Defining Two Chemosensory Arrays in Shewanella oneidensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Construction of Deletion Mutants
2.3. Plasmid Construction
2.4. RT-PCR Experiments
2.4.1. RNA Extraction
2.4.2. cDNA Synthesis
2.4.3. PCR Amplification
2.4.4. Cell Fractionation and Preparation of Protein Extracts
2.4.5. Visible Spectrophotometry of Purified Aer2so
2.4.6. Blue Native Electrophoresis (BNE) Analysis and Immunodetection
2.4.7. Mass Spectrometry-Based Proteomic Analyses
2.4.8. Swimming Plate Assays
2.4.9. Fluorescent Microscopy
3. Results
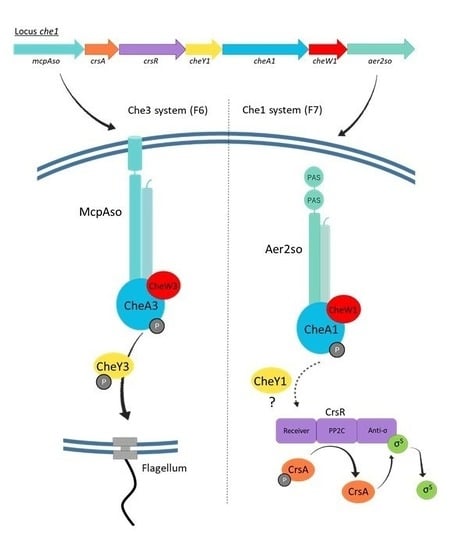
3.1. MCPs and Chemosensory Systems in S. oneidensis
3.2. Aer2so Is Related to the Che1 Machinery
3.3. Che1 Forms Complexes In Vivo
3.4. McpAso Forms Clusters with CheA3
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gumerov, V.M.; Andrianova, E.P.; Zhulin, I.B. Diversity of bacterial chemosensory systems. Curr. Opin. Microbiol. 2021, 61, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Wadhams, G.H.; Armitage, J.P. Making sense of it all: Bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 2004, 5, 1024–1037. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J.S.; Hazelbauer, G.L.; Falke, J.J. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015, 23, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Burt, A.; Cassidy, C.K.; Stansfeld, P.J.; Gutsche, I. Alternative Architecture of the E. coli Chemosensory Array. Biomolecules 2021, 11, 495. [Google Scholar] [CrossRef] [PubMed]
- Arapov, T.D.; Saldaña, R.C.; Sebastian, A.L.; Ray, W.K.; Helm, R.F.; Scharf, B.E. Cellular Stoichiometry of Chemotaxis Proteins in Sinorhizobium meliloti. J. Bacteriol. 2020, 202, e00141-20. [Google Scholar] [CrossRef]
- Yang, W.; Alvarado, A.; Glatter, T.; Ringgaard, S.; Briegel, A. Baseplate variability of Vibrio cholerae chemoreceptor arrays. Proc. Natl Acad. Sci. USA 2018, 15, 13365–13370. [Google Scholar] [CrossRef]
- Wuichet, K.; Zhulin, I.B. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 2010, 3, ra50. [Google Scholar] [CrossRef]
- Briegel, A.; Ortega, D.R.; Tocheva, E.I.; Wuichet, K.; Li, Z.; Chen, S.; Müller, A.; Iancu, C.V.; Murphy, G.E.; Dobro, M.J.; et al. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. USA 2009, 106, 17181–17186. [Google Scholar] [CrossRef]
- Sourjik, V.; Wingreen, N.S. Responding to chemical gradients: Bacterial chemotaxis. Curr. Opin. Cell Biol. 2012, 24, 262–268. [Google Scholar] [CrossRef]
- Matilla, M.A.; Martín-Mora, D.; Gavira, J.A.; Krell, T. Pseudomonas aeruginosa as a Model To Study Chemosensory Pathway Signaling. Microbiol. Mol. Biol. Rev. 2021, 85, e00151-20. [Google Scholar] [CrossRef]
- Ortega, D.R.; Kjaer, A.; Briegel, A. The chemosensory systems of Vibrio cholerae. Mol. Microbiol. 2020, 114, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Hazelbauer, G.L.; Falke, J.J.; Parkinson, J.S. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem. Sci. 2008, 33, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Briegel, A. Diversity of Bacterial Chemosensory Arrays. Trends Microbiol. 2020, 28, 68–80. [Google Scholar] [CrossRef] [PubMed]
- O’Neal, L.; Gullett, J.M.; Aksenova, A.; Hubler, A.; Briegel, A.; Ortega, D.; Kjær, A.; Jensen, G.; Alexandre, G. Distinct Chemotaxis Protein Paralogs Assemble into Chemoreceptor Signaling Arrays to Coordinate Signaling Output. Available online: https://journals.asm.org/doi/10.1128/mBio.01757–19 (accessed on 28 November 2022).
- Alexander, R.P.; Zhulin, I.B. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA 2007, 104, 2885–2890. [Google Scholar] [CrossRef]
- Salah Ud-Din, A.I.M.; Roujeinikova, A. Methyl-accepting chemotaxis proteins: A core sensing element in prokaryotes and archaea. Cell. Mol. Life Sci. 2017, 74, 3293–3303. [Google Scholar] [CrossRef] [PubMed]
- Ortega, D.R.; Fleetwood, A.D.; Krell, T.; Harwood, C.S.; Jensen, G.J.; Zhulin, I.B. Assigning chemoreceptors to chemosensory pathways in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 12809–12814. [Google Scholar] [CrossRef]
- Ortega, D.R.; Yang, W.; Subramanian, P.; Mann, P.; Kjær, A.; Chen, S.; Watts, K.J.; Pirbadian, S.; Collins, D.A.; Kooger, R.; et al. Repurposing a chemosensory macromolecular machine. Nat. Commun. 2020, 11, 2041. [Google Scholar] [CrossRef] [PubMed]
- Burt, A.; Cassidy, C.K.; Ames, P.; Bacia-Verloop, M.; Baulard, M.; Huard, K.; Luthey-Schulten, Z.; Desfosses, A.; Stansfeld, P.J.; Margolin, W.; et al. Complete structure of the chemosensory array core signalling unit in an E. coli minicell strain. Nat. Commun. 2020, 11, 743. [Google Scholar] [CrossRef]
- Gullett, J.M.; Bible, A.; Alexandre, G. Distinct Domains of CheA Confer Unique Functions in Chemotaxis and Cell Length in Azospirillum brasilense Sp7. J. Bacteriol. 2017, 199, e00189-17. [Google Scholar] [CrossRef]
- Lemaire, O.N.; Méjean, V.; Iobbi-Nivol, C. The Shewanella genus: Ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol. Rev. 2020, 44, 155–170. [Google Scholar] [CrossRef]
- Armitano, J.; Méjean, V.; Jourlin-Castelli, C. Aerotaxis governs floating biofilm formation in Shewanella oneidensis. Environ. Microbiol. 2013, 15, 3108–3118. [Google Scholar] [PubMed]
- Boyeldieu, A.; Ali Chaouche, A.; Ba, M.; Honoré, F.A.; Méjean, V.; Jourlin-Castelli, C. The phosphorylated regulator of chemotaxis is crucial throughout biofilm biogenesis in Shewanella oneidensis. NPJ Biofilms Microbiomes 2020, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Romine, M.F.; Ward, M.J. Identification and analysis of a highly conserved chemotaxis gene cluster in Shewanella species. FEMS Microbiol. Lett. 2007, 273, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Romine, M.F.; Carlson, T.S.; Norbeck, A.D.; McCue, L.A.; Lipton, M.S. Identification of mobile elements and pseudogenes in the Shewanella oneidensis MR-1 genome. Appl. Environ. Microbiol. 2008, 74, 3257–3265. [Google Scholar] [CrossRef]
- Lemaire, O.N.; Honoré, F.A.; Tempel, S.; Fortier, E.M.; Leimkühler, S.; Méjean, V.; Iobbi-Nivol, C. Shewanella decolorationis LDS1 Chromate Resistance. Appl. Environ. Microbiol. 2019, 85, e00777-19. [Google Scholar] [CrossRef]
- Bouillet, S.; Arabet, D.; Jourlin-Castelli, C.; Méjean, V.; Iobbi-Nivol, C. Regulation of σ factors by conserved partner switches controlled by divergent signalling systems. Environ. Microbiol. Rep. 2018, 10, 127–139. [Google Scholar] [CrossRef]
- Bouillet, S.; Genest, O.; Jourlin-Castelli, C.; Fons, M.; Mejean, V.; Iobbi-Nivol, C. The General Stress Response σS Is Regulated by a Partner Switch in the Gram-negative Bacterium Shewanella oneidensis. J. Biol. Chem. 2016, 291, 26151–26163. [Google Scholar] [CrossRef]
- Bouillet, S.; Genest, O.; Méjean, V.; Iobbi-Nivol, C. Protection of the general stress response σS factor by the CrsR regulator allows a rapid and efficient adaptation of Shewanella oneidensis. J. Biol. Chem. 2017, 292, 14921–14928. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harb Lab Press: Long Island, NY, USA, 1972. [Google Scholar]
- Bordi, C.; Iobbi-Nivol, C.; Méjean, V.; Patte, J.C. Effects of ISSo2 insertions in structural and regulatory genes of the trimethylamine oxide reductase of Shewanella oneidensis. J. Bacteriol. 2003, 185, 2042–2045. [Google Scholar] [CrossRef]
- Baraquet, C.; Théraulaz, L.; Iobbi-Nivol, C.; Méjean, V.; Jourlin-Castelli, C. Unexpected chemoreceptors mediate energy taxis towards electron acceptors in Shewanella oneidensis. Mol. Microbiol. 2009, 73, 278–290. [Google Scholar] [CrossRef]
- Herrero, M.; de Lorenzo, V.; Timmis, K.N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1990, 172, 6557–6567. [Google Scholar] [CrossRef] [PubMed]
- Appleyard, R.K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics 1954, 39, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J.S.; Houts, S.E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 1982, 151, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.S.; Saw, J.H.; Hou, S.; Larsen, R.W.; Watts, K.J.; Johnson, M.S.; Zimmer, M.A.; Ordal, G.W.; Taylor, B.L.; Alam, M. Aerotactic responses in bacteria to photoreleased oxygen. FEMS Microbiol. Lett. 2002, 217, 237–242. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Figurski, D.H.; Helinski, D.R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 1979, 76, 1648–1652. [Google Scholar] [CrossRef] [PubMed]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Boyeldieu, A.; Ali Chaouche, A.; Méjean, V.; Jourlin-Castelli, C. Combining two optimized and affordable methods to assign chemoreceptors to a specific signal. Anal. Biochem. 2021, 620, 114139. [Google Scholar] [CrossRef]
- Markwell, J.P.; Lascelles, J. Membrane-bound, pyridine nucleotide-independent L-lactate dehydrogenase of Rhodopseudomonas sphaeroides. J. Bacteriol. 1978, 133, 593–600. [Google Scholar] [CrossRef]
- Schägger, H.; Cramer, W.A.; von Jagow, G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994, 217, 220–230. [Google Scholar] [CrossRef]
- Boughanemi, S.; Lyonnet, J.; Infossi, P.; Bauzan, M.; Kosta, A.; Lignon, S.; Giudici-Orticoni, M.T.; Guiral, M. Microbial oxidative sulfur metabolism: Biochemical evidence of the membrane-bound heterodisulfide reductase-like complex of the bacterium Aquifex aeolicus. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]
- Taylor, B.L.; Watts, K.J.; Johnson, M.S. Oxygen and redox sensing by two-component systems that regulate behavioral responses: Behavioral assays and structural studies of aer using in vivo disulfide cross-linking. Methods Enzymol. 2007, 422, 190–232. [Google Scholar] [PubMed]
- Gumerov, V.M.; Ortega, D.R.; Adebali, O.; Ulrich, L.E.; Zhulin, I.B. MiST 3.0: An updated microbial signal transduction database with an emphasis on chemosensory systems. Nucleic Acids Res. 2020, 48, D459–D644. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. The Clustal Omega Multiple Alignment Package. Methods Mol. Biol. 2021, 2231, 3–16. [Google Scholar] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Iobbi-Nivol, C.; Crooke, H.; Griffiths, L.; Grove, J.; Hussain, H.; Pommier, J.; Mejean, V.; Cole, J.A. A reassessment of the range of c-type cytochromes synthesized by Escherichia coli K-12. FEMS Microbiol. Lett. 1994, 119, 89–94. [Google Scholar] [CrossRef]
- Greer-Phillips, S.E.; Sukomon, N.; Chua, T.K.; Johnson, M.S.; Crane, B.R.; Watts, K.J. 2The Aer2 Receptor from Vibrio Cholerae Is a Dual PAS-heme Oxygen Sensor. Molecular Microbiology. Available online: https://pubmed.ncbi.nlm.nih.gov/29719085/ (accessed on 7 September 2022).
- Wittig, I.; Schägger, H. Features and applications of blue-native and clear-native electrophoresis. Proteomics 2008, 8, 3974–3990. [Google Scholar] [CrossRef]
- Güvener, Z.T.; Tifrea, D.F.; Harwood, C.S. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol. Microbiol. 2006, 61, 106–118. [Google Scholar] [CrossRef]
- Boyeldieu, A.; Poli, J.P.; Ali Chaouche, A.; Fierobe, H.P.; Giudici-Orticoni, M.T.; Méjean, V.; Jourlin-Castelli, C. Multiple detection of both attractants and repellents by the dCache-chemoreceptor SO_1056 of Shewanella oneidensis. FEBS J. 2022, 289, 6752–6766. [Google Scholar] [CrossRef]
- Jones, C.W.; Armitage, J.P. Positioning of bacterial chemoreceptors. Trends Microbiol. 2015, 23, 247–256. [Google Scholar] [CrossRef]
- Mauriello, E.M.F.; Jones, C.; Moine, A.; Armitage, J.P. Cellular targeting and segregation of bacterial chemosensory systems. FEMS Microbiol. Rev. 2018, 42, 462–476. [Google Scholar] [CrossRef]
- Mauriello, E.M.F.; Astling, D.P.; Sliusarenko, O.; Zusman, D.R. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc. Natl. Acad. Sci. USA 2009, 106, 4852–4857. [Google Scholar] [CrossRef] [PubMed]
- Herrera Seitz, M.K.; Frank, V.; Massazza, D.A.; Vaknin, A.; Studdert, C.A. Bacterial chemoreceptors of different length classes signal independently. Mol. Microbiol. 2014, 93, 814–822. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wang, Y.H.; Zhu, H.Z.; Andrianova, E.P.; Jiang, C.Y.; Li, D.; Ma, L.; Feng, J.; Liu, Z.P.; Xiang, H. Cross Talk between Chemosensory Pathways That Modulate Chemotaxis and Biofilm Formation. MBio 2019, 10, e02876-18. [Google Scholar] [CrossRef] [PubMed]







| Strains | Characteristic and Genotypes | References |
|---|---|---|
| S. oneidensis strains | ||
| MR1-R | Shewanella oneidensis, wild-type strain, RifR | [31] |
| ΔcheA1 | MR1-R deleted of cheA1 (so_2121) | [22] |
| ΔcheW1 | MR1-R deleted of cheW1 (so_2122) | This work |
| Δso2123 (Δaer2so) | MR1-R deleted of aer2so (so_2123) | [32] |
| ΔmcpAso | MR1-R deleted of mcpAso (so_2117) | This work |
| ΔcheA3 | MR1-R deleted of cheA3 (so_3207) | [22] |
| ΔcheW3 | MR1-R deleted of cheW3 (so_3202) | [22] |
| E. coli strains | ||
| CC118 λpir | Δ(ara-leu) araDE ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE (Am) recA1 λpir | [33] |
| C600 | F- tonA21 thi-1 thr-1 leuB6 lacY1 glnV44 rfbC1 fhuA1 λ- | [34] |
| RP437 | F- thi thr leu his met eda rpsL, wild type for chemotaxis | [35] |
| BT3388 | E. coli strains RP437 (wild type) deleted of tar, tsr, trg, tap, aer | [36] |
| Plasmids | Descriptions | References |
|---|---|---|
| pKNG101 | R6K-derived suicide plasmid containing StrR and sacB | [33] |
| pBad33 | Vector containing pBAD promoter with the pACYC origin of replication | [38] |
| p33Tac | Derived vector of pBad33 containing ptac promoter inducible to IPTG | [29] |
| pM2-6his | p33Tac containing sequence coding for C-terminal 6His-tagged Aer2so | This work |
| pM2-GFP | Sequence coding for Aer2so-GFP cloned into p33Tac | This work |
| pAWM2-6his | cheA1-cheW1-aer2so-6his sequences cloned into p33Tac | This work |
| pAWM2-GFP | cheA1-cheW1-aer2so-GFP sequences cloned into p33Tac | This work |
| pGFP-cheA1 | GFP-cheA1 sequence cloned into p33tac | This work |
| pGFP-AWM2 | GFP-cheA1-cheW1-aer2so sequences cloned into p33Tac | This work |
| pmcpAso-GFP | so2117-GFP sequence cloned into p33Tac | This work |
| pSO4557 | so_4557 sequence cloned into pBad33 | [39] |
| pSO2240 | so_2240 sequence cloned into pBad33 | [39] |
| pSO1056 | so_1056 sequence cloned into pBad33 | [39] |
| pmcpAso | so_2117 sequence cloned into pBad33 | [32] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fortier, E.M.; Bouillet, S.; Infossi, P.; Ali Chaouche, A.; Espinosa, L.; Giudici-Orticoni, M.-T.; Mauriello, E.M.F.; Iobbi-Nivol, C. Defining Two Chemosensory Arrays in Shewanella oneidensis. Biomolecules 2023, 13, 21. https://doi.org/10.3390/biom13010021
Fortier EM, Bouillet S, Infossi P, Ali Chaouche A, Espinosa L, Giudici-Orticoni M-T, Mauriello EMF, Iobbi-Nivol C. Defining Two Chemosensory Arrays in Shewanella oneidensis. Biomolecules. 2023; 13(1):21. https://doi.org/10.3390/biom13010021
Chicago/Turabian StyleFortier, Emma M., Sophie Bouillet, Pascale Infossi, Amine Ali Chaouche, Leon Espinosa, Marie-Thérèse Giudici-Orticoni, Emilia M. F. Mauriello, and Chantal Iobbi-Nivol. 2023. "Defining Two Chemosensory Arrays in Shewanella oneidensis" Biomolecules 13, no. 1: 21. https://doi.org/10.3390/biom13010021
APA StyleFortier, E. M., Bouillet, S., Infossi, P., Ali Chaouche, A., Espinosa, L., Giudici-Orticoni, M.-T., Mauriello, E. M. F., & Iobbi-Nivol, C. (2023). Defining Two Chemosensory Arrays in Shewanella oneidensis. Biomolecules, 13(1), 21. https://doi.org/10.3390/biom13010021