Proteomics of High-Grade Serous Ovarian Cancer Models Identifies Cancer-Associated Fibroblast Markers Associated with Clinical Outcomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Cell Lysis and Protein Digestion
2.3. N-glycopeptide Enrichment
2.4. Mass Spectrometry-Based Proteomics
2.5. Differential Protein Expression Analysis
2.6. Pathway Analysis
2.7. Comparison to Tissue Proteomics Datasets
2.8. Survival Analysis
3. Results
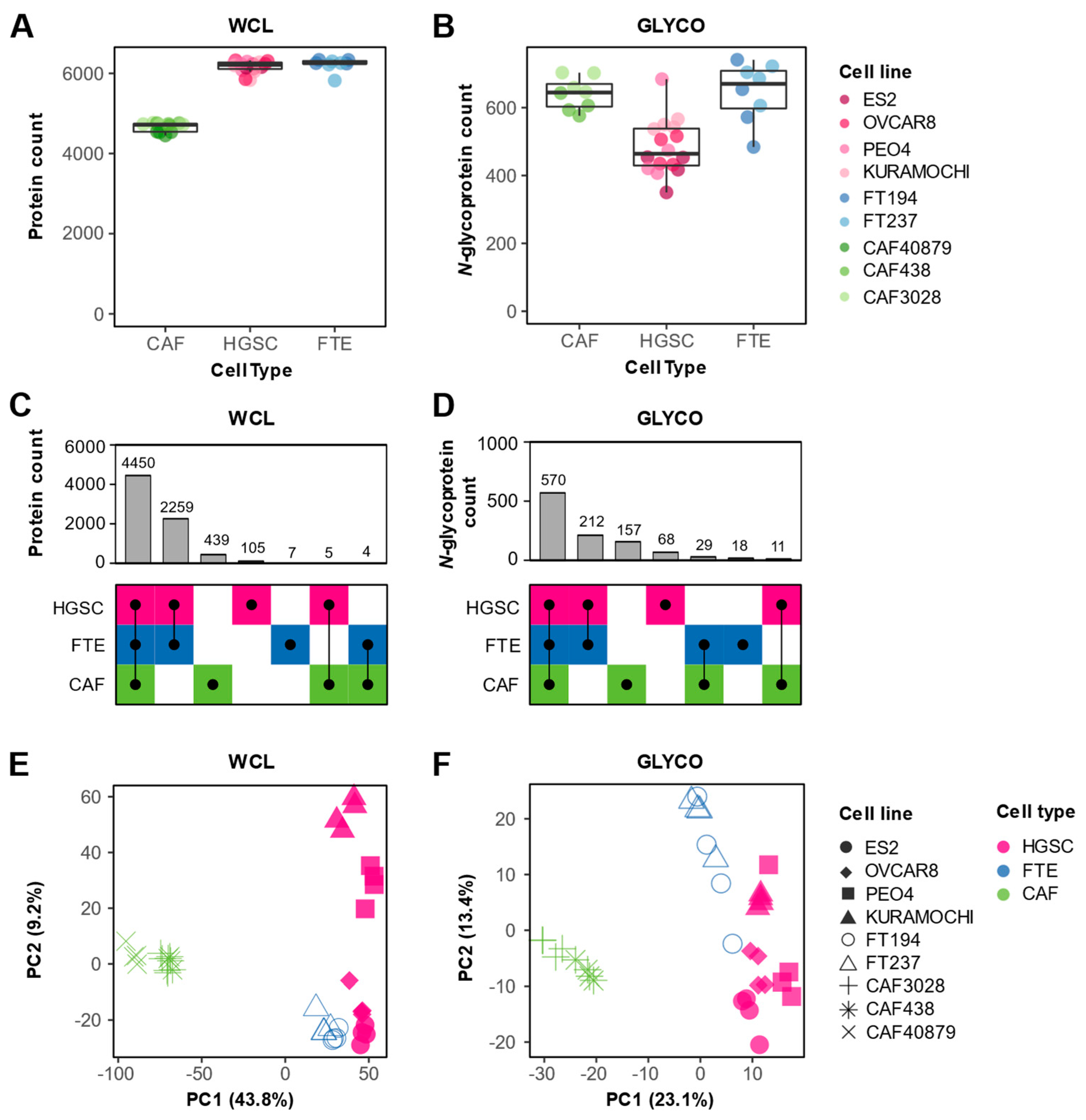
3.1. Proteomic Profiling of In Vitro HGSC Models
3.2. Characterization of Cell Type Elevated and Shared Proteins
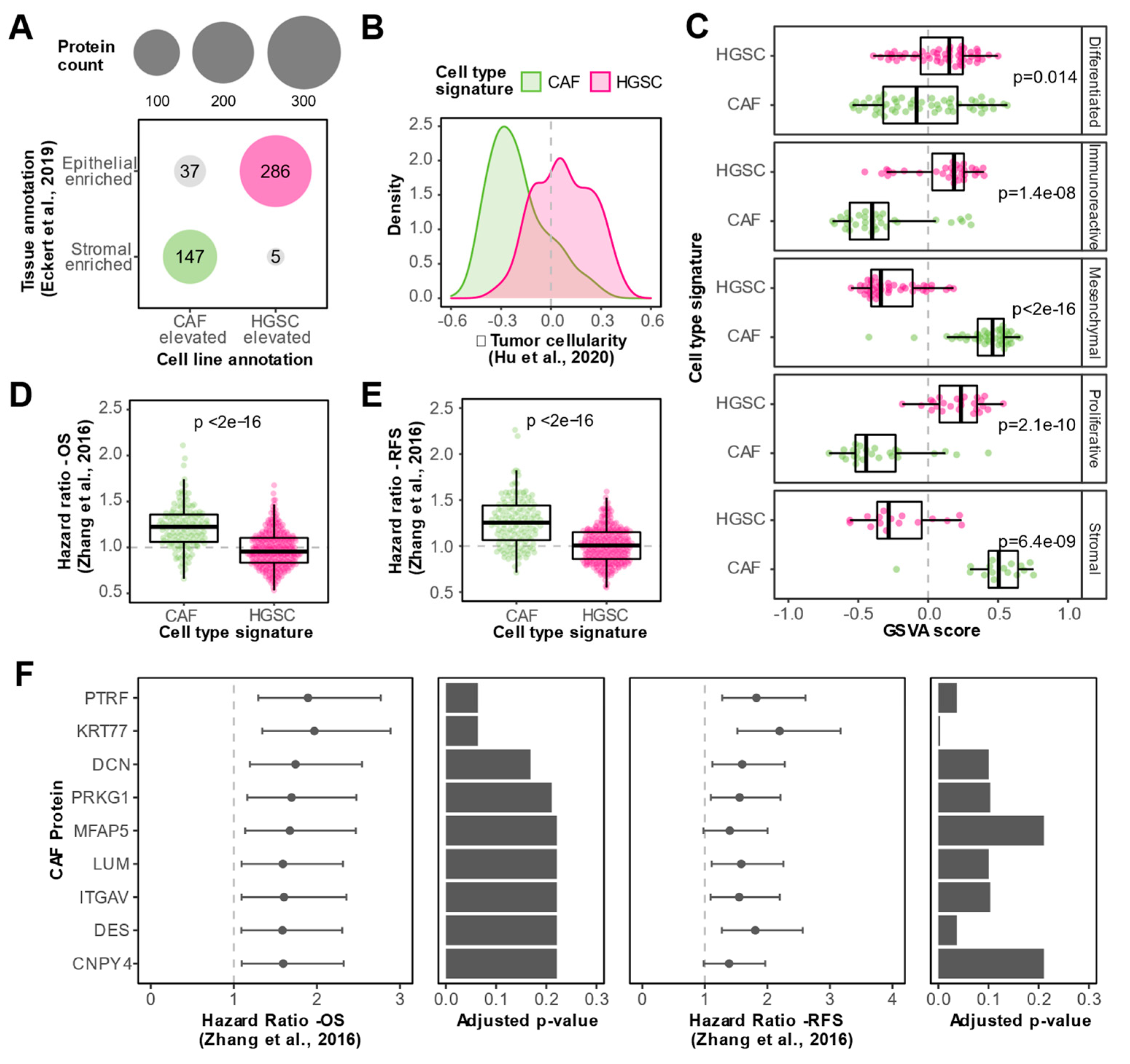
3.3. In Vitro Proteomics Reflects Tissue Proteomic Profiles
3.4. Integration of In Vitro and Tissue Proteomics Data Reveals CAF Enriched Proteins Associated with Clinical Outcomes
3.5. N-glycoproteomics Enables Identification of Additional Prognostic CAF Proteins in HGSC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Canadian Cancer Statistics 2019. Available online: https://cancer.ca/Canadian-Cancer-Statistics-2019-EN (accessed on 1 September 2022).
- ECIS—European Cancer Information. Available online: https://ecis.jrc.ec.europa.eu (accessed on 1 September 2022).
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA A Cancer J. Clin. 2019, 68, 7–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govindarajan, M.; Wohlmuth, C.; Waas, M.; Bernardini, M.Q.; Kislinger, T. High-throughput approaches for precision medicine in high-grade serous ovarian cancer. J. Hematol. Oncol. 2020, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [Green Version]
- Yue, H.; Li, W.; Chen, R.; Wang, J.; Lu, X.; Li, J. Stromal POSTN induced by TGF-β1 facilitates the migration and invasion of ovarian cancer. Gynecol. Oncol. 2021, 160, 530–538. [Google Scholar] [CrossRef]
- Gao, Q.; Yang, Z.; Xu, S.; Li, X.; Yang, X.; Jin, P.; Liu, Y.; Zhou, X.; Zhang, T.; Gong, C.; et al. Heterotypic CAF-tumor spheroids promote early peritoneal metastatis of ovarian cancer. J. Exp. Med. 2019, 216, 688–703. [Google Scholar] [CrossRef] [Green Version]
- Givel, A.-M.; Kieffer, Y.; Scholer-Dahirel, A.; Sirven, P.; Cardon, M.; Pelon, F.; Magagna, I.; Gentric, G.; Costa, A.; Bonneau, C.; et al. miR200-regulated CXCL12β promotes fibroblast heterogeneity and immunosuppression in ovarian cancers. Nat. Commun. 2018, 9, 1056. [Google Scholar] [CrossRef]
- Wang, W.; Kryczek, I.; Dostál, L.; Lin, H.; Tan, L.; Zhao, L.; Lu, F.; Wei, S.; Maj, T.; Peng, D.; et al. Effector T Cells Abrogate Stroma-Mediated Chemoresistance in Ovarian Cancer. Cell 2016, 165, 1092–1105. [Google Scholar] [CrossRef] [Green Version]
- Hussain, A.; Voisin, V.; Poon, S.; Karamboulas, C.; Bui, N.H.B.; Meens, J.; Dmytryshyn, J.; Ho, V.W.; Tang, K.H.; Paterson, J.; et al. Distinct fibroblast functional states drive clinical outcomes in ovarian cancer and are regulated by TCF21. J. Exp. Med. 2020, 217, 2139. [Google Scholar] [CrossRef]
- Olbrecht, S.; Busschaert, P.; Qian, J.; Vanderstichele, A.; Loverix, L.; Van Gorp, T.; Van Nieuwenhuysen, E.; Han, S.; Van den Broeck, A.; Coosemans, A.; et al. High-grade serous tubo-ovarian cancer refined with single-cell RNA sequencing: Specific cell subtypes influence survival and determine molecular subtype classification. Genome Med. 2021, 13, 111. [Google Scholar] [CrossRef]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, A.; Hussain, A.; Ignatchenko, V.; Ignatchenko, A.; Tang, K.H.; Ho, V.W.H.; Neel, B.G.; Clarke, B.; Bernardini, M.Q.; Ailles, L.; et al. N-Glycoproteomics of Patient-Derived Xenografts: A Strategy to Discover Tumor-Associated Proteins in High-Grade Serous Ovarian Cancer. Cell Syst. 2019, 8, 345–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhlmann, L.; Govindarajan, M.; Mejia-Guerrero, S.; Ignatchenko, V.; Liu, L.Y.; Grünwald, B.T.; Cruickshank, J.; Berman, H.; Khokha, R.; Kislinger, T. Glycoproteomics Identifies Plexin-B3 as a Targetable Cell Surface Protein Required for the Growth and Invasion of Triple-Negative Breast Cancer Cells. J. Proteome Res. 2022, 21, 2224–2236. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef] [PubMed]
- Eckert, M.A.; Coscia, F.; Chryplewicz, A.; Chang, J.W.; Hernandez, K.M.; Pan, S.; Tienda, S.M.; Nahotko, D.A.; Li, G.; Blaženović, I.; et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019, 569, 723–728. [Google Scholar] [CrossRef]
- Hu, Y.; Pan, J.; Shah, P.; Ao, M.; Thomas, S.N.; Liu, Y.; Chen, L.; Schnaubelt, M.; Clark, D.J.; Rodriguez, H.; et al. Integrated Proteomic and Glycoproteomic Characterization of Human High-Grade Serous Ovarian Carcinoma. Cell Rep. 2020, 33, 108276. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.; McDermott, J.E.; Zhou, J.-Y.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef] [Green Version]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [Green Version]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef] [Green Version]
- Perets, R.; Wyant, G.A.; Muto, K.W.; Bijron, J.G.; Poole, B.B.; Chin, K.T.; Chen, J.Y.H.; Ohman, A.W.; Stepule, C.D.; Kwak, S.; et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell 2013, 24, 751–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karst, A.M.; Drapkin, R. Primary culture and immortalization of human fallopian tube secretory epithelial cells. Nat. Protoc. 2019, 7, 1755–1764. [Google Scholar] [CrossRef]
- Waas, M.; Littrell, J.; Gundry, R.L. CIRFESS: An Interactive Resource for Querying the Set of Theoretically Detectable Peptides for Cell Surface and Extracellular Enrichment Proteomic Studies. J. Am. Soc. Mass Spectrom. 2020, 31, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Izar, B.; Tirosh, I.; Stover, E.H.; Wakiro, I.; Cuoco, M.S.; Alter, I.; Rodman, C.; Leeson, R.; Su, M.-J.; Shah, P.; et al. A single-cell landscape of high-grade serous ovarian cancer. Nat. Med. 2020, 26, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, Y.; Yang, J.; Zhao, X.; Wei, X. Tumor Microenvironment in Ovarian Cancer: Function and Therapeutic Strategy. Front. Cell Dev. Biol. 2020, 8, 758. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, H.; Cai, J.; Zhang, T.; Guo, J.; Feng, D.; Wang, Z. Ovarian cancer-associated fibroblasts contribute to epithelial ovarian carcinoma metastasis by promoting angiogenesis, lymphangiogenesis and tumor cell invasion. Cancer Lett. 2011, 303, 47–55. [Google Scholar] [CrossRef]
- Curtis, M.; Kenny, H.A.; Ashcroft, B.; Mukherjee, A.; Johnson, A.; Zhang, Y.; Helou, Y.; Batlle, R.; Liu, X.; Gutierrez, N.; et al. Fibroblasts Mobilize Tumor Cell Glycogen to Promote Proliferation and Metastasis. Cell Metab. 2019, 29, 141–155.e9. [Google Scholar] [CrossRef] [Green Version]
- Coscia, F.; Watters, K.M.; Curtis, M.; Eckert, M.A.; Chiang, C.Y.; Tyanova, S.; Montag, A.; Lastra, R.R.; Lengyel, E.; Mann, M. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat. Commun. 2016, 7, 12645. [Google Scholar] [CrossRef]
- Leung, C.S.; Yeung, T.-L.; Yip, K.-P.; Pradeep, S.; Balasubramanian, L.; Liu, J.; Wong, K.-K.; Mangala, L.S.; Armaiz-Pena, G.N.; Lopez-Berestein, G.; et al. Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat. Commun. 2014, 5, 5092. [Google Scholar] [CrossRef]
- Yue, H.; Wang, J.; Chen, R.; Hou, X.; Li, J.; Lu, X. Gene signature characteristic of elevated stromal infiltration and activation is associated with increased risk of hematogenous and lymphatic metastasis in serous ovarian cancer. BMC Cancer 2019, 19, 1266. [Google Scholar] [CrossRef] [Green Version]
- Moon, H.; Lee, C.S.; Inder, K.L.; Sharma, S.; Choi, E.; Black, D.M.; Le Cao, K.A.; Winterford, C.; Coward, J.I.; Ling, M.T.; et al. PTRF/cavin-1 neutralizes non-caveolar caveolin-1 microdomains in prostate cancer. Oncogene 2014, 33, 3561–3570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittka, A.; Ketteler, J.; Bongards, L.; Maier, P.; Herskind, C.; Jendrossek, V.; Klein, D. Stromal Fibroblasts Counteract the Caveolin-1-Dependent Radiation Response of LNCaP Prostate Carcinoma Cells. Front Oncol. 2022, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Biological roles of glycans. Glycobiology 2016, 27, 3–49. [Google Scholar] [CrossRef] [Green Version]
- Peixoto, A.; Relvas-Santos, M.; Azevedo, R.; Santos, L.L.; Ferreira, J.A. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front. Oncol. 2019, 9, 8256. [Google Scholar] [CrossRef] [Green Version]
- Miljkovic-Licina, M.; Hammel, P.; Garrido-Urbani, S.; Lee, B.P.L.; Meguenani, M.; Chaabane, C.; Bochaton-Piallat, M.L.; Imhof, B.A. Targeting olfactomedin-like 3 inhibits tumor growth by impairing angiogenesis and pericyte coverage. Mol Cancer Ther. 2012, 11, 2588–2599. [Google Scholar] [CrossRef] [Green Version]
- Jackson, S.W.; Hoshi, T.; Wu, Y.; Sun, X.; Enjyoji, K.; Cszimadia, E.; Sundberg, C.; Robson, S.C. Disordered purinergic signaling inhibits pathological angiogenesis in cd39/Entpd1-null mice. Am. J. Pathol. 2007, 171, 1395–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulitti, A.; Andreuzzi, E.; Bizzotto, D.; Pellicani, R.; Tarticchio, G.; Marastoni, S.; Pastrello, C.; Jurisica, I.; Ligresti, G.; Bucciotti, F.; et al. The ablation of the matricellular protein EMILIN2 causes defective vascularization due to impaired EGFR-dependent IL-8 production affecting tumor growth. Oncogene 2018, 37, 3399–3414. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Govindarajan, M.; Ignatchenko, V.; Ailles, L.; Kislinger, T. Proteomics of High-Grade Serous Ovarian Cancer Models Identifies Cancer-Associated Fibroblast Markers Associated with Clinical Outcomes. Biomolecules 2023, 13, 75. https://doi.org/10.3390/biom13010075
Govindarajan M, Ignatchenko V, Ailles L, Kislinger T. Proteomics of High-Grade Serous Ovarian Cancer Models Identifies Cancer-Associated Fibroblast Markers Associated with Clinical Outcomes. Biomolecules. 2023; 13(1):75. https://doi.org/10.3390/biom13010075
Chicago/Turabian StyleGovindarajan, Meinusha, Vladimir Ignatchenko, Laurie Ailles, and Thomas Kislinger. 2023. "Proteomics of High-Grade Serous Ovarian Cancer Models Identifies Cancer-Associated Fibroblast Markers Associated with Clinical Outcomes" Biomolecules 13, no. 1: 75. https://doi.org/10.3390/biom13010075





