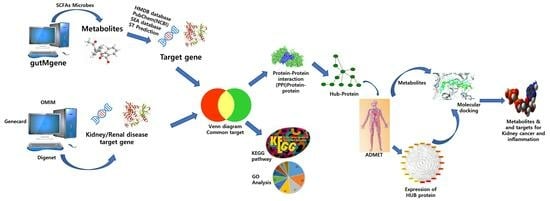
A Network Pharmacology and Molecular-Docking-Based Approach to Identify the Probable Targets of Short-Chain Fatty-Acid-Producing Microbial Metabolites against Kidney Cancer and Inflammation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Target Gene Prediction of SCFAs Microbial Metabolites and Diseases (Kidney Cancer and Inflammation)
2.2. Target Gene Location in Chromosomes and Tissues
2.3. Analysis of Target Gene Pathways Using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Databases
2.4. Protein−Protein Interaction (PPI) Network Analysis of Targeted Gene
2.5. Analysis of the Physiochemical and ADMET Characteristics of Microbial Compounds
2.6. Validation of the Expression of the Hub Targets
2.7. Protein and Ligand Preparation
2.8. Binding Site Identification and Grid Box Generation
2.9. Molecular Docking Simulation
3. Results
3.1. Retrieve Metabolites and Potential Target Proteins Linked to Kidney Cancer and Kidney Inflammation
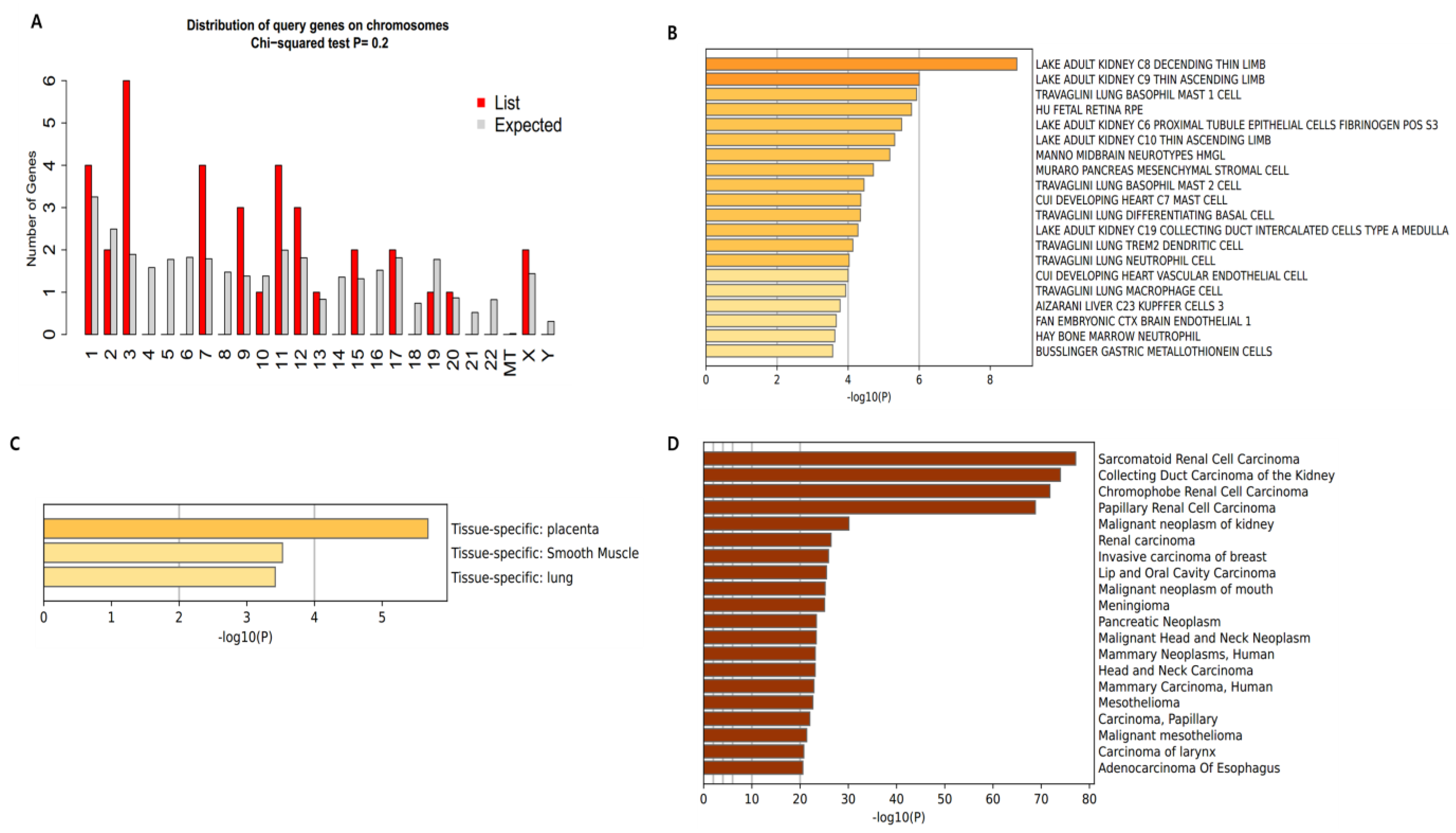
3.2. Distribution and Location of Genes
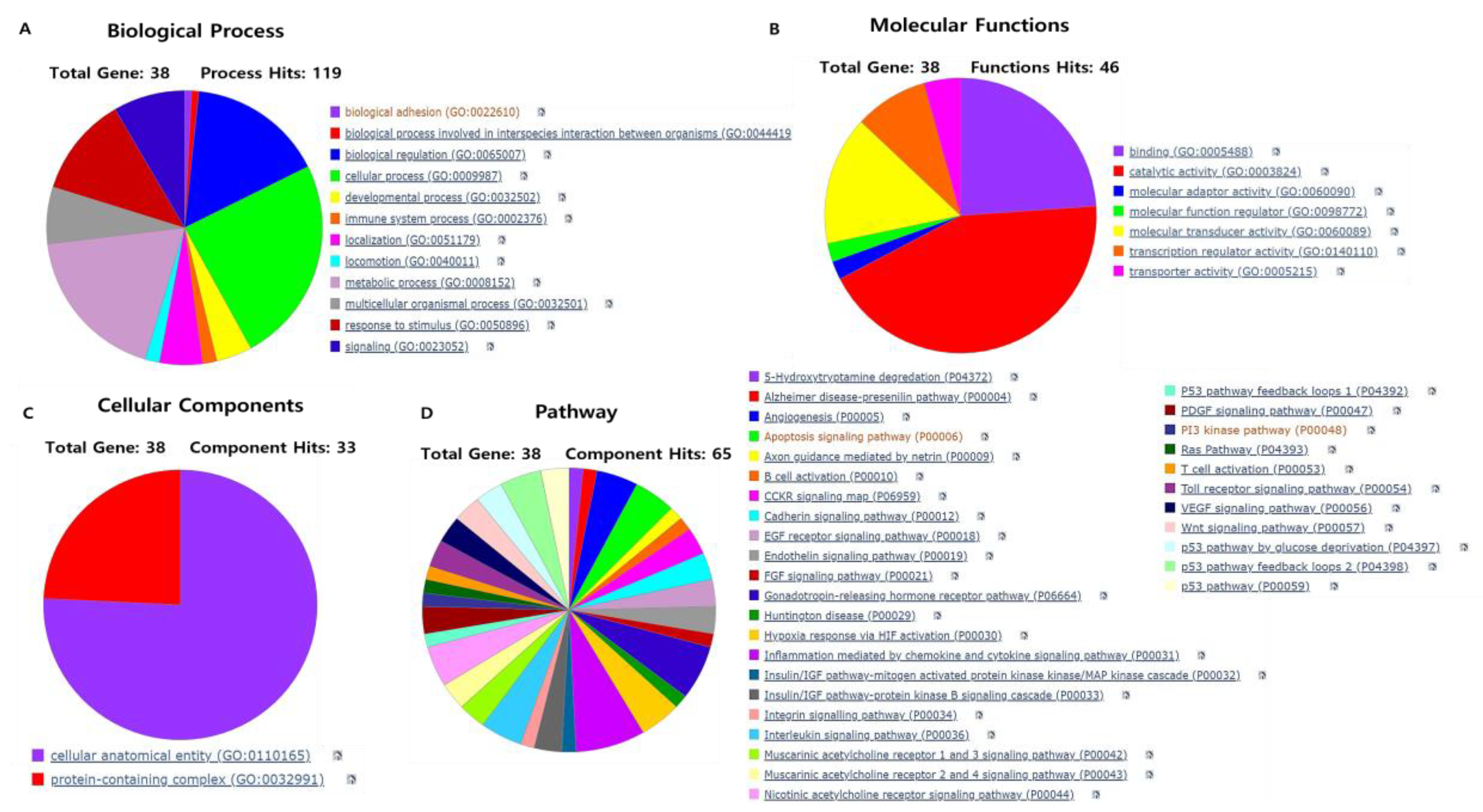
3.3. Gene Ontology and Pathway Analysis of Gene Targets
3.4. Screening of Hub Targets and PPI Network Construction
3.5. Physiochemical and ADMET Property Analysis of Lead Compounds That Control the Hub Targets Expression
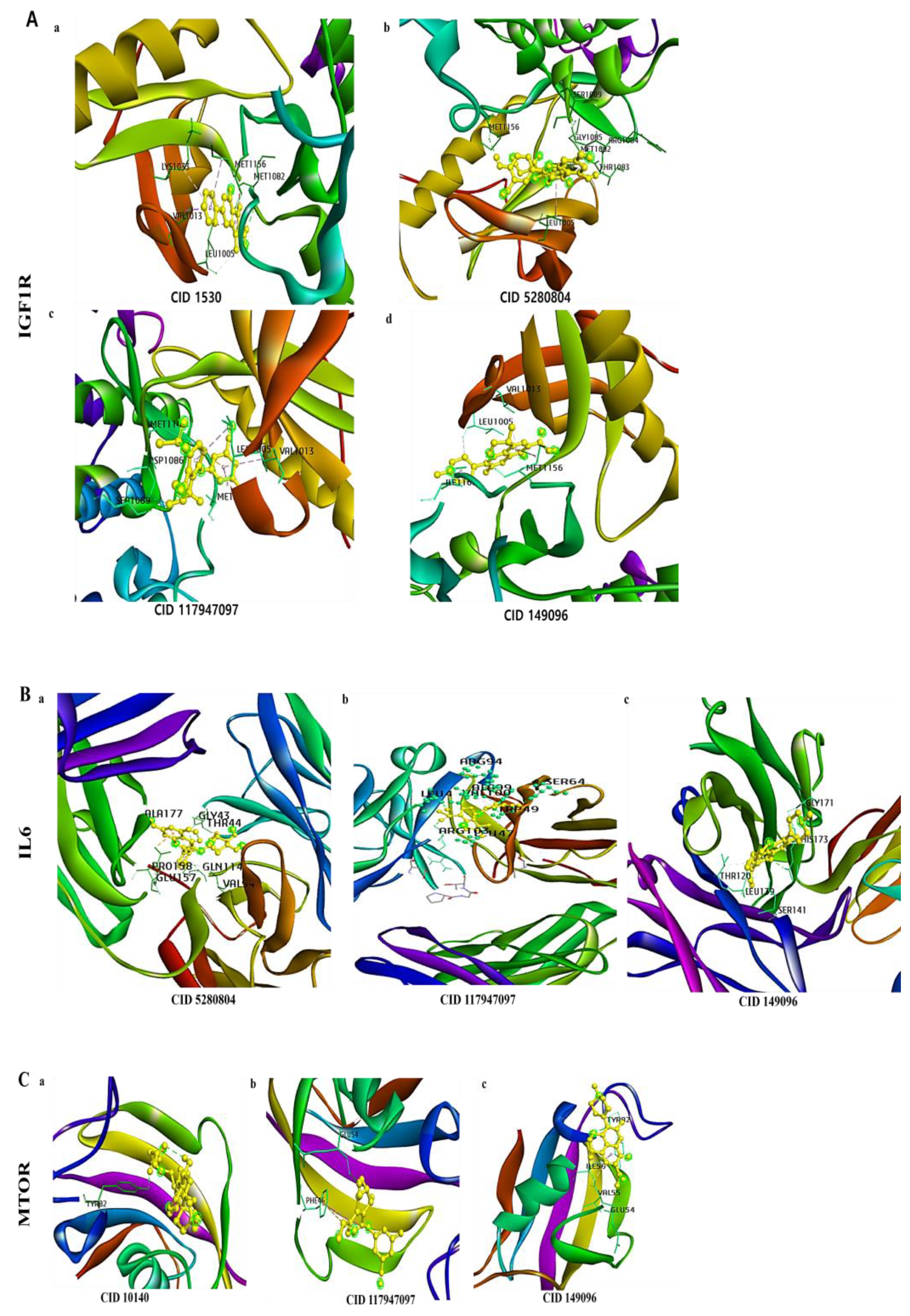
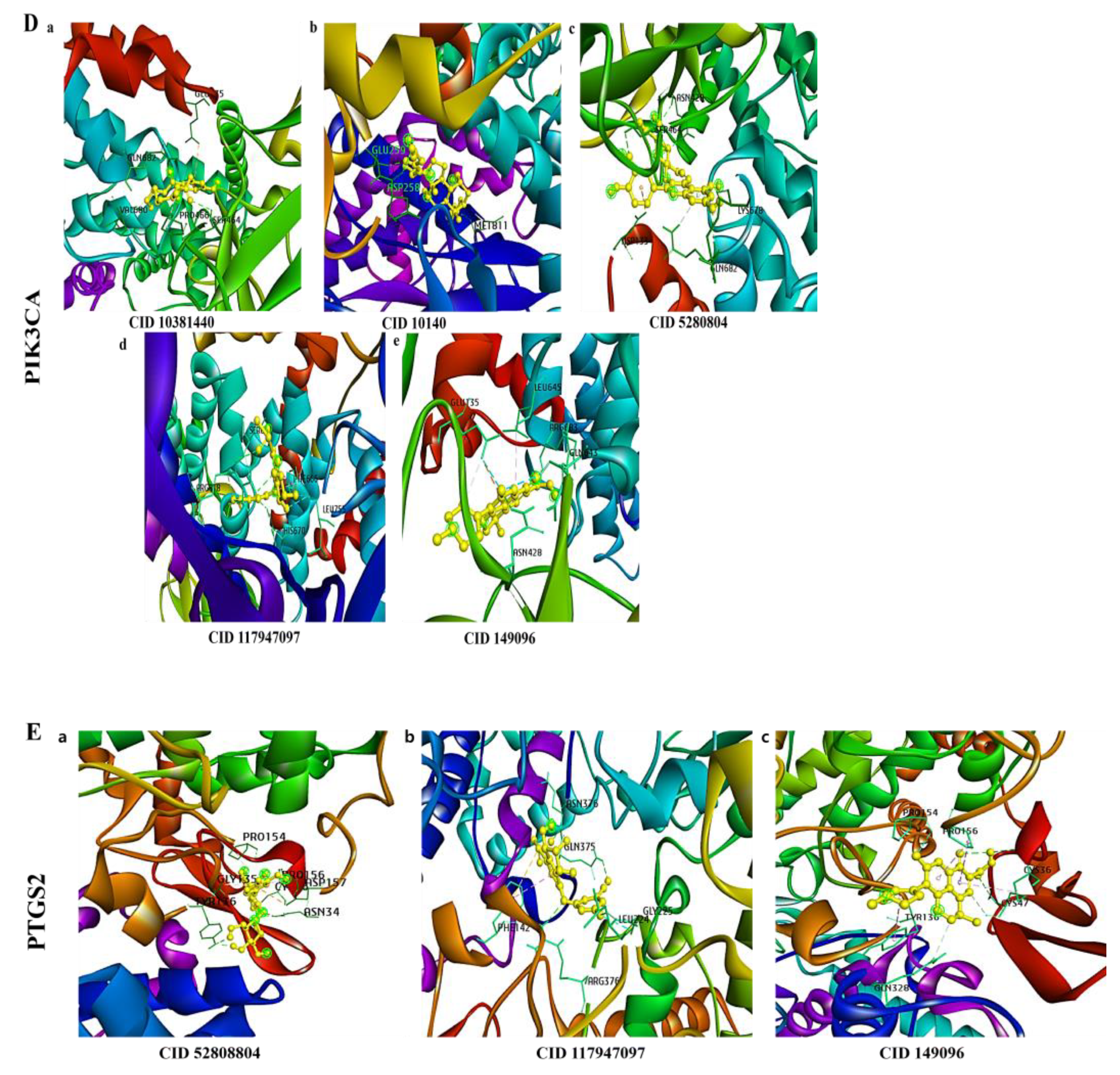
3.6. Molecular Docking of a Bioactive Compound with Its Target
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, J.; Wang, K.; Xiong, Z.; Yuan, C.; Wang, C.; Cao, Q.; Yu, H.; Meng, X.; Xie, K.; Cheng, Z.; et al. Impact of inflammation and immunotherapy in renal cell carcinoma. Oncol. Lett. 2020, 20, 272. [Google Scholar] [CrossRef] [PubMed]
- Sabarwal, A.; Kumar, K.; Singh, R.P. Hazardous effects of chemical pesticides on human health-Cancer and other associated disorders. Environ. Toxicol. Pharmacol. 2018, 63, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Chhikara, B.S.; Parang, K. Global Cancer Statistics 2022: The trends projection analysis. Chem. Biol. Lett. 2022, 10, 451. [Google Scholar]
- Galdiero, M.R.; Marone, G.; Mantovani, A. Cancer Inflammation and Cytokines. Cold Spring Harb. Perspect Biol. 2018, 10, a028662. [Google Scholar] [CrossRef] [PubMed]
- Kay, J.; Thadhani, E.; Samson, L.; Engelward, B. Inflammation-induced DNA damage, mutations and cancer. DNA Repair 2019, 83, 102673. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.; Debnath, B.; Neamati, N. Role of the CXCL8-CXCR1/2 Axis in Cancer and Inflammatory Diseases. Theranostics 2017, 7, 1543–1588. [Google Scholar] [CrossRef]
- Brighi, N.; Farolfi, A.; Conteduca, V.; Gurioli, G.; Gargiulo, S.; Gallà, V.; Schepisi, G.; Lolli, C.; Casadei, C.; De Giorgi, U. The Interplay between Inflammation, Anti-Angiogenic Agents, and Immune Checkpoint Inhibitors: Perspectives for Renal Cell Cancer Treatment. Cancers 2019, 11, 1935. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Smyth, M.J. Targeting cancer-related inflammation in the era of immunotherapy. Immunol. Cell Biol. 2017, 95, 325–332. [Google Scholar] [CrossRef]
- Hammers, H.J.; Plimack, E.R.; Infante, J.R.; Rini, B.I.; McDermott, D.F.; Lewis, L.D.; Voss, M.H.; Sharma, P.; Pal, S.K.; Razak, A.R.A.; et al. Safety and Efficacy of Nivolumab in Combination with Ipilimumab in Metastatic Renal Cell Carcinoma: The CheckMate 016 Study. J. Clin. Oncol. 2017, 35, 3851–3858. [Google Scholar] [CrossRef]
- Hsu, C.N.; Tain, Y.L. Chronic Kidney Disease and Gut Microbiota: What Is Their Connection in Early Life? Int. J. Mol. Sci. 2022, 23, 3954. [Google Scholar] [CrossRef]
- Yang, T.; Richards, E.M.; Pepine, C.J.; Raizada, M.K. The gut microbiota and the brain-gut-kidney axis in hypertension and chronic kidney disease. Nat. Rev. Nephrol. 2018, 14, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Hobby, G.P.; Karaduta, O.; Dusio, G.F.; Singh, M.; Zybailov, B.L.; Arthur, J.M. Chronic kidney disease and the gut microbiome. Am. J. Physiol. Renal Physiol. 2019, 316, F1211–F1217. [Google Scholar] [CrossRef]
- Al Khodor, S.; Shatat, I.F. Gut microbiome and kidney disease: A bidirectional relationship. Pediatr. Nephrol. 2017, 32, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, L.; Fu, P. Gut microbiota-derived short-chain fatty acids and kidney diseases. Drug Des. Devel. Ther. 2017, 11, 3531–3542. [Google Scholar] [CrossRef]
- Diaz Heijtz, R.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Uronis, J.M.; Mühlbauer, M.; Herfarth, H.H.; Rubinas, T.C.; Jones, G.S.; Jobin, C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE 2009, 4, e6026. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Cheng, L.; Qi, C.; Yang, H.; Lu, M.; Cai, Y.; Fu, T.; Ren, J.; Jin, Q.; Zhang, X. gutMGene: A comprehensive database for target genes of gut microbes and microbial metabolites. Nucleic Acids Res. 2022, 50, D795–D800. [Google Scholar] [CrossRef]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005, 33, D514–D517. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.31–31.30.33. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Shi, L.; Yu, L.; Zou, F.; Hu, H.; Liu, K.; Lin, Z. Gene expression profiling and functional analysis reveals that p53 pathway-related gene expression is highly activated in cancer cells treated by cold atmospheric plasma-activated medium. PeerJ 2017, 5, e3751. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Sikić, M.; Tomić, S.; Vlahovicek, K. Prediction of protein-protein interaction sites in sequences and 3D structures by random forests. PLoS Comput. Biol. 2009, 5, e1000278. [Google Scholar] [CrossRef]
- Rao, V.S.; Srinivas, K.; Sujini, G.N.; Kumar, G.N. Protein-protein interaction detection: Methods and analysis. Int. J. Proteom. 2014, 2014, 147648. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [CrossRef]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef]
- Opo, F.A.; Rahman, M.M.; Ahammad, F.; Ahmed, I.; Bhuiyan, M.A.; Asiri, A.M. Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci. Rep. 2021, 11, 4049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, X.; Zhou, W.; Cheng, G.; Wu, J.; Guo, S.; Jia, S.; Liu, Y.; Li, B.; Zhang, X.; et al. A bioinformatics investigation into molecular mechanism of Yinzhihuang granules for treating hepatitis B by network pharmacology and molecular docking verification. Sci. Rep. 2020, 10, 11448. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Kemmish, H.; Fasnacht, M.; Yan, L. Fully automated antibody structure prediction using BIOVIA tools: Validation study. PLoS ONE 2017, 12, e0177923. [Google Scholar] [CrossRef] [PubMed]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Toutain, P.L.; Bousquet-Melou, A. Free drug fraction vs. free drug concentration: A matter of frequent confusion. J. Vet. Pharmacol. Ther. 2002, 25, 460–463. [Google Scholar] [CrossRef]
- Zhou, J.; Gong, K. Belzutifan: A novel therapy for von Hippel–Lindau disease. Nat. Rev. Nephrol. 2022, 18, 205–206. [Google Scholar] [CrossRef]
- McGregor, J.C.; Allen, G.P.; Bearden, D.T. Levofloxacin in the treatment of complicated urinary tract infections and acute pyelonephritis. Ther. Clin. Risk Manag. 2008, 4, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Bukavina, L.; Bensalah, K.; Bray, F.; Carlo, M.; Challacombe, B.; Karam, J.A.; Kassouf, W.; Mitchell, T.; Montironi, R.; O’Brien, T.; et al. Epidemiology of Renal Cell Carcinoma: 2022 Update. Eur. Urol. 2022, 82, 529–542. [Google Scholar] [CrossRef]
- Kibble, M.; Saarinen, N.; Tang, J.; Wennerberg, K.; Mäkelä, S.; Aittokallio, T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat. Prod. Rep. 2015, 32, 1249–1266. [Google Scholar] [CrossRef] [PubMed]
- Athanasios, A.; Charalampos, V.; Vasileios, T.; Ashraf, G.M. Protein-Protein Interaction (PPI) Network: Recent Advances in Drug Discovery. Curr. Drug Metab. 2017, 18, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Balani, S.K.; Miwa, G.T.; Gan, L.S.; Wu, J.T.; Lee, F.W. Strategy of utilizing in vitro and in vivo ADME tools for lead optimization and drug candidate selection. Curr. Top. Med. Chem. 2005, 5, 1033–1038. [Google Scholar] [CrossRef]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef]
- Mostashari-Rad, T.; Arian, R.; Sadri, H.; Mehridehnavi, A.; Mokhtari, M.; Ghasemi, F.; Fassihi, A. Study of CXCR4 chemokine receptor inhibitors using QSPR and molecular docking methodologies. J. Theor. Comput. Chem. 2019, 18, 1950018. [Google Scholar] [CrossRef]
- Chen, F.; Chen, X.; Yang, D.; Che, X.; Wang, J.; Li, X.; Zhang, Z.; Wang, Q.; Zheng, W.; Wang, L.; et al. Isoquercitrin inhibits bladder cancer progression in vivo and in vitro by regulating the PI3K/Akt and PKC signaling pathways. Oncol. Rep. 2016, 36, 165–172. [Google Scholar] [CrossRef]
- Chen, Q.; Li, P.; Li, P.; Xu, Y.; Li, Y.; Tang, B. Isoquercitrin inhibits the progression of pancreatic cancer in vivo and in vitro by regulating opioid receptors and the mitogen-activated protein kinase signalling pathway. Oncol. Rep. 2015, 33, 840–848. [Google Scholar] [CrossRef]
- Huang, G.; Tang, B.; Tang, K.; Dong, X.; Deng, J.; Liao, L.; Liao, Z.; Yang, H.; He, S. Isoquercitrin inhibits the progression of liver cancer in vivo and in vitro via the MAPK signalling pathway. Oncol. Rep. 2014, 31, 2377–2384. [Google Scholar] [CrossRef]
- Lee, E.-H.; Park, H.-J.; Jung, H.-Y.; Kang, I.-K.; Kim, B.-O.; Cho, Y.-J. Isoquercitrin isolated from newly bred Green ball aapple peel in lipopolysaccharide-stimulated macrophage regulates NF-κB inflammatory pathways and cytokines. 3 Biotech 2022, 12, 100. [Google Scholar] [CrossRef]
- Ge, X.; Huang, S.; Ren, C.; Zhao, L. Taurocholic Acid and Glycocholic Acid Inhibit Inflammation and Activate Farnesoid X Receptor Expression in LPS-Stimulated Zebrafish and Macrophages. Molecules 2023, 28, 2005. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.M.; Ko, W.; Lee, S.J.; Kim, Y.-C.; Son, J.-Y.; Sohn, J.H.; Yim, J.H.; Oh, H. Anti-Inflammatory Effects of Curvularin-Type Metabolites from a Marine-Derived Fungal Strain Penicillium sp. SF-5859 in Lipopolysaccharide-Induced RAW264.7 Macrophages. Mar. Drugs 2017, 15, 282. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.J.; Basnakian, A.G.; Orloff, M.S.; Ning, B.; Yao-Borengasser, A.; Raj, V.; Kadlubar, S. 2-amino-1-methyl-6-phenylimidazo(4,5-b)pyridine (PhIP) induces gene expression changes in JAK/STAT and MAPK pathways related to inflammation, diabetes and cancer. Nutr. Metab. 2016, 13, 54. [Google Scholar] [CrossRef] [PubMed]
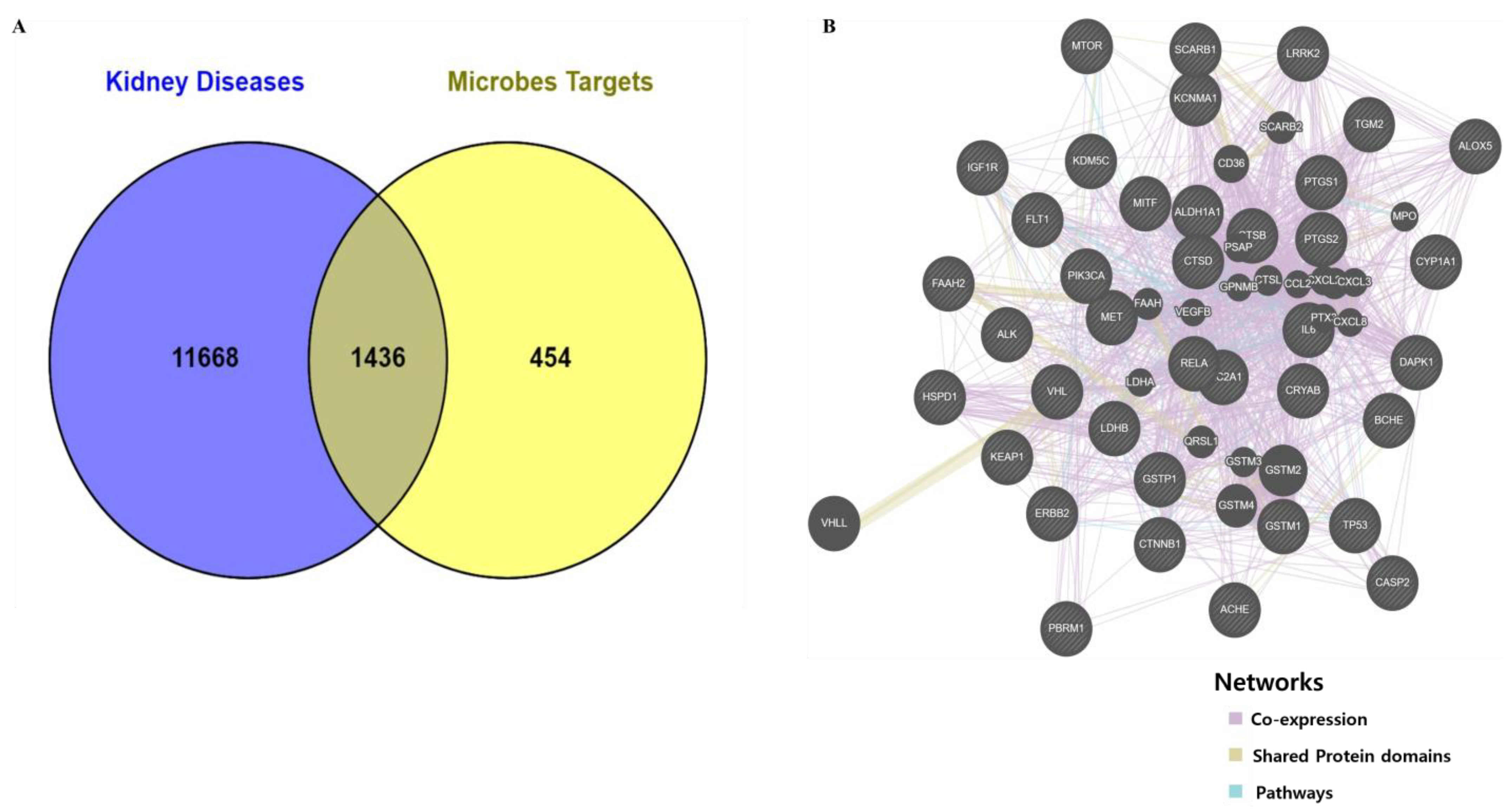

| Name | Betweenness Centrality | Closeness Centrality | Degree | Number of Directed Edges |
|---|---|---|---|---|
| TP53 | 0.319071552 | 0.872340426 | 35 | 2 |
| CTNNB1 | 0.108958047 | 0.732142857 | 26 | 20 |
| IL6 | 0.156313791 | 0.683333333 | 23 | 23 |
| MTOR | 0.054536656 | 0.672131148 | 22 | 5 |
| PTGS2 | 0.064988518 | 0.650793651 | 20 | 35 |
| PIK3CA | 0.03684383 | 0.630769231 | 18 | 17 |
| ERBB2 | 0.0341558 | 0.630769231 | 17 | 10 |
| NFKBIA | 0.010607996 | 0.594202899 | 14 | 26 |
| KEAP1 | 0.012401937 | 0.585714286 | 13 | 7 |
| SIRT1 | 0.020993867 | 0.585714286 | 13 | 6 |
| RELA | 0.003634516 | 0.569444444 | 11 | 18 |
| GSTP1 | 0.01601626 | 0.554054054 | 11 | 22 |
| MET | 0.005245046 | 0.561643836 | 10 | 4 |
| SLC2A1 | 0.007293932 | 0.561643836 | 10 | 3 |
| IGF1R | 0.000513 | 0.554054054 | 9 | 6 |
| VHL | 0.003383222 | 0.525641026 | 8 | 7 |
| LRRK2 | 0.006013884 | 0.532467532 | 8 | 14 |
| CTSD | 0.006634727 | 0.518987342 | 8 | 8 |
| CYP1A1 | 0.005259476 | 0.539473684 | 7 | 1 |
| FLT1 | 0.001178862 | 0.5125 | 7 | 1 |
| Hub Target Genes | Gene Targeted Compounds and ID | Target Microbes [19] |
|---|---|---|
| TP53 | Bile acid (439520, CHEBI:22868) | Bacteroides distasonis, Clostridium scindens, Faecalibacterium prausnitzii, Haemophilus parainfluenzae. |
| CTNNB1 | 2-Hydroxy-3-(5-hydroxy-1H-indol-3-yl)propanoic acid (192215) | Clostridium sporogenes. |
| 3-Hydroxy-4-methoxybenzenepropanoic acid (2752054, HMDB0131138) | Clostridium orbiscindens, Eubacterium ramulus, | |
| Dihydrocaffeic acid (348154, HMDB0000423, CHEBI:48400) | Bifidobacterium, Bifidobacterium longum, Clostridium orbiscindens, Clostridium sporogenes, Eubacterium ramulus, Faecalibacterium prausnitzii, Lactobacillus mucosae, Lactobacillus zeae. | |
| Indole-3-lactic acid (92904, CHEBI:24813) | Clostridium sporogenes. | |
| MTOR | 11-Methoxycurvularin (10381440) | Bacillus sp. |
| Dihydrodaidzein (176907, HMDB0005760, CHEBI:75842) | Blautia producta, Bacillus sp., Clostridium sp., Lactobacillus mucosae, Lactococcus sp., | |
| Glycocholic acid (10140, HMDB0000138, CHEBI:17687) | Bacteroides fragilis, Butyricicoccus pullicaecorum, Ruminococcus flavefaciens. | |
| PIK3CA | 11-Methoxycurvularin (10381440) | Bacillus sp. |
| 3-Hydroxyphenethyl alcohol (83404) | Bifidobacterium. | |
| Caffeic acid (689043, HMDB0001964, CHEBI:16433) | Bifidobacterium, Bifidobacterium animalis. | |
| Dihydrodaidzein (176907, HMDB0005760, CHEBI:75842) | Blautia producta, Bacillus sp., Clostridium sp., Lactobacillus mucosae, Lactococcus sp., | |
| Dihydroglycitein (101101166, CHEBI:174736) | Eubacterium limosum. | |
| Glycocholic acid (10140, HMDB0000138, CHEBI:17687) | Bacteroides fragilis, Butyricicoccus pullicaecorum, Ruminococcus flavefaciens. | |
| Isoquercitrin (5280804, HMDB0037362, CHEBI:68352) | Bacillus sp., Bacteroides sp., Eubacterium ramulus. | |
| IL6 | Acetate (175, CHEBI:30089) | Bacteroides thetaiotaomicron, Bacteroidetes, Bifidobacterium dentium, Bifidobacterium longum, Blautia faecis, Clostridium asparagiforme, Clostridium pasteurianum, Clostridium scindens, Clostridium sp. L2-50, Eubacterium limosum, Eubacterium ramulus, Eubacterium rectale, Lawsonibacter asaccharolyticus, Ruminococcus champanellensis, Succinivibrio dextrinosolvens, |
| Butyrate (104775, CHEBI:17968) | Butyricimonas synergistica, Butyricimonas virosa, Clostridium, Clostridium butyricum, Clostridium pasteurianum, Clostridium tyrobutyricum, Eubacterium hallii, Eubacterium limosum, Eubacterium ramulus, Eubacterium rectale, Faecalibacterium prausnitzii, Firmicutes, Fusobacteriia, Lawsonibacter asaccharolyticus, Prevotella copri, Roseburia inulinivorans. | |
| Isoquercitrin (5280804, HMDB0037362, CHEBI:68352) | Bacillus sp., Bacteroides sp., Eubacterium ramulus. | |
| Propionate (104745, CHEBI:17272) | Bacteroides, Bacteroides thetaiotaomicron, Eubacterium limosum, Haemophilus parainfluenzae, Parasutterella excrementihominis, Phascolarctobacterium succinatutens, Propionibacterium avidum, Roseburia inulinivorans, Ruminococcus bromii, Veillonella, Veillonella ratti. | |
| ERBB2 | 2,3-Dihydroxypropyl (E)-3-(3,4-dihydroxyphenyl)prop-2-enoate (5315606) | Bifidobacterium. |
| 2-Hydroxy-3-(5-hydroxy-1H-indol-3-yl)propanoic acid (192215) | Clostridium sporogenes. | |
| 3-Hydroxy-4-methoxybenzenepropanoic acid (2752054, HMDB0131138) | Clostridium orbiscindens, Eubacterium ramulus. | |
| 4-Hydroxy-(3′,4′-dihydroxyphenyl)-valeric acid (52920332, HMDB0041679, CHEBI:137478) | Lactobacillus plantarum. | |
| 5-(3,4-Dihydroxyphenyl)-valerolactone (45093073) | Lactobacillus plantarum. | |
| Caffeic acid (689043, HMDB0001964, CHEBI:16433) | Bifidobacterium, Bifidobacterium animalis. | |
| Ethyl phenyllactate, (-)- (9877619, HMDB0032618) | Bacteroides caccae, Clostridium sp. | |
| Hydroquinone (785, HMDB0002434, CHEBI:17594) | Bacteroides, Bifidobacterium, Bifidobacterium longum, Eubacterium. | |
| Indole-3-lactic acid (92904, CHEBI:24813) | Clostridium sporogenes. | |
| PTGS2 | (R)-3-(4-Hydroxyphenyl)lactate (9548632, CHEBI:10980) | Bacteroides caccae, Clostridium sp. |
| 2-(4-Hydroxyphenyl)propionic acid, (2S)- (6971268) | Eubacterium ramulus. | |
| 2,3-Dihydroxypropyl (E)-3-(3,4-dihydroxyphenyl)prop-2-enoate (5315606) | Bifidobacterium, | |
| 2-Hydroxy-3-(4-hydroxyphenyl)propanoic acid (9378, HMDB0000755, CHEBI:17385) | Clostridium sporogenes. | |
| 2-Hydroxy-3-(5-hydroxy-1H-indol-3-yl)propanoic acid (192215) | Clostridium sporogenes. | |
| 3-(3,4-Dihydroxyphenyl)-2-hydroxypropanoic acid (439435, HMDB0003503, CHEBI:17807) | Clostridium sporogenes. | |
| 3-(3-Hydroxyphenyl)propanoic acid (91, HMDB0000375, CHEBI:1427) | Bifidobacterium. | |
| 3-(4-Hydroxyphenyl)propionic acid (10394, HMDB0002199, CHEBI:32980) | Clostridium orbiscindens, Eubacterium ramulus. | |
| 3,4-Dihydroxybenzoic acid (72, CHEBI:36062) | Bacteroides sp. | |
| 3,4-Dihydroxyphenylacetic acid (547, HMDB0001336, CHEBI:41941) | Clostridium orbiscindens, Eubacterium ramulus, | |
| 3-Hydroxy-4-methoxybenzenepropanoic acid (2752054, HMDB0131138) | Clostridium orbiscindens, Eubacterium ramulus, | |
| 3-Hydroxybenzoic acid (7420, HMDB0002466, CHEBI:30764) | Eubacterium. | |
| 3-Hydroxyphenethyl alcohol (83404) | Bifidobacterium. | |
| 3-Phenylpropionic acid (107, CHEBI:28631) | Clostridium sporogenes | |
| 4-Hydroxy-(3′,4′-dihydroxyphenyl)-valeric acid (52920332, HMDB0041679, CHEBI:137478) | Lactobacillus plantarum. | |
| 4-Hydroxybenzoic acid (135, HMDB0000500, CHEBI:30763) | Eubacterium. | |
| 4-Hydroxyphenylacetic acid (127, HMDB0000020, CHEBI:18101) | Eubacterium ramulus, | |
| Caffeic acid (689043, HMDB0001964, CHEBI:16433) | Bifidobacterium, Bifidobacterium animalis. | |
| Dihydrocaffeic acid (348154, HMDB0000423, CHEBI:48400) | Bifidobacterium, Bifidobacterium longum, Clostridium orbiscindens, Clostridium sporogenes, Eubacterium ramulus, Faecalibacterium prausnitzii, Lactobacillus mucosae, Lactobacillus zeae. | |
| D-Lactic acid (61503, HMDB0001311, CHEBI:42111) | Faecalibacterium prausnitzii | |
| Ethyl phenyllactate, (-)- (9877619, HMDB0032618) | Bacteroides caccae, Clostridium sp. | |
| Isobutyric acid (6590, HMDB0001873, CHEBI:16135) | Butyricimonas synergistica, Butyricimonas virosa | |
| Isoquercitrin (5280804, HMDB0037362, CHEBI:68352) | Bacillus sp., Bacteroides sp., Eubacterium ramulus. | |
| Leucine (6106, HMDB0000687, CHEBI:15603) | Blautia, Faecalibacterium prausnitzii, Ruminococcus | |
| Phenolic acid (CHEBI:166890) | Eubacterium ramulus. | |
| Phenylacetic acid (999, HMDB0000209, CHEBI:30745) | Bifidobacterium. | |
| Pipecolic acid (849, HMDB0000070, CHEBI:17964) | Lactobacillus casei. | |
| Proline (145742, HMDB0000162, CHEBI:17203) | Blautia, Ruminococcus. | |
| Quinic acid (6508, HMDB0003072, CHEBI:17521) | Bifidobacterium animalis. | |
| IGF1R | 2-Amino-1-methyl-6-phenylimidazo[4,5-b] pyridine (1530, CHEBI:76290) | Blautia obeum, Faecalibacterium prausnitzii, Lactobacillus reuteri. |
| 3-(3,4-Dihydroxyphenyl)-2-hydroxypropanoic acid (439435, HMDB0003503, CHEBI:17807) | Clostridium sporogenes | |
| 3-(3-Hydroxyphenyl)propanoic acid (91, HMDB0000375, CHEBI:1427) | Bifidobacterium. | |
| 3-(4-Hydroxyphenyl)propionic acid (10394, HMDB0002199, CHEBI:32980) | Clostridium orbiscindens, Eubacterium ramulus. | |
| 3,4-Dihydroxybenzoic acid (72, CHEBI:36062) | Bacteroides sp. | |
| 3,4-Dihydroxyphenylacetic acid (547, HMDB0001336, CHEBI:41941) | Clostridium orbiscindens, Eubacterium ramulus, | |
| 3-Hydroxy-4-methoxybenzenepropanoic acid (2752054, HMDB0131138) | Clostridium orbiscindens, Eubacterium ramulus, | |
| 3-Hydroxybenzoic acid (7420, HMDB0002466, CHEBI:30764) | Eubacterium. | |
| 4-Hydroxy-(3′,4′-dihydroxyphenyl)-valeric acid (52920332, HMDB0041679, CHEBI:137478) | Lactobacillus plantarum. | |
| 4-Hydroxybenzoic acid (135, HMDB0000500, CHEBI:30763) | Eubacterium. | |
| Dihydrocaffeic acid (348154, HMDB0000423, CHEBI:48400) | Bifidobacterium, Bifidobacterium longum, Clostridium orbiscindens, Clostridium sporogenes, Eubacterium ramulus, Faecalibacterium prausnitzii, Lactobacillus mucosae, Lactobacillus zeae. | |
| Dihydrodaidzein (176907, HMDB0005760, CHEBI:75842) | Blautia producta, Bacillus sp., Clostridium sp., Lactobacillus mucosae, Lactococcus sp., | |
| Ethyl phenyllactate, (-)- (9877619, HMDB0032618) | Bacteroides caccae, Clostridium sp. | |
| Glutathione (124886, HMDB0000125, CHEBI:16856) | Bacteroides thetaiotaomicron. | |
| Isoquercitrin (5280804, HMDB0037362, CHEBI:68352) | Bacillus sp., Bacteroides sp., Eubacterium ramulus. | |
| Phenolic acid (CHEBI:166890) | Eubacterium ramulus. | |
| RELA | 2,3-Dihydroxypropyl (E)-3-(3,4-dihydroxyphenyl)prop-2-enoate (5315606) | Bifidobacterium. |
| Caffeic acid (689043, HMDB0001964, CHEBI:16433) | Bifidobacterium, Bifidobacterium animalis. |
| Targets | Compound | Binging Energy | Hydrogen Bond | Other Bonds | Grid Box Center | Dimension |
|---|---|---|---|---|---|---|
| IGF1R | 2-Amino-1-methyl-6-phenylimidazo(4,5-b) pyridine_CID_1530 | −7.4 | MET | LEU, VAL, LYS, MET | x= 20.89, y = 3.76, z= 45.48 | x = 74.703, y = 55.85, z = 62.013 |
| Isoquercetin_COMPOUND_CID_5280804 | −7.9 | MET, THR | GLY, ARG, LEU, SER | |||
| Control_Belzutifan_Cancer_CID_117947097 | −7.3 | SER | LEU, ASP, MET, VAL | |||
| Control_levofloxacin__inflammation_CID_149096 | −7.6 | LEU | MET, VAL, ILE | |||
| IL6 | Isoquercetin_CID_5280804 | −7.9 | ALA, GLN, GLY, THR | VAL, GLU, PRO | x = 16.06, y = 14.52, z = 22.37 | x = 64.69, y = 83.398, z = 56.699 |
| Belzutifan_Cancer_CID_117947097 | −7.5 | ARG, LEU, SER, TRP | ALA, LEU, ARG, VAL | |||
| levofloxacin__inflammation_CID_149096 | −7.5 | HIS, LEU | SER, GLY, THR | |||
| MTOR | 2fap_Glycocholic acid_COMPOUND_CID_10140 | −6.5 | TYR | x = −6.45, y = 21.63, z = 43.68 | x = 35.723, y = 45.28, z = 33.53 | |
| 2fap_Belzutifan_Cancer_COMPOUND_CID_117947097 | −7.4 | GLU | PHE | |||
| 2fap_levofloxacin__inflammation_COMPOUND_CID_149096 | −6.3 | GLU | TYR, ILE, VAL | |||
| PIK3CA | 5dxt.p_11-Methoxycurvularin_COMPOUND_CID_10381440 | −8.2 | GLN, SER | VAL, GLU, PRO | x = −1.83, y = 5.71, z = 17.04 | x = 79.48, y = 96.48, z = 89.84 |
| 5dxt.p_Glycocholic acid_COMPOUND_CID_10140 | −9.3 | MET, GLU, ASP | ||||
| 5dxt.p_Isoquercetin_CID_5280804 | −8.4 | GLN, LYN, SER, ASN | ASP | |||
| 5dxt.p_Belzutifan__CID_117947097 | −7.3 | ARG, HIS | SER, PHE, LEU | |||
| 5dxt.p_levofloxacin__inflammation_COMPOUND_CID_149096 | −8.1 | ARG, ASN | GLU, LEU, GLN | |||
| PTGS2 | 5ikq.pp_Isoquercetin_COMPOUND_CID_5280804 | −9.5 | TYR, ASN | PRO, CYS, ASP | x = 27.15, y = 38.38, z= 41.79 | x = 82.98, y = 83.28, z = 104.06 |
| 5ikq.pp_Belzutifan_Cancer_COMPOUND_CID_117947097 | −9.1 | ARG, GLN, ASN | LEU, GLY, PHE | |||
| 5ikq.pp_levofloxacin__inflammation_COMPOUND_CID_149096 | −9 | CYS | PRO, TYR, GLN |
| Final Target Gene | Gene Targeted Compounds and ID | Target Microbes [19] |
|---|---|---|
| MTOR | 11-Methoxycurvularin (10381440) | Bacillus sp. |
| Glycocholic acid (10140, HMDB0000138, CHEBI:17687) | Bacteroides fragilis, Butyricicoccus pullicaecorum, Ruminococcus flavefaciens. | |
| PIK3CA | 11-Methoxycurvularin (10381440) | Bacillus sp. |
| Glycocholic acid (10140, HMDB0000138, CHEBI:17687) | Bacteroides fragilis, Butyricicoccus pullicaecorum, Ruminococcus flavefaciens. | |
| Isoquercitrin (5280804, HMDB0037362, CHEBI:68352) | Bacillus sp., Bacteroides sp., Eubacterium ramulus. | |
| IL6 | Isoquercitrin (5280804, HMDB0037362, CHEBI:68352) | Bacillus sp., Bacteroides sp., Eubacterium ramulus. |
| PTGS2 | Isoquercitrin (5280804, HMDB0037362, CHEBI:68352) | Bacillus sp., Bacteroides sp., Eubacterium ramulus. |
| IGF1R | 2-Amino-1-methyl-6-phenylimidazo[4,5-b] pyridine (1530, CHEBI:76290) | Blautia obeum, Faecalibacterium prausnitzii, Lactobacillus reuteri. |
| Isoquercitrin (5280804, HMDB0037362, CHEBI:68352) | Bacillus sp., Bacteroides sp., Eubacterium ramulus. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karim, M.R.; Morshed, M.N.; Iqbal, S.; Mohammad, S.; Mathiyalagan, R.; Yang, D.C.; Kim, Y.J.; Song, J.H.; Yang, D.U. A Network Pharmacology and Molecular-Docking-Based Approach to Identify the Probable Targets of Short-Chain Fatty-Acid-Producing Microbial Metabolites against Kidney Cancer and Inflammation. Biomolecules 2023, 13, 1678. https://doi.org/10.3390/biom13111678
Karim MR, Morshed MN, Iqbal S, Mohammad S, Mathiyalagan R, Yang DC, Kim YJ, Song JH, Yang DU. A Network Pharmacology and Molecular-Docking-Based Approach to Identify the Probable Targets of Short-Chain Fatty-Acid-Producing Microbial Metabolites against Kidney Cancer and Inflammation. Biomolecules. 2023; 13(11):1678. https://doi.org/10.3390/biom13111678
Chicago/Turabian StyleKarim, Md. Rezaul, Md. Niaj Morshed, Safia Iqbal, Shahnawaz Mohammad, Ramya Mathiyalagan, Deok Chun Yang, Yeon Ju Kim, Joon Hyun Song, and Dong Uk Yang. 2023. "A Network Pharmacology and Molecular-Docking-Based Approach to Identify the Probable Targets of Short-Chain Fatty-Acid-Producing Microbial Metabolites against Kidney Cancer and Inflammation" Biomolecules 13, no. 11: 1678. https://doi.org/10.3390/biom13111678
APA StyleKarim, M. R., Morshed, M. N., Iqbal, S., Mohammad, S., Mathiyalagan, R., Yang, D. C., Kim, Y. J., Song, J. H., & Yang, D. U. (2023). A Network Pharmacology and Molecular-Docking-Based Approach to Identify the Probable Targets of Short-Chain Fatty-Acid-Producing Microbial Metabolites against Kidney Cancer and Inflammation. Biomolecules, 13(11), 1678. https://doi.org/10.3390/biom13111678