Expression of a Salt-Tolerant Pseudolysin in Yeast for Efficient Protein Hydrolysis under High-Salt Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids and Strains
| Plasmids | Description | Source |
|---|---|---|
| CPOTud | 2 μm, AmpR, TPI1p, TPI1t, POT1 gene from S. pombe as a selection marker. | [28] |
| pAlphaAmyCPOT | CPOTud with amylase gene driven by α-factor prepro leader | [28] |
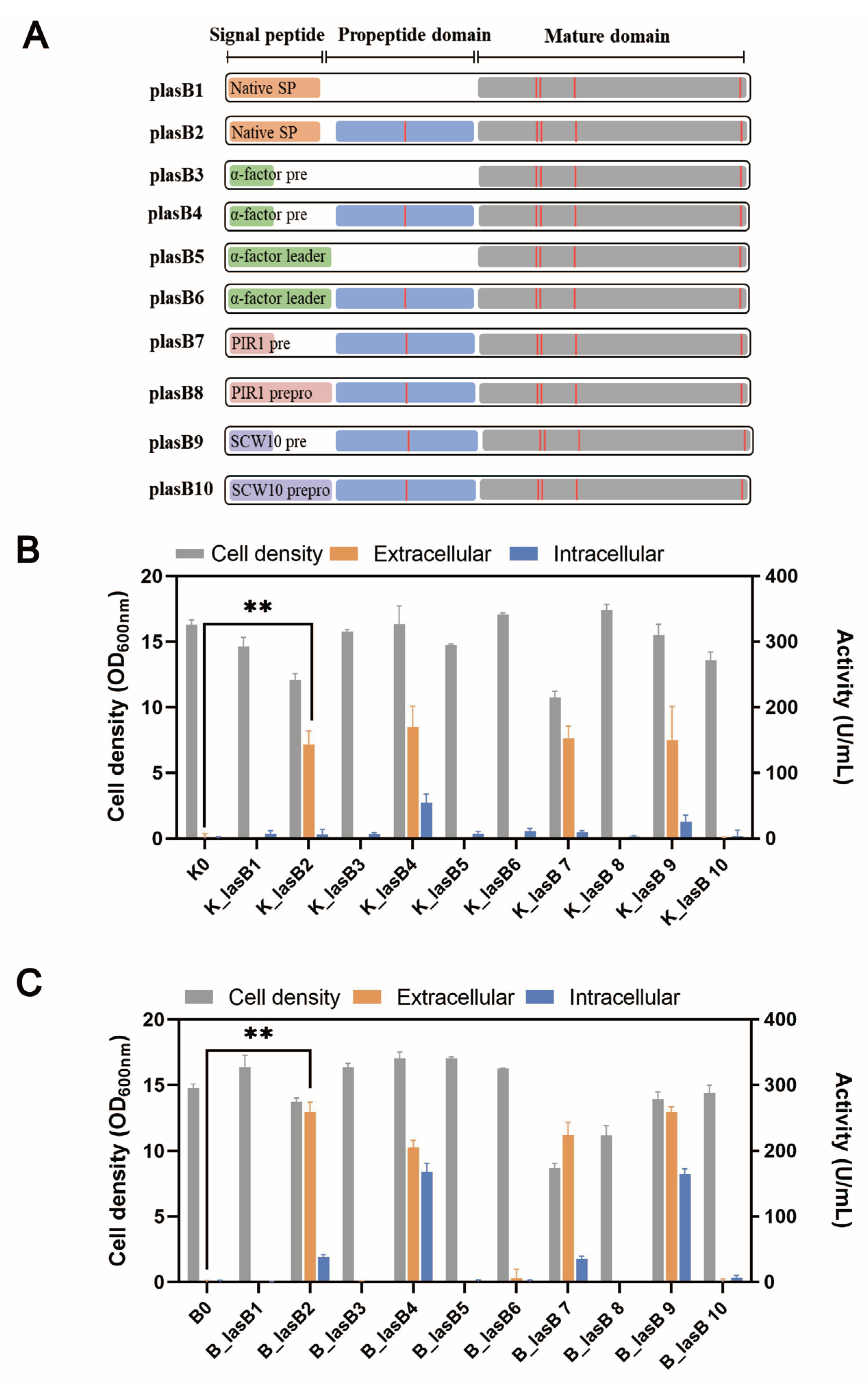
| plasB1 | CPOTud- (native signal peptide + mature domain of mutant pseudolysin) | This study |
| plasB2 | CPOTud- (native signal peptide + propeptide + mature domain of mutant pseudolysin) | This study |
| plasB3 | CPOTud- (α-factor pre leader + mature domain of mutant pseudolysin) | This study |
| plasB4 | CPOTud- (α-factor pre leader + propeptide + mature domain of mutant pseudolysin) | This study |
| plasB5 | CPOTud- (α-factor prepro leader + mature domain of mutant pseudolysin) | This study |
| plasB6 | CPOTud- (α-factor prepro leader + propeptide + mature domain of mutant pseudolysin) | This study |
| plasB7 | CPOTud- (PIR1 pre leader + propeptide + mature domain of mutant pseudolysin) | This study |
| plasB8 | CPOTud- (PIR1 prepro leader + propeptide + mature domain of mutant pseudolysin) | This study |
| PlasB9 | CPOTud- (SCW10 pre leader + propeptide + mature domain of mutant pseudolysin) | This study |
| PlasB10 | CPOTud- (SCW10 prepro leader + propeptide + mature domain of mutant pseudolysin) | This study |
| Strains | Description | Source |
|---|---|---|
| P. aeruginosa SWJSS3 | Strain-secreting mutant pseudolysin | [8] |
| E. coli DH5α | F- Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17(rk-, mk+) phoA supE44 thi-1 gyrA96 relA1λ- | AngYuBio Co., Ltd. |
| S. cerevisiae CEN.PK 530-1C | MATa tpi1(41-707)::loxP-KanMX4-loxP | [29] |
| B184M | The UV mutant strain obtained from CEN.PK 530-1C | [30] |
| K0 | CEN.PK 530-1C/CPOTud | This study |
| K_lasB1 | CEN.PK 530-1C/plasB1 | This study |
| K_lasB2 | CEN.PK 530-1C/plasB2 | This study |
| K_lasB3 | CEN.PK 530-1C/plasB3 | This study |
| K_lasB4 | CEN.PK 530-1C/plasB4 | This study |
| K_lasB5 | CEN.PK 530-1C/plasB5 | This study |
| K_lasB6 | CEN.PK 530-1C/plasB6 | This study |
| K_lasB7 | CEN.PK 530-1C/plasB7 | This study |
| K_lasB8 | CEN.PK 530-1C/plasB8 | This study |
| K_lasB9 | CEN.PK 530-1C/plasB9 | This study |
| K_lasB10 | CEN.PK 530-1C/plasB10 | This study |
| B0 | B184M /CPOTud | This study |
| B_lasB1 | B184M/plasB1 | This study |
| B_lasB2 | B184M/plasB2 | This study |
| B_lasB3 | B184M/plasB3 | This study |
| B_lasB4 | B184M/plasB4 | This study |
| B_lasB5 | B184M/plasB5 | This study |
| B_lasB6 | B184M/plasB6 | This study |
| B_lasB7 | B184M/plasB7 | This study |
| B_lasB8 | B184M/plasB8 | This study |
| B_lasB9 | B184M/plasB9 | This study |
| B_lasB10 | B184M/plasB10 | This study |
2.2. Media and Cultivations
2.3. Pseudolysin Activity Assay
2.4. Protein Purification
2.5. Western Blot of Recombinant Pseudolysin
2.6. Enzyme Kinetics and General Characteristics
2.7. Soy Protein Isolate Hydrolysis
3. Results and Discussion
3.1. Bioinformatic Analysis of the lasB Gene from P. aeruginosa SWJSS3
3.2. Heterologous Expression of Pseudolysin by S. cerevisiae
3.3. Native Propeptide Is Essential for Pseudolysin Secretion in S. cerevisiae
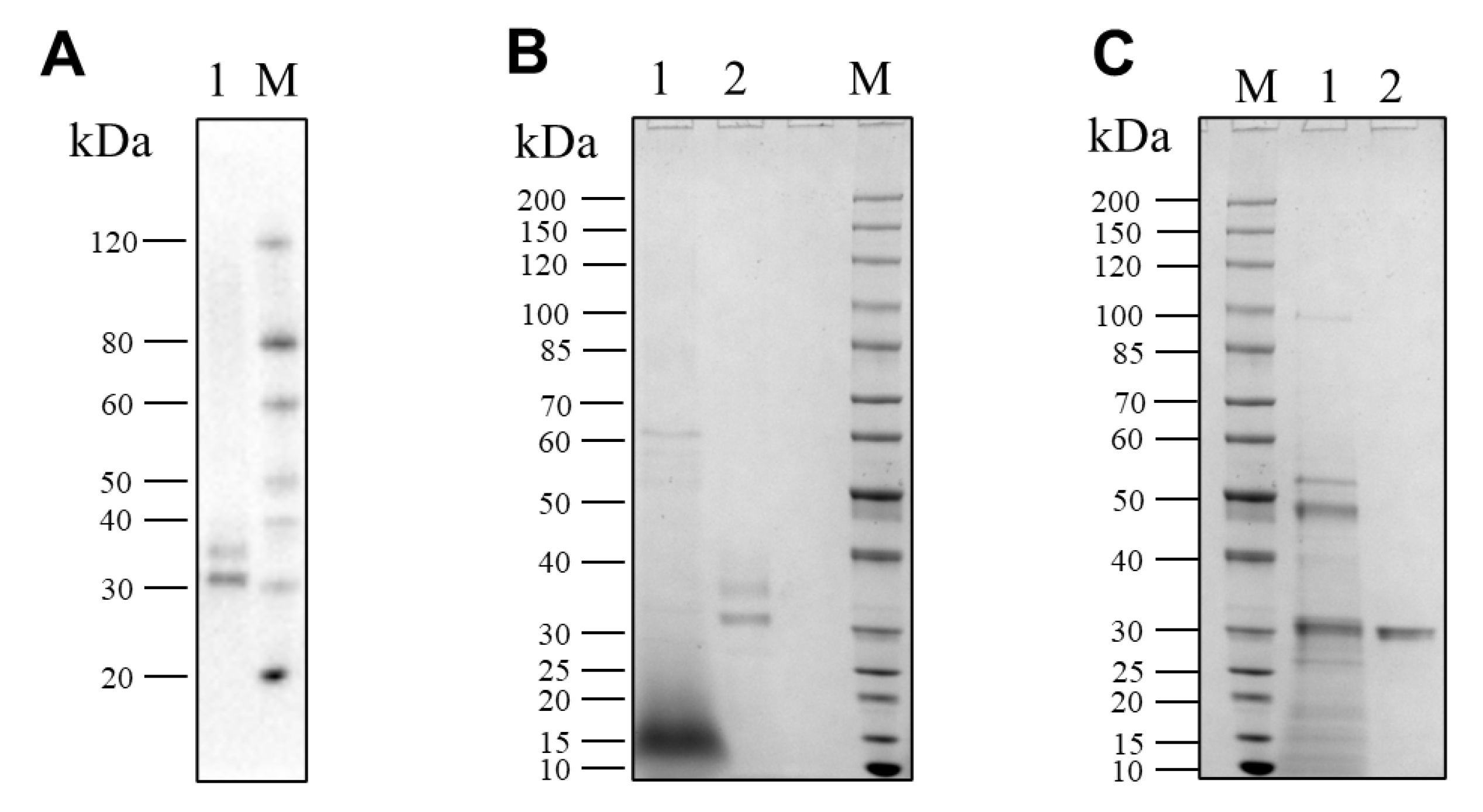
3.4. Purification of Pseudolysin
3.5. Evaluation of Enzymatic Characteristics
3.6. Hydrolyzation of LD-SPI and HD-SPI in a High-Salt Environment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Naveed, M.; Nadeem, F.; Mehmood, T.; Bilal, M.; Anwar, Z.; Amjad, F. Protease—A versatile and ecofriendly biocatalyst with multi-industrial applications: An updated review. Catal. Lett. 2021, 151, 307–323. [Google Scholar] [CrossRef]
- Gupta, A.; Roy, I.; Patel, R.K.; Singh, S.P.; Khare, S.K.; Gupta, M.N. One-step purification and characterization of an alkaline protease from haloalkaliphilic Bacillus sp. J. Chromatogr. A 2005, 1075, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Roccatano, D.; Lorenz, M.; Martinez, R.; Schwaneberg, U. Insights on activity and stability of subtilisin E towards guanidinium chloride and sodium dodecylsulfate. J. Biotechnol. 2014, 169, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Ding, L.; Yao, Y.; Cao, Y.; Pan, Z.; Kong, D. Extracellular proteome analysis and flavor formation during soy sauce fermentation. Front. Microbiol. 2018, 9, 1872. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.W.; Roccatano, D.; Lorenz, M.; Schwaneberg, U. Directed evolution of subtilisin E into a highly active and guanidinium chloride-and sodium dodecylsulfate-tolerant protease. Chembiochem 2012, 13, 691–699. [Google Scholar] [CrossRef]
- Fang, Z.; Zhang, J.; Du, G.; Chen, J. Improved catalytic efficiency, thermophilicity, anti-salt and detergent tolerance of keratinase KerSMD by partially truncation of PPC domain. Sci. Rep. 2016, 6, 27953. [Google Scholar] [CrossRef] [Green Version]
- Narhi, L.O.; Stabinsky, Y.; Levitt, M.; Miller, L.; Sachdev, R.; Finley, S.; Park, S.; Kolvenbach, C.; Arakawa, T.; Zukowski, M. Enhanced stability of subtilisin by three point mutations. Biotechnol. Appl. Biochem. 1991, 13, 12–24. [Google Scholar] [CrossRef]
- Zhao, M.; Shu, H.; Cui, C.; Lei, F.; Tang, X. The isolation and identification of deep-sea bacteria that produce salt-tolerant proteases. Mod. Food Sci. Technol. 2015, 31, 50–54. [Google Scholar]
- Han, M. Isolation and characterization of a keratinolytic protease from a feather-degrading bacterium Pseudomonas aeruginosa C11. Afr. J. Microbiol. Res. 2012, 6, 2211–2221. [Google Scholar] [CrossRef]
- Ghorbel-Bellaaj, O.; Hayet, B.K.; Bayoudh, A.; Younes, I.; Hmidet, N.; Jellouli, K.; Nasri, M. Pseudomonas aeruginosa A2 elastase: Purification, characterization and biotechnological applications. Int. J. Biol. Macromol. 2012, 50, 679–686. [Google Scholar] [CrossRef]
- Pandeeti, E.V.P.; Pitchika, G.K.; Jotshi, J.; Nilegaonkar, S.S.; Kanekar, P.P.; Siddavattam, D. Enzymatic depilation of animal hide: Identification of elastase (LasB) from Pseudomonas aeruginosa MCM B-327 as a depilating protease. PLoS ONE 2011, 6, e16742. [Google Scholar] [CrossRef]
- Ghorbel-Bellaaj, O.; Jellouli, K.; Younes, I.; Manni, L.; Salem, M.O.; Nasri, M. A Solvent-Stable Metalloprotease Produced by Pseudomonas aeruginosa A2 Grown on Shrimp Shell Waste and Its Application in Chitin Extraction. Appl. Biochem. Biotechnol. 2011, 164, 410–425. [Google Scholar] [CrossRef]
- Tang, X.; Pan, Y.; Li, S.; He, B. Screening and isolation of an organic solvent-tolerant bacterium for high-yield production of organic solvent-stable protease. Bioresour. Technol. 2008, 99, 7388–7392. [Google Scholar] [CrossRef]
- Ogino, H.; Yokoo, J.; Watanabe, F.; Ishikawa, H. Cloning and sequencing of a gene of organic solvent-stable protease secreted from Pseudomonas aeruginosa PST-01 and its expression in Escherichia coli. Biochem. Eng. J. 2000, 5, 191–200. [Google Scholar] [CrossRef]
- Waters, C.M.; Goldberg, J.B. Pseudomonas aeruginosa in cystic fibrosis: A chronic cheater. Proc. Natl. Acad. Sci. USA 2019, 116, 6525–6527. [Google Scholar] [CrossRef] [Green Version]
- McIver, K.; Kessler, E.; Ohman, D.E. Substitution of active-site His-223 in Pseudomonas aeruginosa elastase and expression of the mutated lasB alleles in Escherichia coli show evidence for autoproteolytic processing of proelastase. J. Bacteriol. 1991, 173, 7781–7789. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Zhu, F.; Zhu, L.; Chen, G.; He, B. Highly efficient enzymatic synthesis of Z-aspartame in aqueous medium via in situ product removal. Biochem. Eng. J. 2015, 98, 63–67. [Google Scholar] [CrossRef]
- Shinji, T.; Moe, Y.; Yukihiro, K.; Yoko, Y.; Hitoshi, A. Characterization of an organic-solvent-stable elastase from Pseudomonas indica and its potential use in eggshell membrane hydrolysis. Process Biochem. 2019, 85, 156–163. [Google Scholar] [CrossRef]
- Odunuga, O.O.; Adekoya, O.A.; Sylte, I. High-level expression of pseudolysin, the extracellular elastase of Pseudomonas aeruginosa, in Escherichia coli and its purification. Protein Expr. Purif. 2015, 113, 79–84. [Google Scholar] [CrossRef]
- Takenaka, S.; Hano, S.; Cheng, M.; Yoshida, K.; Aoki, K. Organic solvent-tolerant elastase efficiently hydrolyzes insoluble, cross-linked, protein fiber of eggshell membranes. Biotechnol. Lett. 2012, 34, 949–955. [Google Scholar] [CrossRef]
- Lin, X.; Xu, W.; Huang, K.; Mei, X.; Liang, Z.; Li, Z.; Guo, J.; Luo, Y. Cloning, expression and characterization of recombinant elastase from Pseudomonas aeruginosa in Picha pastoris. Protein Expr. Purif. 2009, 63, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Ding, H.; Wang, J.; Jin, M.; Yu, X. Expression of the lasB gene encoding an organic solvent-stable elastase in Pichia pastoris and potential applications of the recombinant enzymes in peptide synthesis. Biochem. Eng. J. 2013, 77, 154–160. [Google Scholar] [CrossRef]
- Han, M.; Wang, W.; Jiang, G.; Wang, X.; Liu, X.; Cao, H.; Tao, Y.; Yu, X. Enhanced expression of recombinant elastase in Pichia pastoris through addition of N-glycosylation sites to the propeptide. Biotechnol. Lett. 2014, 36, 2467–2471. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Wang, W.; Zhou, J.; Gong, X.; Xu, C.; Li, Y.; Li, Q. Activation of the unfolded protein response via co-expression of the HAC1i gene enhances expression of recombinant elastase in Pichia pastoris. Biotechnol. Bioprocess Eng. 2020, 25, 302–307. [Google Scholar] [CrossRef]
- Shusta, E.V.; Raines, R.T.; Plückthun, A.; Wittrup, K.D. Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat. Biotechnol. 1998, 16, 773–777. [Google Scholar] [CrossRef]
- den Haan, R.; Rose, S.H.; Cripwell, R.A.; Trollope, K.M.; Myburgh, M.W.; Viljoen-Bloom, M.; van Zyl, W.H. Heterologous production of cellulose- and starch-degrading hydrolases to expand Saccharomyces cerevisiae substrate utilization: Lessons learnt. Biotechnol. Adv. 2021, 53, 107859. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 1–4. [Google Scholar] [CrossRef]
- Liu, Z.; Tyo, K.E.J.; Martínez, J.L.; Petranovic, D.; Nielsen, J. Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2012, 109, 1259–1268. [Google Scholar] [CrossRef] [Green Version]
- Overkamp, K.M.; Bakker, B.M.; Kötter, P.; Luttik, M.A.; Van Dijken, J.P.; Pronk, J.T. Metabolic engineering of glycerol production in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2002, 68, 2814–2821. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Bai, Y.; Sjostrom, S.L.; Hallström, B.M.; Liu, Z.; Petranovic, D.; Uhlén, M.; Joensson, H.N.; Andersson-Svahn, H.; Nielsen, J. Microfluidic screening and whole-genome sequencing identifies mutations associated with improved protein secretion by yeast. Proc. Natl. Acad. Sci. USA 2015, 112, E4689–E4696. [Google Scholar] [CrossRef] [Green Version]
- Zhu, F.; Liu, F.; Wu, B.; He, B. Efficient Extracellular Expression of Metalloprotease for Z-Aspartame Synthesis. J. Agric. Food Chem. 2016, 64, 9631–9638. [Google Scholar] [CrossRef]
- Han, M.; Wang, X.; Ding, H.; Jin, M.; Yu, L.; Wang, J.; Yu, X. The role of N-glycosylation sites in the activity, stability, and expression of the recombinant elastase expressed by Pichia pastoris. Enzym. Microb. Technol. 2014, 54, 32–37. [Google Scholar] [CrossRef]
- Shen, P.; Zhou, F.; Zhang, Y.; Yuan, D.; Zhao, Q.; Zhao, M. Formation and characterization of soy protein nanoparticles by controlled partial enzymatic hydrolysis. Food Hydrocoll. 2020, 105, 105844. [Google Scholar] [CrossRef]
- Ogino, H.; Yasui, K.; Shiotani, T.; Ishihara, T.; Ishikawa, H. Organic solvent-tolerant bacterium which secretes an organic solvent-stable proteolytic enzyme. Appl. Environ. Microbiol. 1995, 61, 4258–4262. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Bao, J.; Hallström, B.M.; Petranovic, D.; Nielsen, J. Efficient protein production by yeast requires global tuning of metabolism. Nat. Commun. 2017, 8, 1131. [Google Scholar] [CrossRef] [Green Version]
- Gast, V.; Sandegren, A.; Dunås, F.; Ekblad, S.; Güler, R.; Thorén, S.; Mohedano, M.T.; Molin, M.; Engqvist, M.K.; Siewers, V. Engineering Saccharomyces cerevisiae for the production and secretion of Affibody molecules. Microb. Cell Fact. 2022, 21, 36. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Chen, X.; Nielsen, J.; Petranovic, D.; Siewers, V. Expression of antibody fragments in Saccharomyces cerevisiae strains evolved for enhanced protein secretion. Microb. Cell Fact. 2021, 20, 134. [Google Scholar] [CrossRef]
- Kessler, E.; Ohman, D.E. Chapter 120—Pseudolysin. In Handbook of Proteolytic Enzymes, 3rd ed.; Rawlings, N.D., Salvesen, G., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 582–592. [Google Scholar] [CrossRef]
- Wei, G.; Tian, N.; Valery, A.C.; Zhong, Y.; Schuppan, D.; Helmerhorst, E.J. Identification of pseudolysin (lasB) as an aciduric gluten-degrading enzyme with high therapeutic potential for celiac disease. Am. J. Gastroenterol. Suppl. 2015, 110, 899–908. [Google Scholar] [CrossRef] [Green Version]
- Xiao, H.; Liu, X.; Feng, Y.; Zheng, L.; Zhao, M.; Huang, M. Secretion of collagenases by Saccharomyces cerevisiae for collagen degradation. Biotechnol. Biofuels Bioprod. 2022, 15, 89. [Google Scholar] [CrossRef]
- Eide, D.J. The molecular biology of metal ion transport in Saccharomyces cerevisiae. Annu. Rev. Nutr. 1998, 18, 441–469. [Google Scholar] [CrossRef]
- Zhao, X.; Bai, F. Zinc and yeast stress tolerance: Micronutrient plays a big role. J. Biotechnol. 2012, 158, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Shet, A.R.; Patil, L.R.; Hombalimath, V.S.; Yaraguppi, D.A.; Udapudi, B.B. Enrichment of Saccharomyces cerevisiae with zinc and their impact on cell growth. Biotechnol. Bioinforma. Bioeng. 2011, 1, 523–527. [Google Scholar]
- Mrvčić, J.; Stanzer, D.; Stehlik-Tomas, V.; Škevin, D.; Grba, S. Optimization of bioprocess for production of copper-enriched biomass of industrially important microorganism Saccharomyces cerevisiae. J. Biosci. Bioeng. 2007, 103, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cao, C.; Liu, Y.; Wang, J.; Li, J.; Li, S.; Deng, Y. Identification of the genetic requirements for zinc tolerance and toxicity in Saccharomyces cerevisiae. G3: Genes Genomes Genet. 2020, 10, 479–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagani, M.A.; Casamayor, A.; Serrano, R.; Atrian, S.; Ariño, J. Disruption of iron homeostasis in Saccharomyces cerevisiae by high zinc levels: A genome-wide study. Mol. Microbiol. 2007, 65, 521–537. [Google Scholar] [CrossRef]
- Kamizono, A.; Nishizawa, M.; Teranishi, Y.; Murata, K.; Kimura, A. Identification of a gene conferring resistance to zinc and cadmium ions in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 1989, 219, 161–167. [Google Scholar] [CrossRef]
- Li, X.; Qian, J.; Wang, C.; Zheng, K.; Ye, L.; Fu, Y.; Han, N.; Bian, H.; Pan, J.; Wang, J.; et al. Regulating cytoplasmic calcium homeostasis can reduce aluminum toxicity in yeast. PLoS ONE 2011, 6, e21148. [Google Scholar] [CrossRef] [Green Version]
- Aza, P.; Molpeceres, G.; de Salas, F.; Camarero, S. Design of an improved universal signal peptide based on the α-factor mating secretion signal for enzyme production in yeast. Cell. Mol. Life Sci. 2021, 78, 3691–3707. [Google Scholar] [CrossRef]
- Werner, N.; Zibek, S. Expression of a codon-optimized Carica papaya papain sequence in the methylotrophic yeast Pichia pastoris. J. Microb. Biochem. Technol. 2015, 7, 313–317. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.-P.; Ma, Y. High-level expression, purification and characterization of recombinant Aspergillus oryzae alkaline protease in Pichia pastoris. Protein Expr. Purif. 2008, 58, 301–308. [Google Scholar] [CrossRef]
- Aggarwal, S.; Mishra, S. Differential role of segments of α-mating factor secretion signal in Pichia pastoris towards granulocyte colony-stimulating factor emerging from a wild type or codon optimized copy of the gene. Microb. Cell Fact. 2020, 19, 199. [Google Scholar] [CrossRef]
- Barrero, J.J.; Casler, J.C.; Valero, F.; Ferrer, P.; Glick, B.S. An improved secretion signal enhances the secretion of model proteins from Pichia pastoris. Microb. Cell Fact. 2018, 17, 161. [Google Scholar] [CrossRef]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar]
- Chen, M.M.; Bartlett, A.I.; Nerenberg, P.S.; Friel, C.T.; Hackenberger, C.P.R.; Stultz, C.M.; Radford, S.E.; Imperiali, B. Perturbing the folding energy landscape of the bacterial immunity protein Im7 by site-specific N-linked glycosylation. Proc. Natl. Acad. Sci. USA 2010, 107, 22528–22533. [Google Scholar] [CrossRef] [Green Version]
- Culyba, E.K.; Price, J.L.; Hanson, S.R.; Dhar, A.; Wong, C.-H.; Gruebele, M.; Powers, E.T.; Kelly, J.W. Protein native-state stabilization by placing aromatic side chains in N-glycosylated reverse turns. Science 2011, 331, 571–575. [Google Scholar] [CrossRef] [Green Version]
- Tan, N.Y.; Bailey, U.-M.; Jamaluddin, M.F.; Mahmud, S.H.B.; Raman, S.C.; Schulz, B.L. Sequence-based protein stabilization in the absence of glycosylation. Nat. Commun. 2014, 5, 3099. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Yin, Y.; Yan, J.; Zhang, J.; Ma, H.; Zhou, C. Separation, biochemical characterization and salt-tolerant mechanisms of alkaline protease from Aspergillus oryzae. J. Sci. Food Agric. 2019, 99, 3359–3366. [Google Scholar] [CrossRef]
- Warden, A.C.; Williams, M.; Peat, T.S.; Seabrook, S.A.; Newman, J.; Dojchinov, G.; Haritos, V.S. Rational engineering of a mesohalophilic carbonic anhydrase to an extreme halotolerant biocatalyst. Nat. Commun. 2015, 6, 10278. [Google Scholar] [CrossRef] [Green Version]
- Bhuyan, A.K. On the mechanism of SDS-induced protein denaturation. Biopolymers 2010, 93, 186–199. [Google Scholar] [CrossRef]
- Fuguet, E.; Ràfols, C.; Rosés, M.; Bosch, E. Critical micelle concentration of surfactants in aqueous buffered and unbuffered systems. Anal. Chim. Acta 2005, 548, 95–100. [Google Scholar] [CrossRef]
- Jellouli, K.; Bayoudh, A.; Manni, L.; Agrebi, R.; Nasri, M. Purification, biochemical and molecular characterization of a metalloprotease from Pseudomonas aeruginosa MN7 grown on shrimp wastes. Appl. Microbiol. Biotechnol. 2008, 79, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Ogino, H.; Uchiho, T.; Yokoo, J.; Kobayashi, R.; Ichise, R.; Ishikawa, H. Role of intermolecular disulfide bonds of the organic solvent-stable PST-01 protease in its organic solvent stability. Appl. Environ. Microbiol. 2001, 67, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Ogino, H.; Uchiho, T.; Doukyu, N.; Yasuda, M.; Ishimi, K.; Ishikawa, H. Effect of exchange of amino acid residues of the surface region of the PST-01 protease on its organic solvent-stability. Biochem. Biophys. Res. Commun. 2007, 358, 1028–1033. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Guo, G.; Fu, X.; Yao, Y.; Yuan, L.; Xiang, A. Fabrication, properties and applications of soy-protein-based materials: A review. Int. J. Biol. Macromol. 2018, 120, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Qin, G.X.; Sun, Z.W.; Zhao, Y. Advances of research on glycinin and β-conglycinin: A review of two major soybean allergenic proteins. Crit. Rev. Food Sci. Nutr. 2014, 54, 850–862. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Wang, C. Formation and characterization of amyloid-like fibrils from soy β-conglycinin and glycinin. J. Agric. Food Chem. 2010, 58, 11058–11066. [Google Scholar] [CrossRef]
- Kitamura, K. Genetic improvement of nutritional and food processing quality in soybean. Jpn. Agric. Res. Q. 1995, 29, 1. [Google Scholar]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef] [Green Version]
- de Castro, R.J.S.; Sato, H.H. Antioxidant activities and functional properties of soy protein isolate hydrolysates obtained using microbial proteases. Int. J. Food Sci. Technol. 2014, 49, 317–328. [Google Scholar] [CrossRef]
- Mukhia, S.; Kumar, A.; Kumar, R. Generation of antioxidant peptides from soy protein isolate through psychrotrophic Chryseobacterium sp. derived alkaline broad temperature active protease. LWT 2021, 143, 111152. [Google Scholar] [CrossRef]




| Enzyme | Concentration (mg/mL) | Activity (U/mL) | Specific Enzyme Activity (U/mg) | Vmax (μg•mL−1•min−1) | Km (g•L−1) | Kcat (min−1) | Kcat/Km (L•g−1•min−1) | R2 |
|---|---|---|---|---|---|---|---|---|
| SC-lasB | 0.097 | 996.49 | 10269.18 | 2.95 | 5.08 | 5.15 | 1.01 | 0.991 1 |
| PA-lasB | 0.108 | 1563.16 | 14503.53 | 3.37 | 4.78 | 8.38 | 1.75 | 0.972 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Lu, Q.; Xiao, H.; Feng, Y.; Su, G.; Zhao, M.; Huang, M. Expression of a Salt-Tolerant Pseudolysin in Yeast for Efficient Protein Hydrolysis under High-Salt Conditions. Biomolecules 2023, 13, 83. https://doi.org/10.3390/biom13010083
Liu X, Lu Q, Xiao H, Feng Y, Su G, Zhao M, Huang M. Expression of a Salt-Tolerant Pseudolysin in Yeast for Efficient Protein Hydrolysis under High-Salt Conditions. Biomolecules. 2023; 13(1):83. https://doi.org/10.3390/biom13010083
Chicago/Turabian StyleLiu, Xiufang, Qian Lu, Han Xiao, Yunzi Feng, Guowan Su, Mouming Zhao, and Mingtao Huang. 2023. "Expression of a Salt-Tolerant Pseudolysin in Yeast for Efficient Protein Hydrolysis under High-Salt Conditions" Biomolecules 13, no. 1: 83. https://doi.org/10.3390/biom13010083
APA StyleLiu, X., Lu, Q., Xiao, H., Feng, Y., Su, G., Zhao, M., & Huang, M. (2023). Expression of a Salt-Tolerant Pseudolysin in Yeast for Efficient Protein Hydrolysis under High-Salt Conditions. Biomolecules, 13(1), 83. https://doi.org/10.3390/biom13010083






