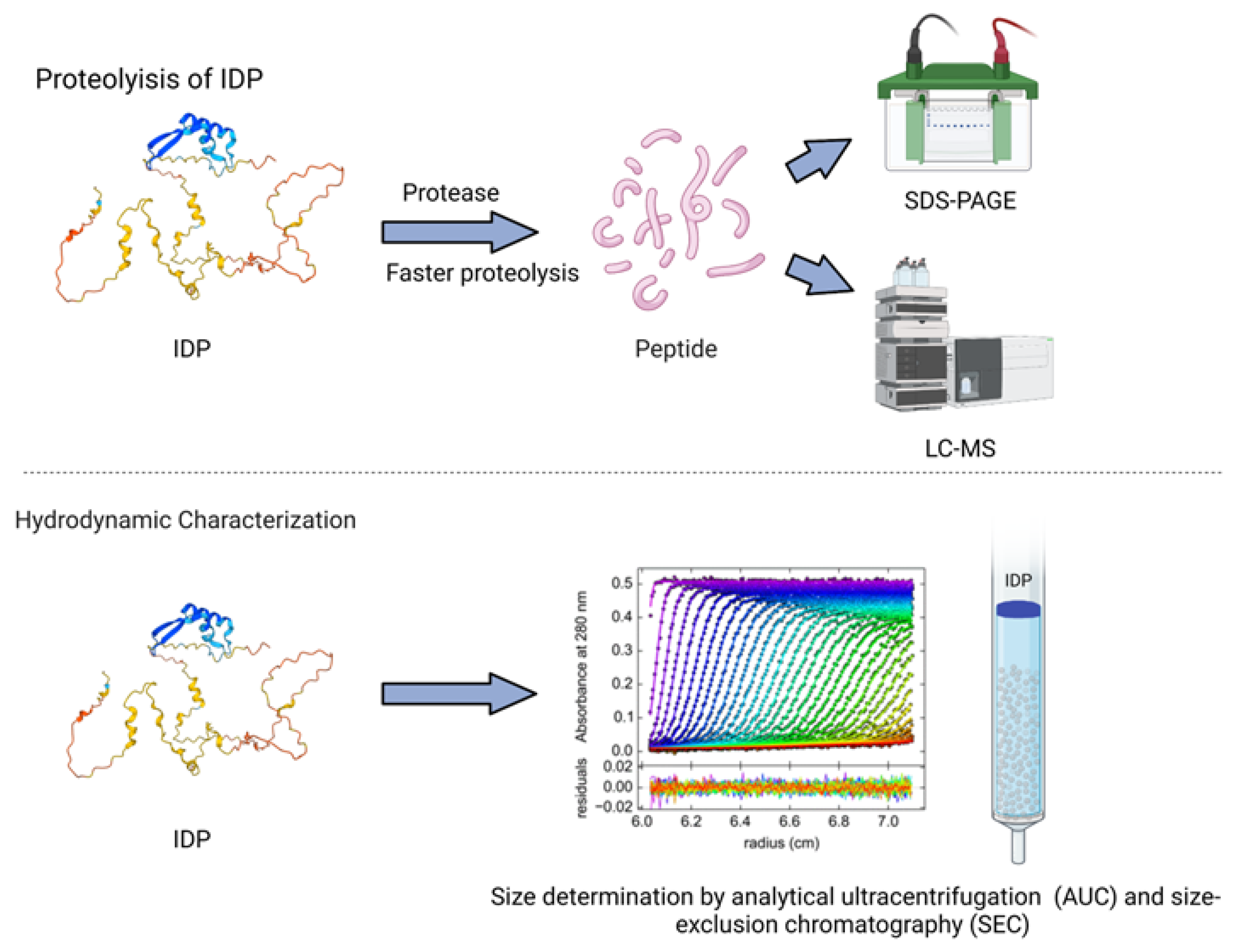
2.7.3. Proteolysis
2.8. Spectroscopies
2.8.1. Circular Dichroism
2.8.2. Fourier Transform Infrared Spectroscopy
2.8.3. Raman Spectroscopy
2.8.4. Mass Spectrometry
2.9. Hydrodynamic Characterizations
2.10. Computational Methods
3. The Integrative Structural Biology Approach to IDPs and Examples
3.1. Ubiquitin
3.2. Nucleoporins
3.3. Aggregation-Prone Synaptic Proteins
3.4. Sic1
3.5. N-WASP
3.6. SNAP-25
3.7. p27
3.8. PAGE4
4. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, Z.; Yan, J.; Fan, X.; Mizianty, M.J.; Xue, B.; Wang, K.; Hu, G.; Uversky, V.N.; Kurgan, L. Exceptionally Abundant Exceptions: Comprehensive Characterization of Intrinsic Disorder in All Domains of Life. Cell. Mol. Life Sci. 2015, 72, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunker, A.K.; Uversky, V.N. Orderly Order in Protein Intrinsic Disorder Distribution: Disorder in 3500 Proteomes from Viruses and the Three Domains of Life. J. Biomol. Struct. Dyn. 2012, 30, 137–149. [Google Scholar] [CrossRef]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Obradovic, Z.; Romero, P.; Garner, E.C.; Brown, C.J. Intrinsic Protein Disorder in Complete Genomes. Genome Inform. Ser. Workshop Genome Inform. 2000, 11, 161–171. [Google Scholar] [PubMed]
- DeForte, S.; Uversky, V.N. Intrinsically Disordered Proteins in PubMed: What Can the Tip of the Iceberg Tell Us about What Lies Below? RSC Adv. 2016, 6, 11513–11521. [Google Scholar] [CrossRef]
- Chao, L.-P.; Roboz Einstein, E. Estimation of the Molecular Weight of Flexible Disordered Proteins by Exclusion Chromatography. J. Chromatogr. A 1969, 42, 485–492. [Google Scholar] [CrossRef]
- Boesch, C.; Bundi, A.; Oppliger, M.; Wuthrich, K. 1H Nuclear-Magnetic-Resonance Studies of the Molecular Conformation of Monomeric Glucagon in Aqueous Solution. Eur. J. Biochem. 1978, 91, 209–214. [Google Scholar] [CrossRef]
- Boublik, M.; Bradbury, E.M.; Crane-Robinson, C.; Johns, E.W. An Investigation of the Conformational Changes of Histone F2b by High Resolution Nuclear Magnetic Resonance. Eur. J. Biochem. 1970, 17, 151–159. [Google Scholar] [CrossRef]
- Huber, R.; Bennett, W.S. Functional Significance of Flexibility in Proteins. Biopolymers 1983, 22, 261–279. [Google Scholar] [CrossRef]
- Sigler, P.B. Acid Blobs and Negative Noodles. Nature 1988, 333, 210–212. [Google Scholar] [CrossRef]
- Nishikawa, K. Natively Unfolded Proteins: An Overview. Biophysics 2009, 5, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. Understanding Protein Non-Folding. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Intrinsically Unstructured Proteins and Their Functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.; Uversky, V.N. Intrinsically Disordered Proteins: The Dark Horse of the Dark Proteome. Proteomics 2018, 18, 1800061. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically Unstructured Proteins: Re-Assessing the Protein Structure-Function Paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why Are “Natively Unfolded” Proteins Unstructured under Physiologic Conditions? Proteins Struct. Funct. Genet. 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically Unstructured Proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically Disordered Protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Ferrie, J.J.; Karr, J.P.; Tjian, R.; Darzacq, X. “Structure”-Function Relationships in Eukaryotic Transcription Factors: The Role of Intrinsically Disordered Regions in Gene Regulation. Mol. Cell 2022, 82, 3970–3984. [Google Scholar] [CrossRef]
- Trnka, M.J.; Pellarin, R.; Robinson, P.J. Role of Integrative Structural Biology in Understanding Transcriptional Initiation. Methods 2019, 159–160, 4–22. [Google Scholar] [CrossRef]
- Choi, U.B.; Kazi, R.; Stenzoski, N.; Wollmuth, L.P.; Uversky, V.N.; Bowen, M.E. Modulating the Intrinsic Disorder in the Cytoplasmic Domain Alters the Biological Activity of the N-Methyl-D-Aspartatesensitive Glutamate Receptor. J. Biol. Chem. 2013, 288, 22506–22515. [Google Scholar] [CrossRef]
- Hu, G.; Wu, Z.; Uversky, V.; Kurgan, L. Functional Analysis of Human Hub Proteins and Their Interactors Involved in the Intrinsic Disorder-Enriched Interactions. Int. J. Mol. Sci. 2017, 18, 2761. [Google Scholar] [CrossRef] [PubMed]
- Deiana, A.; Forcelloni, S.; Porrello, A.; Giansanti, A. Intrinsically Disordered Proteins and Structured Proteins with Intrinsically Disordered Regions Have Different Functional Roles in the Cell. PLoS ONE 2019, 14, e0217889. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically Disordered Proteins in Cellular Signalling and Regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.; Leite, V.B.P.; Roy, S.; Bhattacharyya, S.; Mohanty, A.; Achuthan, S.; Singh, D.; Appadurai, R.; Rangarajan, G.; Weninger, K.; et al. Intrinsically Disordered Proteins: Ensembles at the Limits of Anfinsen’s Dogma. Biophys. Rev. 2022, 3, 011306. [Google Scholar] [CrossRef]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible Nets. The Roles of Intrinsic Disorder in Protein Interaction Networks. FEBS J. 2005, 272, 5129–5148. [Google Scholar] [CrossRef]
- Csermely, P.; Palotai, R.; Nussinov, R. Induced Fit, Conformational Selection and Independent Dynamic Segments: An Extended View of Binding Events. Nat. Preced. 2010, 35, 539–546. [Google Scholar] [CrossRef]
- Berlow, R.B.; Dyson, H.J.; Wright, P.E. Expanding the Paradigm: Intrinsically Disordered Proteins and Allosteric Regulation. J. Mol. Biol. 2018, 430, 2309–2320. [Google Scholar] [CrossRef]
- Fung, H.Y.J.; Birol, M.; Rhoades, E. IDPs in Macromolecular Complexes: The Roles of Multivalent Interactions in Diverse Assemblies. Curr. Opin. Struct. Biol. 2018, 49, 36–43. [Google Scholar] [CrossRef]
- Coskuner-Weber, O.; Mirzanli, O.; Uversky, V.N. Intrinsically Disordered Proteins and Proteins with Intrinsically Disordered Regions in Neurodegenerative Diseases. Biophys. Rev. 2022, 14, 679–707. [Google Scholar] [CrossRef]
- Martinelli, A.; Lopes, F.; John, E.; Carlini, C.; Ligabue-Braun, R. Modulation of Disordered Proteins with a Focus on Neurodegenerative Diseases and Other Pathologies. Int. J. Mol. Sci. 2019, 20, 1322. [Google Scholar] [CrossRef] [PubMed]
- Dignon, G.L.; Best, R.B.; Mittal, J. Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem. 2020, 71, 53–75. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Tompa, P.; Pappu, R.v. Polymer Physics of Intracellular Phase Transitions. Nat. Phys. 2015, 11, 899–904. [Google Scholar] [CrossRef]
- Rout, M.P.; Sali, A. Principles for Integrative Structural Biology Studies. Cell 2019, 177, 1384–1403. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.B.; Sali, A.; Wilson, I.A. Integrative Structural Biology. Science 2013, 339, 913–915. [Google Scholar] [CrossRef] [PubMed]
- Masrati, G.; Landau, M.; Ben-Tal, N.; Lupas, A.; Kosloff, M.; Kosinski, J. Integrative Structural Biology in the Era of Accurate Structure Prediction: The Era of Accurate Structure Prediction. J. Mol. Biol. 2021, 433, 167127. [Google Scholar] [CrossRef]
- Cohen, I.R.; Lajtha, A.; Lambris, J.D.; Paoletti, R. Intrinsically Disordered Proteins Studied by NMR Spectroscopy; Felli, I.C., Pierattelli, R., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2015; Volume 870, ISBN 978-3-319-20163-4. [Google Scholar]
- Musacchio, A. On the Role of Phase Separation in the Biogenesis of Membraneless Compartments. EMBO J. 2022, 41, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Saunders, M.; Wishnia, A.; Kirkwood, J.G. The Nuclear Magnetic Resonance Spectrum of Ribonuclease. J. Am. Chem. Soc. 1957, 79, 3289–3290. [Google Scholar] [CrossRef]
- Kowalsky, A. Nuclear Magnetic Resonance Studies of Proteins. J. Biol. Chem. 1962, 237, 1807–1819. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. NMR Illuminates Intrinsic Disorder. Curr. Opin. Struct. Biol. 2021, 70, 44–52. [Google Scholar] [CrossRef]
- Mureddu, L.; Vuister, G.W. Simple High-Resolution NMR Spectroscopy as a Tool in Molecular Biology. FEBS J. 2019, 286, 2035–2042. [Google Scholar] [CrossRef] [PubMed]
- Konrat, R. NMR Contributions to Structural Dynamics Studies of Intrinsically Disordered Proteins. J. Magn. Reson. 2014, 241, 74–85. [Google Scholar] [CrossRef]
- Prestel, A.; Bugge, K.; Staby, L.; Hendus-Altenburger, R.; Kragelund, B.B. Characterization of Dynamic IDP Complexes by NMR Spectroscopy, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 611, ISBN 9780128156490. [Google Scholar]
- Drescher, M. EPR in Protein Science; Springer: Berlin/Heidelberg, Germany, 2011; pp. 91–119. [Google Scholar]
- Fontana, A.; de Laureto, P.P.; Spolaore, B.; Frare, E. Intrinsically Disordered Protein Analysis; Uversky, V.N., Dunker, A.K., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2012; Volume 896, ISBN 978-1-4614-3703-1. [Google Scholar]
- Bernadó, P.; Svergun, D.I. Structural Analysis of Intrinsically Disordered Proteins by Small-Angle X-ray Scattering. Mol. BioSyst. 2012, 8, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Pauw, B.R. Corrigendum: Everything SAXS: Small-Angle Scattering Pattern Collection and Correction (2013 J. Phys.: Condens. Matter 25 383201). J. Phys. Condens. Matter 2014, 26, 239501. [Google Scholar] [CrossRef] [PubMed]
- Fuertes, G.; Banterle, N.; Ruff, K.M.; Chowdhury, A.; Mercadante, D.; Koehler, C.; Kachala, M.; Estrada Girona, G.; Milles, S.; Mishra, A.; et al. Decoupling of Size and Shape Fluctuations in Heteropolymeric Sequences Reconciles Discrepancies in SAXS vs. FRET Measurements. Proc. Natl. Acad. Sci. USA 2017, 114, E6342–E6351. [Google Scholar] [CrossRef] [PubMed]
- Luitz, M.P.; Barth, A.; Crevenna, A.H.; Bomblies, R.; Lamb, D.C.; Zacharias, M. Covalent Dye Attachment Influences the Dynamics and Conformational Properties of Flexible Peptides. PLoS ONE 2017, 12, e0177139. [Google Scholar] [CrossRef] [PubMed]
- Nasir, I.; Onuchic, P.L.; Labra, S.R.; Deniz, A.A. Single-Molecule Fluorescence Studies of Intrinsically Disordered Proteins and Liquid Phase Separation. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2019, 1867, 980–987. [Google Scholar] [CrossRef]
- Yu, L.; Lei, Y.; Ma, Y.; Liu, M.; Zheng, J.; Dan, D.; Gao, P. A Comprehensive Review of Fluorescence Correlation Spectroscopy. Front. Phys. 2021, 9, 644450. [Google Scholar] [CrossRef]
- Haustein, E.; Schwille, P. Fluorescence Correlation Spectroscopy: Novel Variations of an Established Technique. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 151–169. [Google Scholar] [CrossRef]
- Li, X.-H.; Culver, J.A.; Rhoades, E. Tau Binds to Multiple Tubulin Dimers with Helical Structure. J. Am. Chem. Soc. 2015, 137, 9218–9221. [Google Scholar] [CrossRef]
- Drescher, M.; Jeschke, G. (Eds.) EPR Spectroscopy; Topics in Current Chemistry; Springer: Berlin/Heidelberg, Germany, 2012; Volume 321, ISBN 978-3-642-28346-8. [Google Scholar]
- Le Breton, N.; Martinho, M.; Mileo, E.; Etienne, E.; Gerbaud, G.; Guigliarelli, B.; Belle, V. Exploring Intrinsically Disordered Proteins Using Site-Directed Spin Labeling Electron Paramagnetic Resonance Spectroscopy. Front. Mol. Biosci. 2015, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, M.; Sylvi, L.; Gerbaud, G.; Mileo, E.; Halgand, F.; Walburger, A.; Vezin, H.; Belle, V.; Guigliarelli, B.; Magalon, A. Conformational Selection Underlies Recognition of a Molybdoenzyme by Its Dedicated Chaperone. PLoS ONE 2012, 7, e49523. [Google Scholar] [CrossRef] [PubMed]
- Klare, J.P.; Steinhoff, H.-J. Spin Labeling EPR. Photosynth. Res. 2009, 102, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Longhi, S.; Belle, V.; Fournel, A.; Guigliarelli, B.; Carrière, F. Probing Structural Transitions in Both Structured and Disordered Proteins Using Site-Directed Spin-Labeling EPR Spectroscopy. J. Pept. Sci. 2011, 17, 315–328. [Google Scholar] [CrossRef]
- Van Doorslaer, S.; Murphy, D.M. EPR Spectroscopy in Catalysis. In EPR Spectroscopy; Springer: Berlin, Germany, 2011; pp. 1–39. [Google Scholar]
- Peter, M.F.; Gebhardt, C.; Mächtel, R.; Muñoz, G.G.M.; Glaenzer, J.; Narducci, A.; Thomas, G.H.; Cordes, T.; Hagelueken, G. Cross-Validation of Distance Measurements in Proteins by PELDOR/DEER and Single-Molecule FRET. Nat. Commun. 2022, 13, 4396. [Google Scholar] [CrossRef]
- Widengren, J.; Kudryavtsev, V.; Antonik, M.; Berger, S.; Gerken, M.; Seidel, C.A.M. Single-Molecule Detection and Identification of Multiple Species by Multiparameter Fluorescence Detection. Anal. Chem. 2006, 78, 2039–2050. [Google Scholar] [CrossRef]
- Ma, J.; Yanez-Orozco, I.S.; Rezaei Adariani, S.; Dolino, D.; Jayaraman, V.; Sanabria, H. High Precision FRET at Single-Molecule Level for Biomolecule Structure Determination. J. Vis. Exp. 2017, 123, e55623. [Google Scholar] [CrossRef]
- Margittai, M.; Widengren, J.; Schweinberger, E.; Schröder, G.F.; Felekyan, S.; Haustein, E.; König, M.; Fasshauer, D.; Grubmüller, H.; Jahn, R.; et al. Single-Molecule Fluorescence Resonance Energy Transfer Reveals a Dynamic Equilibrium between Closed and Open Conformations of Syntaxin 1. Proc. Natl. Acad. Sci. USA 2003, 100, 15516–15521. [Google Scholar] [CrossRef]
- Rothwell, P.J.; Berger, S.; Kensch, O.; Felekyan, S.; Antonik, M.; Wöhrl, B.M.; Restle, T.; Goody, R.S.; Seidel, C.A.M. Multiparameter Single-Molecule Fluorescence Spectroscopy Reveals Heterogeneity of HIV-1 Reverse Transcriptase:Primer/Template Complexes. Proc. Natl. Acad. Sci. USA 2003, 100, 1655–1660. [Google Scholar] [CrossRef]
- Eggeling, C.; Berger, S.; Brand, L.; Fries, J.R.; Schaffer, J.; Volkmer, A.; Seidel, C.A.M. Data Registration and Selective Single-Molecule Analysis Using Multi-Parameter Fluorescence Detection. J. Biotechnol. 2001, 86, 163–180. [Google Scholar] [CrossRef]
- Hamilton, G.; Sanabria, H. Multiparameter Fluorescence Spectroscopy of Single Molecules. In Spectroscopy and Dynamics of Single Molecules; Elsevier: Amsterdam, The Netherlands, 2019; pp. 269–333. [Google Scholar]
- Sisamakis, E.; Valeri, A.; Kalinin, S.; Rothwell, P.J.; Seidel, C.A.M. Accurate Single-Molecule FRET Studies Using Multiparameter Fluorescence Detection. In Methods in Enzymology; Academic Press: New York, NY, USA, 2010; pp. 455–514. [Google Scholar]
- LeBlanc, S.; Kulkarni, P.; Weninger, K. Single Molecule FRET: A Powerful Tool to Study Intrinsically Disordered Proteins. Biomolecules 2018, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Holmstrom, E.D.; Holla, A.; Zheng, W.; Nettels, D.; Best, R.B.; Schuler, B. Accurate Transfer Efficiencies, Distance Distributions, and Ensembles of Unfolded and Intrinsically Disordered Proteins from Single-Molecule FRET. In Methods in Enzymology; Academic Press: New York, NY, USA, 2018; Volume 611, pp. 287–325. ISBN 9780128156490. [Google Scholar]
- Barth, A.; Opanasyuk, O.; Peulen, T.O.; Felekyan, S.; Kalinin, S.; Sanabria, H.; Seidel, C.A.M. Unraveling Multi-State Molecular Dynamics in Single-Molecule FRET Experiments. I. Theory of FRET-Lines. J. Chem. Phys. 2022, 156, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Metskas, L.A.; Rhoades, E. Single-Molecule FRET of Intrinsically Disordered Proteins. Annu. Rev. Phys. Chem. 2020, 71, 391–414. [Google Scholar] [CrossRef]
- Hofmann, H. Understanding Disordered and Unfolded Proteins Using Single-Molecule FRET and Polymer Theory. Methods Appl. Fluoresc. 2016, 4, 042003. [Google Scholar] [CrossRef] [PubMed]
- Schuler, B.; Soranno, A.; Hofmann, H.; Nettels, D. Single-Molecule FRET Spectroscopy and the Polymer Physics of Unfolded and Intrinsically Disordered Proteins. Annu. Rev. Biophys. 2016, 45, 207–231. [Google Scholar] [CrossRef]
- Tan, P.S.; Lemke, E.A. Probing Differential Binding Mechanisms of Phenylalanine-Glycine-Rich Nucleoporins by Single-Molecule FRET. Methods Enzymol. 2018, 611, 327–346. [Google Scholar] [CrossRef] [PubMed]
- Borgia, A.; Borgia, M.B.; Bugge, K.; Kissling, V.M.; Heidarsson, P.O.; Fernandes, C.B.; Sottini, A.; Soranno, A.; Buholzer, K.J.; Nettels, D.; et al. Extreme Disorder in an Ultrahigh-Affinity Protein Complex. Nature 2018, 555, 61–66. [Google Scholar] [CrossRef]
- Sakon, J.J.; Weninger, K.R. Detecting the Conformation of Individual Proteins in Live Cells. Nat. Methods 2010, 7, 203–205. [Google Scholar] [CrossRef]
- Choi, U.B.; McCann, J.J.; Weninger, K.R.; Bowen, M.E. Beyond the Random Coil: Stochastic Conformational Switching in Intrinsically Disordered Proteins. Structure 2011, 19, 566–576. [Google Scholar] [CrossRef]
- Mooney, S.M.; Qiu, R.; Kim, J.J.; Sacho, E.J.; Rajagopalan, K.; Johng, D.; Shiraishi, T.; Kulkarni, P.; Weninger, K.R. Cancer/Testis Antigen PAGE4, a Regulator of c-Jun Transactivation, Is Phosphorylated by Homeodomain-Interacting Protein Kinase 1, a Component of the Stress-Response Pathway. Biochemistry 2014, 53, 1670–1679. [Google Scholar] [CrossRef]
- He, Y.; Chen, Y.; Mooney, S.M.; Rajagopalan, K.; Bhargava, A.; Sacho, E.; Weninger, K.; Bryan, P.N.; Kulkarni, P.; Orban, J. Phosphorylation-Induced Conformational Ensemble Switching in an Intrinsically Disordered Cancer/Testis Antigen. J. Biol. Chem. 2015, 290, 25090–25102. [Google Scholar] [CrossRef] [PubMed]
- Gomes, G.-N.; Gradinaru, C.C. Insights into the Conformations and Dynamics of Intrinsically Disordered Proteins Using Single-Molecule Fluorescence. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2017, 1865, 1696–1706. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.B.; Sanabria, H.; Smirnova, T.; Bowen, M.E.; Weninger, K.R. Spontaneous Switching among Conformational Ensembles in Intrinsically Disordered Proteins. Biomolecules 2019, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, K.; Qiu, R.; Mooney, S.M.; Rao, S.; Shiraishi, T.; Sacho, E.; Huang, H.; Shapiro, E.; Weninger, K.R.; Kulkarni, P. The Stress-Response Protein Prostate-Associated Gene 4, Interacts with c-Jun and Potentiates Its Transactivation. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2014, 1842, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, H.; Soranno, A.; Borgia, A.; Gast, K.; Nettels, D.; Schuler, B. Polymer Scaling Laws of Unfolded and Intrinsically Disordered Proteins Quantified with Single-Molecule Spectroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 16155–16160. [Google Scholar] [CrossRef] [PubMed]
- Soranno, A.; Buchli, B.; Nettels, D.; Cheng, R.R.; Müller-Späth, S.; Pfeil, S.H.; Hoffmann, A.; Lipman, E.A.; Makarov, D.E.; Schuler, B. Quantifying Internal Friction in Unfolded and Intrinsically Disordered Proteins with Single-Molecule Spectroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 17800–17806. [Google Scholar] [CrossRef]
- Brucale, M.; Schuler, B.; Samorì, B. Single-Molecule Studies of Intrinsically Disordered Proteins. Chem. Rev. 2014, 114, 3281–3317. [Google Scholar] [CrossRef]
- Weninger, K.; Bowen, M.E.; Choi, U.B.; Chu, S.; Brunger, A.T. Accessory Proteins Stabilize the Acceptor Complex for Synaptobrevin, the 1:1 Syntaxin/SNAP-25 Complex. Structure 2008, 16, 308–320. [Google Scholar] [CrossRef]
- Miyagi, A.; Tsunaka, Y.; Uchihashi, T.; Mayanagi, K.; Hirose, S.; Morikawa, K.; Ando, T. Visualization of Intrinsically Disordered Regions of Proteins by High-Speed Atomic Force Microscopy. ChemPhysChem 2008, 9, 1859–1866. [Google Scholar] [CrossRef]
- Kodera, N.; Ando, T. Guide to Studying Intrinsically Disordered Proteins by High-Speed Atomic Force Microscopy. Methods 2022, 207, 44–56. [Google Scholar] [CrossRef]
- Kodera, N.; Ando, T. Visualization of Intrinsically Disordered Proteins by High-Speed Atomic Force Microscopy. Curr. Opin. Struct. Biol. 2022, 72, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Nwanochie, E.; Uversky, V.N. Structure Determination by Single-Particle Cryo-Electron Microscopy: Only the Sky (and Intrinsic Disorder) Is the Limit. Int. J. Mol. Sci. 2019, 20, 4186. [Google Scholar] [CrossRef] [PubMed]
- Benjin, X.; Ling, L. Developments, Applications, and Prospects of Cryo-electron Microscopy. Protein Sci. 2020, 29, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Abriata, L.A.; Dal Peraro, M. Will Cryo-Electron Microscopy Shift the Current Paradigm in Protein Structure Prediction? J. Chem. Inf. Model. 2020, 60, 2443–2447. [Google Scholar] [CrossRef] [PubMed]
- Musselman, C.A.; Kutateladze, T.G. Characterization of Functional Disordered Regions within Chromatin-Associated Proteins. iScience 2021, 24, 102070. [Google Scholar] [CrossRef]
- Bonomi, M.; Vendruscolo, M. Determination of Protein Structural Ensembles Using Cryo-Electron Microscopy. Curr. Opin. Struct. Biol. 2019, 56, 37–45. [Google Scholar] [CrossRef]
- Schmidt, M. Macromolecular Movies, Storybooks Written by Nature. Biophys. Rev. 2021, 13, 1191–1197. [Google Scholar] [CrossRef]
- Brändén, G.; Neutze, R. Advances and Challenges in Time-Resolved Macromolecular Crystallography. Science 2021, 373, eaba0954. [Google Scholar] [CrossRef]
- Malla, T.N.; Schmidt, M. Transient State Measurements on Proteins by Time-Resolved Crystallography. Curr. Opin. Struct. Biol. 2022, 74, 102376. [Google Scholar] [CrossRef]
- Frank, J. Time-Resolved Cryo-Electron Microscopy: Recent Progress. J. Struct. Biol. 2017, 200, 303–306. [Google Scholar] [CrossRef]
- Dandey, V.P.; Budell, W.C.; Wei, H.; Bobe, D.; Maruthi, K.; Kopylov, M.; Eng, E.T.; Kahn, P.A.; Hinshaw, J.E.; Kundu, N.; et al. Time-Resolved Cryo-EM Using Spotiton. Nat. Methods 2020, 17, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Hassan, M.; Islam, A.; Ahmad, F. A Review of Methods Available to Estimate Solvent-Accessible Surface Areas of Soluble Proteins in the Folded and Unfolded States. Curr. Protein Pept. Sci. 2014, 15, 456–476. [Google Scholar] [CrossRef] [PubMed]
- Hodge, E.A.; Benhaim, M.A.; Lee, K.K. Bridging Protein Structure, Dynamics, and Function Using Hydrogen/Deuterium-exchange Mass Spectrometry. Protein Sci. 2020, 29, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rempel, D.L.; Zhang, J.; Sharma, A.K.; Mirica, L.M.; Gross, M.L. Pulsed Hydrogen–Deuterium Exchange Mass Spectrometry Probes Conformational Changes in Amyloid Beta (Aβ) Peptide Aggregation. Proc. Natl. Acad. Sci. USA 2013, 110, 14604–14609. [Google Scholar] [CrossRef]
- Beveridge, R.; Calabrese, A.N. Structural Proteomics Methods to Interrogate the Conformations and Dynamics of Intrinsically Disordered Proteins. Front. Chem. 2021, 9, 603639. [Google Scholar] [CrossRef]
- Illes-Toth, E.; Rempel, D.L.; Gross, M.L. Pulsed Hydrogen–Deuterium Exchange Illuminates the Aggregation Kinetics of α-Synuclein, the Causative Agent for Parkinson’s Disease. ACS Chem. Neurosci. 2018, 9, 1469–1476. [Google Scholar] [CrossRef]
- Piersimoni, L.; Kastritis, P.L.; Arlt, C.; Sinz, A. Cross-Linking Mass Spectrometry for Investigating Protein Conformations and Protein–Protein Interactions─A Method for All Seasons. Chem. Rev. 2022, 122, 7500–7531. [Google Scholar] [CrossRef]
- Ubbiali, D.; Fratini, M.; Piersimoni, L.; Ihling, C.H.; Kipping, M.; Heilmann, I.; Iacobucci, C.; Sinz, A. Direct Observation of “Elongated” Conformational States in A-Synuclein upon Liquid-Liquid Phase Separation. Angew. Chem. Int. Ed. 2022, 134, e202205726. [Google Scholar] [CrossRef]
- Chen, D.; Drombosky, K.W.; Hou, Z.; Sari, L.; Kashmer, O.M.; Ryder, B.D.; Perez, V.A.; Woodard, D.R.; Lin, M.M.; Diamond, M.I.; et al. Tau Local Structure Shields an Amyloid-Forming Motif and Controls Aggregation Propensity. Nat. Commun 2019, 10, 2493. [Google Scholar] [CrossRef]
- Niemeyer, M.; Moreno Castillo, E.; Ihling, C.H.; Iacobucci, C.; Wilde, V.; Hellmuth, A.; Hoehenwarter, W.; Samodelov, S.L.; Zurbriggen, M.D.; Kastritis, P.L.; et al. Flexibility of Intrinsically Disordered Degrons in AUX/IAA Proteins Reinforces Auxin Co-Receptor Assemblies. Nat. Commun 2020, 11, 2277. [Google Scholar] [CrossRef]
- Suskiewicz, M.J.; Sussman, J.L.; Silman, I.; Shaul, Y. Context-Dependent Resistance to Proteolysis of Intrinsically Disordered Proteins. Protein Sci. 2011, 20, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Xue, B.; Sickmeier, M.D.; Meng, J.; Cortese, M.S.; Oldfield, C.J.; le Gall, T.; Dunker, A.K.; Uversky, V.N. High-Throughput Characterization of Intrinsic Disorder in Proteins from the Protein Structure Initiative. J. Struct. Biol. 2012, 180, 201–215. [Google Scholar] [CrossRef]
- Baker, E.S.; Luckner, S.R.; Krause, K.L.; Lambden, P.R.; Clarke, I.N.; Ward, V.K. Inherent Structural Disorder and Dimerisation of Murine Norovirus NS1-2 Protein. PLoS ONE 2012, 7, e30534. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, K.; Salladini, E.; O’Brien, D.P.; Brier, S.; Chenal, A.; Yacoubi, I.; Longhi, S. Structural Disorder and Induced Folding within Two Cereal, ABA Stress and Ripening (ASR) Proteins. Sci. Rep. 2017, 7, 15544. [Google Scholar] [CrossRef] [PubMed]
- Chemes, L.B.; Alonso, L.G.; Noval, M.G.; de Prat-Gay, G. Circular Dichroism Techniques for the Analysis of Intrinsically Disordered Proteins and Domains. In Intrinsically Disordered Protein Analysis; Humana Press: Totowa, NJ, USA, 2012; pp. 387–404. [Google Scholar]
- Micsonai, A.; Moussong, É.; Murvai, N.; Tantos, Á.; Tőke, O.; Réfrégiers, M.; Wien, F.; Kardos, J. Disordered–Ordered Protein Binary Classification by Circular Dichroism Spectroscopy. Front. Mol. Biosci. 2022, 9, 863141. [Google Scholar] [CrossRef] [PubMed]
- Ezerski, J.C.; Zhang, P.; Jennings, N.C.; Waxham, M.N.; Cheung, M.S. Molecular Dynamics Ensemble Refinement of Intrinsically Disordered Peptides According to Deconvoluted Spectra from Circular Dichroism. Biophys. J. 2020, 118, 1665–1678. [Google Scholar] [CrossRef]
- Uversky, V.N. Natively Unfolded Proteins: A Point Where Biology Waits for Physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef]
- Na, J.-H.; Lee, W.-K.; Yu, Y. How Do We Study the Dynamic Structure of Unstructured Proteins: A Case Study on Nopp140 as an Example of a Large, Intrinsically Disordered Protein. Int. J. Mol. Sci. 2018, 19, 381. [Google Scholar] [CrossRef]
- Auvray, F.; Dennetiere, D.; Giuliani, A.; Jamme, F.; Wien, F.; Nay, B.; Zirah, S.; Polack, F.; Menneglier, C.; Lagarde, B.; et al. Time Resolved Transient Circular Dichroism Spectroscopy Using Synchrotron Natural Polarization. Struct. Dyn. 2019, 6, 054307. [Google Scholar] [CrossRef]
- Natalello, A.; Ami, D.; Doglia, S.M. Fourier transform infrared spectroscopy of intrinsically disordered proteins: Measurement procedures and data analyses. In Intrinsically Disordered Protein Analysis; Humana Press: Totowa, NJ, USA, 2012; pp. 229–244. [Google Scholar]
- Uversky, V.N. Biophysical Methods to Investigate Intrinsically Disordered Proteins: Avoiding an “Elephant and Blind Men” Situation. Adv. Exp. Med. Biol. 2015, 870, 215–260. [Google Scholar]
- Sethi, A.; Anunciado, D.; Tian, J.; Vu, D.M.; Gnanakaran, S. Deducing Conformational Variability of Intrinsically Disordered Proteins from Infrared Spectroscopy with Bayesian Statistics. Chem. Phys. 2013, 422, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Takekiyo, T.; Yamada, N.; Nakazawa, C.T.; Amo, T.; Asano, A.; Yoshimura, Y. Formation of A-synuclein Aggregates in Aqueous Ethylammonium Nitrate Solutions. Biopolymers 2020, 111, e23352. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Isaacs, N.W.; Hecht, L.; Barron, L.D. Raman Optical Activity: A Tool for Protein Structure Analysis. Structure 2005, 13, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Sane, S.U.; Cramer, S.M.; Przybycien, T.M. A Holistic Approach to Protein Secondary Structure Characterization Using Amide I Band Raman Spectroscopy. Anal. Biochem. 1999, 269, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Berjot, M.; Marx, J.; Alix, A.J.P. Determination of the Secondary Structure of Proteins from the Raman Amide I Band: The Reference Intensity Profiles Method. J. Raman Spectrosc. 1987, 18, 289–300. [Google Scholar] [CrossRef]
- Maiti, N.C.; Apetri, M.M.; Zagorski, M.G.; Carey, P.R.; Anderson, V.E. Raman Spectroscopic Characterization of Secondary Structure in Natively Unfolded Proteins: α-Synuclein. J. Am. Chem. Soc. 2004, 126, 2399–2408. [Google Scholar] [CrossRef]
- Syme, C.D.; Blanch, E.W.; Holt, C.; Jakes, R.; Goedert, M.; Hecht, L.; Barron, L.D. A Raman Optical Activity Study of Rheomorphism in Caseins, Synucleins and Tau: New Insight into the Structure and Behaviour of Natively Unfolded Proteins. Eur. J. Biochem. 2002, 269, 148–156. [Google Scholar] [CrossRef]
- Stuchfield, D.; France, A.P.; Migas, L.G.; Thalhammer, A.; Bremer, A.; Bellina, B.; Barran, P.E. The Use of Mass Spectrometry to Examine IDPs: Unique Insights and Caveats, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 611, ISBN 9780128156490. [Google Scholar]
- Santambrogio, C.; Natalello, A.; Brocca, S.; Ponzini, E.; Grandori, R. Conformational Characterization and Classification of Intrinsically Disordered Proteins by Native Mass Spectrometry and Charge-State Distribution Analysis. Proteomics 2019, 19, e1800060. [Google Scholar] [CrossRef]
- Lento, C.; Wilson, D.J. Subsecond Time-Resolved Mass Spectrometry in Dynamic Structural Biology. Chem. Rev. 2022, 122, 7624–7646. [Google Scholar] [CrossRef]
- Salvay, A.G.; Communie, G.; Ebel, C. Sedimentation Velocity Analytical Ultracentrifugation for Intrinsically Disordered Proteins; John Wiely & Sons: New York, NY, USA, 2012; pp. 91–105. [Google Scholar]
- Gast, K.; Fiedler, C. Dynamic and Static Light Scattering of Intrinsically Disordered Proteins. In Intrinsically Disordered Protein Analysis; Springer: New York, NY, USA, 2012; pp. 137–161. [Google Scholar]
- Al-Ghobashy, M.A.; Mostafa, M.M.; Abed, H.S.; Fathalla, F.A.; Salem, M.Y. Correlation between Dynamic Light Scattering and Size Exclusion High Performance Liquid Chromatography for Monitoring the Effect of PH on Stability of Biopharmaceuticals. J. Chromatogr. B 2017, 1060, 1–9. [Google Scholar] [CrossRef]
- Leite, J.P.; Gimeno, A.; Taboada, P.; Jiménez-Barbero, J.J.; Gales, L. Dissection of the Key Steps of Amyloid-β Peptide 1–40 Fibrillogenesis. Int. J. Biol. Macromol. 2020, 164, 2240–2246. [Google Scholar] [CrossRef] [PubMed]
- Hochmair, J.; Exner, C.; Betzel, C.; Mandelkow, E.; Wegmann, S. Light Microscopy and Dynamic Light Scattering to Study Liquid-Liquid Phase Separation of Tau Proteins In Vitro. In Protein Aggregation; Humana: New York, NY, USA, 2023; pp. 225–243. [Google Scholar]
- Tomasso, M.E.; Tarver, M.J.; Devarajan, D.; Whitten, S.T. Hydrodynamic Radii of Intrinsically Disordered Proteins Determined from Experimental Polyproline II Propensities. PLoS Comput. Biol. 2016, 12, e1004686. [Google Scholar] [CrossRef] [PubMed]
- Wang, W. Recent Advances in Atomic Molecular Dynamics Simulation of Intrinsically Disordered Proteins. Phys. Chem. Chem. Phys. 2021, 23, 777–784. [Google Scholar] [CrossRef]
- Dokholyan, N.V. Experimentally-Driven Protein Structure Modeling. J. Proteom. 2020, 220, 103777. [Google Scholar] [CrossRef]
- Hsu, C.C.; Buehler, M.J.; Tarakanova, A. The Order-Disorder Continuum: Linking Predictions of Protein Structure and Disorder through Molecular Simulation. Sci. Rep. 2020, 10, 2068. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B. Computational and Theoretical Advances in Studies of Intrinsically Disordered Proteins. Curr. Opin. Struct. Biol. 2017, 42, 147–154. [Google Scholar] [CrossRef]
- Lin, X.; Roy, S.; Jolly, M.K.; Bocci, F.; Schafer, N.P.; Tsai, M.-Y.; Chen, Y.; He, Y.; Grishaev, A.; Weninger, K.; et al. PAGE4 and Conformational Switching: Insights from Molecular Dynamics Simulations and Implications for Prostate Cancer. J. Mol. Biol. 2018, 430, 2422–2438. [Google Scholar] [CrossRef]
- Kasahara, K.; Terazawa, H.; Takahashi, T.; Higo, J. Studies on Molecular Dynamics of Intrinsically Disordered Proteins and Their Fuzzy Complexes: A Mini-Review. Comput. Struct. Biotechnol. J. 2019, 17, 712–720. [Google Scholar] [CrossRef]
- Lin, X.; Kulkarni, P.; Bocci, F.; Schafer, N.P.; Roy, S.; Tsai, M.Y.; He, Y.; Chen, Y.; Rajagopalan, K.; Mooney, S.M.; et al. Structural and Dynamical Order of a Disordered Protein: Molecular Insights into Conformational Switching of Page4 at the Systems Level. Biomolecules 2019, 9, 77. [Google Scholar] [CrossRef]
- Blackledge, M.; Ferrage, F.; Kadeřávek, P.; Salvi, N.; Zapletal, V.; Jaseňáková, Z.; Zachrdla, M.; Padrta, P.; Narasimhan, S.; Marquardsen, T.; et al. Convergent Views on Disordered Protein Dynamics from NMR and Computational Approaches. Biophys. J. 2022, 121, 3785–3794. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence Complexity of Disordered Protein. Proteins Struct. Funct. Genet. 2001, 42, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting Heterogeneous Sequence Properties Improves Prediction of Protein Disorder. Proteins Struct. Funct. Bioinform. 2005, 61, 176–182. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A Meta-Predictor of Intrinsically Disordered Amino Acids. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-Dependent Prediction of Protein Intrinsic Disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Sirovetz, B.J.; Schafer, N.P.; Wolynes, P.G. Protein Structure Prediction: Making AWSEM AWSEM-ER by Adding Evolutionary Restraints. Proteins Struct. Funct. Bioinform. 2017, 85, 2127–2142. [Google Scholar] [CrossRef]
- Kmiecik, S.; Gront, D.; Kolinski, M.; Wieteska, L.; Dawid, A.E.; Kolinski, A. Coarse-Grained Protein Models and Their Applications. Chem. Rev. 2016, 116, 7898–7936. [Google Scholar] [CrossRef]
- Chen, M.; Lin, X.; Zheng, W.; Onuchic, J.N.; Wolynes, P.G. Protein Folding and Structure Prediction from the Ground Up: The Atomistic Associative Memory, Water Mediated, Structure and Energy Model. J. Phys. Chem. B 2016, 120, 8557–8565. [Google Scholar] [CrossRef]
- Chen, M.; Lin, X.; Lu, W.; Onuchic, J.N.; Wolynes, P.G. Protein Folding and Structure Prediction from the Ground Up II: AAWSEM for α/β Proteins. J. Phys. Chem. B 2017, 121, 3473–3482. [Google Scholar] [CrossRef]
- Ruff, K.M.; Pappu, R.v. AlphaFold and Implications for Intrinsically Disordered Proteins. J. Mol. Biol. 2021, 433, 167208. [Google Scholar] [CrossRef]
- Ehm, T.; Shinar, H.; Meir, S.; Sekhon, A.; Sethi, V.; Morgan, I.L.; Rahamim, G.; Saleh, O.A.; Beck, R. Intrinsically Disordered Proteins at the Nano-Scale. Nano Futures 2021, 5, 1–15. [Google Scholar] [CrossRef]
- Aznauryan, M.; Delgado, L.; Soranno, A.; Nettels, D.; Huang, J.; Labhardt, A.M.; Grzesiek, S.; Schuler, B. Comprehensive Structural and Dynamical View of an Unfolded Protein from the Combination of Single-Molecule FRET, NMR, and SAXS. Proc. Natl. Acad. Sci. USA 2016, 113, E5389–E5398. [Google Scholar] [CrossRef] [PubMed]
- Dedmon, M.M.; Lindorff-Larsen, K.; Christodoulou, J.; Vendruscolo, M.; Dobson, C.M. Mapping Long-Range Interactions in α-Synuclein Using Spin-Label NMR and Ensemble Molecular Dynamics Simulations. J. Am. Chem. Soc. 2005, 127, 476–477. [Google Scholar] [CrossRef] [PubMed]
- Ferrie, J.J.; Haney, C.M.; Yoon, J.; Pan, B.; Lin, Y.-C.; Fakhraai, Z.; Rhoades, E.; Nath, A.; Petersson, E.J. Using a FRET Library with Multiple Probe Pairs To Drive Monte Carlo Simulations of α-Synuclein. Biophys. J. 2018, 114, 53–64. [Google Scholar] [CrossRef]
- Hamilton, G.L.; Saikia, N.; Basak, S.; Welcome, F.S.; Wu, F.; Kubiak, J.; Zhang, C.; Hao, Y.; Seidel, C.A.; Ding, F.; et al. Fuzzy Supertertiary Interactions within PSD-95 Enable Ligand Binding. eLife 2022, 11, e77242. [Google Scholar] [CrossRef]
- Thomasen, F.E.; Lindorff-Larsen, K. Conformational Ensembles of Intrinsically Disordered Proteins and Flexible Multidomain Proteins. Biochem. Soc. Trans. 2022, 50, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Saikia, N.; Yanez-Orozco, I.S.; Qiu, R.; Hao, P.; Milikisiyants, S.; Ou, E.; Hamilton, G.L.; Weninger, K.R.; Smirnova, T.I.; Sanabria, H.; et al. Integrative Structural Dynamics Probing of the Conformational Heterogeneity in Synaptosomal-Associated Protein 25. Cell Rep. Phys. Sci. 2021, 2, 100616. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.B.; Xiao, S.; Wollmuth, L.P.; Bowen, M.E. Effect of Src Kinase Phosphorylation on Disordered C-Terminal Domain of N-Methyl-D-Aspartic Acid (NMDA) Receptor Subunit GluN2B Protein. J. Biol. Chem. 2011, 286, 29904–29912. [Google Scholar] [CrossRef]
- Meng, F.; Bellaiche, M.M.J.; Kim, J.-Y.; Zerze, G.H.; Best, R.B.; Chung, H.S. Highly Disordered Amyloid-β Monomer Probed by Single-Molecule FRET and MD Simulation. Biophys. J. 2018, 114, 870–884. [Google Scholar] [CrossRef]
- Brunger, A.T.; Strop, P.; Vrljic, M.; Chu, S.; Weninger, K.R. Three-Dimensional Molecular Modeling with Single Molecule FRET. J. Struct. Biol. 2011, 173, 497–505. [Google Scholar] [CrossRef]
- Choi, U.B.; Strop, P.; Vrljic, M.; Chu, S.; Brunger, A.T.; Weninger, K.R. Single-Molecule FRET–Derived Model of the Synaptotagmin 1–SNARE Fusion Complex. Nat. Struct. Mol. Biol. 2010, 17, 318–324. [Google Scholar] [CrossRef]
- Lerner, E.; Barth, A.; Hendrix, J.; Ambrose, B.; Birkedal, V.; Blanchard, S.C.; Börner, R.; Sung Chung, H.; Cordes, T.; Craggs, T.D.; et al. FRET-Based Dynamic Structural Biology: Challenges, Perspectives and an Appeal for Open-Science Practices. eLife 2021, 10, e60416. [Google Scholar] [CrossRef] [PubMed]
- Mercadante, D.; Milles, S.; Fuertes, G.; Svergun, D.I.; Lemke, E.A.; Gräter, F. Kirkwood–Buff Approach Rescues Overcollapse of a Disordered Protein in Canonical Protein Force Fields. J. Phys. Chem. B 2015, 119, 7975–7984. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Yagi, N.; Nakatani, R.; Sekiguchi, H.; So, M.; Yagi, H.; Ohta, N.; Nagai, Y.; Goto, Y.; Mochizuki, H. A Small-Angle X-ray Scattering Study of Alpha-Synuclein from Human Red Blood Cells. Sci. Rep. 2016, 6, 30473. [Google Scholar] [CrossRef]
- Schweers, O.; Schönbrunn-Hanebeck, E.; Marx, A.; Mandelkow, E. Structural Studies of Tau Protein and Alzheimer Paired Helical Filaments Show No Evidence for Beta-Structure. J. Biol. Chem. 1994, 269, 24290–24297. [Google Scholar] [CrossRef]
- Gomes, G.-N.W.; Krzeminski, M.; Namini, A.; Martin, E.W.; Mittag, T.; Head-Gordon, T.; Forman-Kay, J.D.; Gradinaru, C.C. Conformational Ensembles of an Intrinsically Disordered Protein Consistent with NMR, SAXS, and Single-Molecule FRET. J. Am. Chem. Soc. 2020, 142, 15697–15710. [Google Scholar] [CrossRef]
- Liu, B.; Chia, D.; Csizmok, V.; Farber, P.; Forman-Kay, J.D.; Gradinaru, C.C. The Effect of Intrachain Electrostatic Repulsion on Conformational Disorder and Dynamics of the Sic1 Protein. J. Phys. Chem. B 2014, 118, 4088–4097. [Google Scholar] [CrossRef] [PubMed]
- Gomes, G.-N.W.; Namini, A.; Gradinaru, C.C. Integrative Conformational Ensembles of Sic1 Using Different Initial Pools and Optimization Methods. Front. Mol. Biosci. 2022, 9, 910956. [Google Scholar] [CrossRef]
- Sala, D.; Cosentino, U.; Ranaudo, A.; Greco, C.; Moro, G. Dynamical Behavior and Conformational Selection Mechanism of the Intrinsically Disordered Sic1 Kinase-Inhibitor Domain. Life 2020, 10, 110. [Google Scholar] [CrossRef]
- Nash, P.; Tang, X.; Orlicky, S.; Chen, Q.; Gertler, F.B.; Mendenhall, M.D.; Sicheri, F.; Pawson, T.; Tyers, M. Multisite Phosphorylation of a CDK Inhibitor Sets a Threshold for the Onset of DNA Replication. Nature 2001, 414, 514–521. [Google Scholar] [CrossRef]
- Mittag, T.; Orlicky, S.; Choy, W.; Tang, X.; Lin, H.; Sicheri, F.; Kay, L.E.; Tyers, M.; Forman-Kay, J.D. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl. Acad. Sci. USA 2008, 105, 17772–17777. [Google Scholar] [CrossRef]
- Chan-Yao-Chong, M.; Deville, C.; Pinet, L.; van Heijenoort, C.; Durand, D.; Ha-Duong, T. Structural Characterization of N-WASP Domain V Using MD Simulations with NMR and SAXS Data. Biophys. J. 2019, 116, 1216–1227. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.D.H.; Kwiatkowski, A.V. Control of Actin Dynamics by Allosteric Regulation of Actin Binding Proteins. Int. Rev. Cell Mol. Biol. 2013, 303, 1–25. [Google Scholar]
- Robustelli, P.; Piana, S.; Shaw, D.E. Developing a Molecular Dynamics Force Field for Both Folded and Disordered Protein States. Proc. Natl. Acad. Sci. USA 2018, 115, E4758–E4766. [Google Scholar] [CrossRef] [PubMed]
- Chan-Yao-Chong, M.; Durand, D.; Ha-Duong, T. Molecular Dynamics Simulations Combined with Nuclear Magnetic Resonance and/or Small-Angle X-ray Scattering Data for Characterizing Intrinsically Disordered Protein Conformational Ensembles. J. Chem. Inf. Model. 2019, 59, 1743–1758. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, S.; Gapsys, V.; Gajda, M.J.; Zweckstetter, M.; de Groot, B.L.; Grubmüller, H. Structural Ensembles of Intrinsically Disordered Proteins Depend Strongly on Force Field: A Comparison to Experiment. J. Chem. Theory Comput. 2015, 11, 5513–5524. [Google Scholar] [CrossRef] [PubMed]
- Ou, L.; Waddell, M.B.; Kriwacki, R.W. Mechanism of Cell Cycle Entry Mediated by the Intrinsically Disordered Protein P27 Kip1. ACS Chem. Biol. 2012, 7, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Tsytlonok, M.; Hemmen, K.; Hamilton, G.; Kolimi, N.; Felekyan, S.; Seidel, C.A.M.; Tompa, P.; Sanabria, H. Specific Conformational Dynamics and Expansion Underpin a Multi-Step Mechanism for Specific Binding of P27 with Cdk2/Cyclin A. J. Mol. Biol. 2020, 432, 2998–3017. [Google Scholar] [CrossRef] [PubMed]
- Tsytlonok, M.; Sanabria, H.; Wang, Y.; Felekyan, S.; Hemmen, K.; Phillips, A.H.; Yun, M.-K.; Waddell, M.B.; Park, C.-G.; Vaithiyalingam, S.; et al. Dynamic Anticipation by Cdk2/Cyclin A-Bound P27 Mediates Signal Integration in Cell Cycle Regulation. Nat. Commun. 2019, 10, 1676. [Google Scholar] [CrossRef]
- Das, R.K.; Huang, Y.; Phillips, A.H.; Kriwacki, R.W.; Pappu, R.v. Cryptic Sequence Features within the Disordered Protein P27 Kip1 Regulate Cell Cycle Signaling. Proc. Natl. Acad. Sci. USA 2016, 113, 5616–5621. [Google Scholar] [CrossRef]
- Zeng, Y.; He, Y.; Yang, F.; Mooney, S.M.; Getzenberg, R.H.; Orban, J.; Kulkarni, P. The Cancer/Testis Antigen Prostate-Associated Gene 4 (PAGE4) Is a Highly Intrinsically Disordered Protein. J. Biol. Chem. 2011, 286, 13985–13994. [Google Scholar] [CrossRef]
- Kulkarni, P.; Jolly, M.K.; Jia, D.; Mooney, S.M.; Bhargava, A.; Kagohara, L.T.; Chen, Y.; Hao, P.; He, Y.; Veltri, R.W.; et al. Phosphorylation-Induced Conformational Dynamics in an Intrinsically Disordered Protein and Potential Role in Phenotypic Heterogeneity. Proc. Natl. Acad. Sci. USA 2017, 114, E2644–E2653. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.; Dunker, A.; Weninger, K.; Orban, J. Prostate-Associated Gene 4 (PAGE4), an Intrinsically Disordered Cancer/Testis Antigen, Is a Novel Therapeutic Target for Prostate Cancer. Asian J. Androl. 2016, 18, 695. [Google Scholar] [CrossRef] [PubMed]
- Salgia, R.; Jolly, M.; Dorff, T.; Lau, C.; Weninger, K.; Orban, J.; Kulkarni, P. Prostate-Associated Gene 4 (PAGE4): Leveraging the Conformational Dynamics of a Dancing Protein Cloud as a Therapeutic Target. J. Clin. Med. 2018, 7, 156. [Google Scholar] [CrossRef]
- Jolly, M.K.; Kulkarni, P.; Weninger, K.; Orban, J.; Levine, H. Phenotypic Plasticity, Bet-Hedging, and Androgen Independence in Prostate Cancer: Role of Non-Genetic Heterogeneity. Front. Oncol. 2018, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Kulkarni, P. Intrinsically Disordered Proteins: Chronology of a Discovery. Biophys. Chem. 2021, 279, 106694. [Google Scholar] [CrossRef]
- Uversky, V.N. Unusual Biophysics of Intrinsically Disordered Proteins. Biochim. Biophys. Acta 2013, 1834, 932–951. [Google Scholar] [CrossRef]
- Kulkarni, P.; Bhattacharya, S.; Achuthan, S.; Behal, A.; Jolly, M.K.; Kotnala, S.; Mohanty, A.; Rangarajan, G.; Salgia, R.; Uversky, V. Intrinsically Disordered Proteins: Critical Components of the Wetware. Chem. Rev. 2022, 122, 6614–6633. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evans, R.; Ramisetty, S.; Kulkarni, P.; Weninger, K. Illuminating Intrinsically Disordered Proteins with Integrative Structural Biology. Biomolecules 2023, 13, 124. https://doi.org/10.3390/biom13010124
Evans R, Ramisetty S, Kulkarni P, Weninger K. Illuminating Intrinsically Disordered Proteins with Integrative Structural Biology. Biomolecules. 2023; 13(1):124. https://doi.org/10.3390/biom13010124
Chicago/Turabian StyleEvans, Rachel, Sravani Ramisetty, Prakash Kulkarni, and Keith Weninger. 2023. "Illuminating Intrinsically Disordered Proteins with Integrative Structural Biology" Biomolecules 13, no. 1: 124. https://doi.org/10.3390/biom13010124
APA StyleEvans, R., Ramisetty, S., Kulkarni, P., & Weninger, K. (2023). Illuminating Intrinsically Disordered Proteins with Integrative Structural Biology. Biomolecules, 13(1), 124. https://doi.org/10.3390/biom13010124





