How the Intrinsically Disordered N-Terminus of Cancer/Testis Antigen MAGEA10 Is Responsible for Its Expression, Nuclear Localisation and Aberrant Migration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Cells and Transfections
2.3. Western Blot and Flow Cytometry
2.4. Confocal Microscopy
2.5. Purification of EVs
3. Results
3.1. The N-Terminal Intrinsically Disordered Region of MAGEA10 Is Responsible for Aberrant Migration in SDS-PAGE
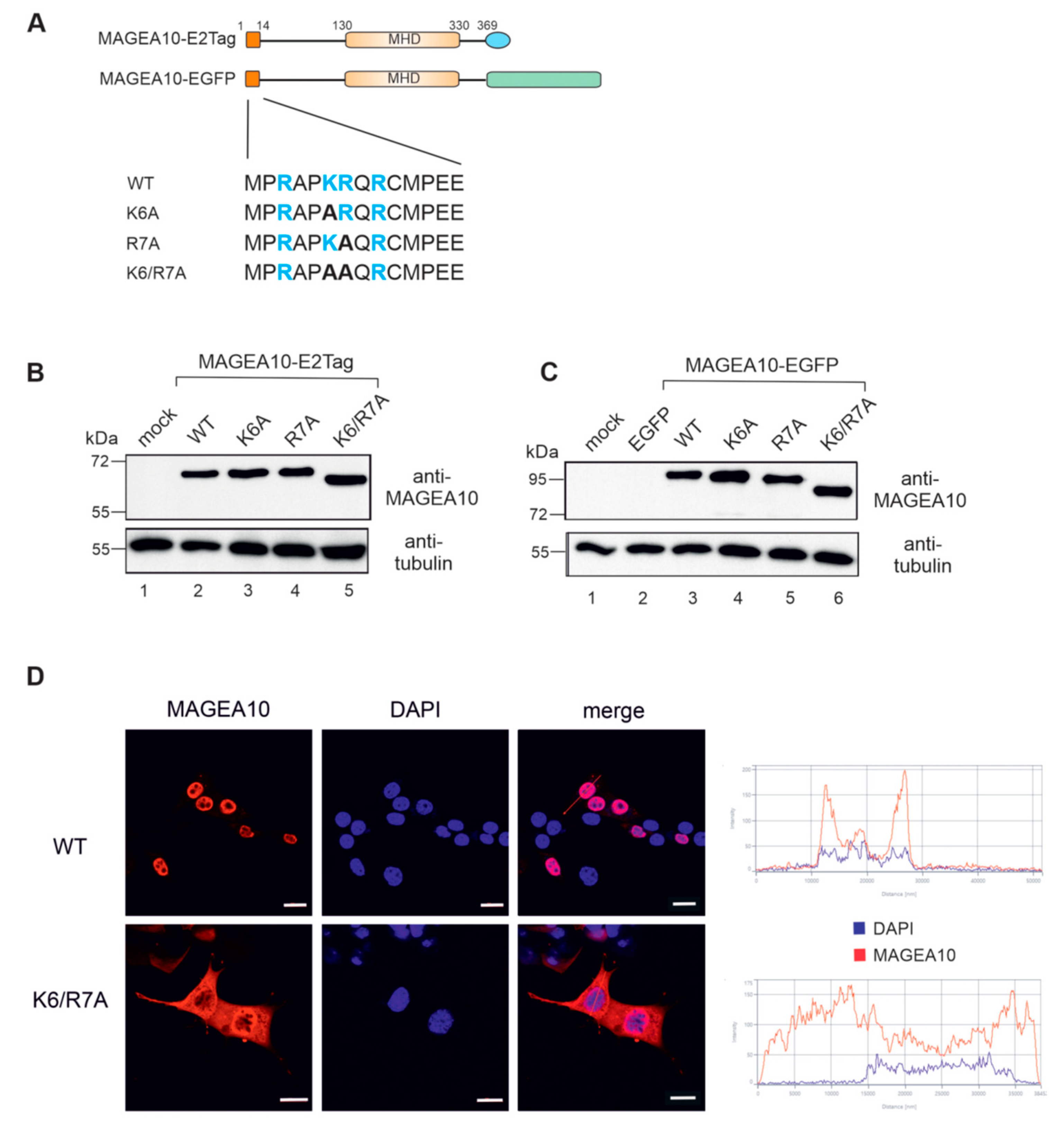
3.2. Amino Acids K6 and R7 Are Required for the Nuclear Localisation of MAGEA10
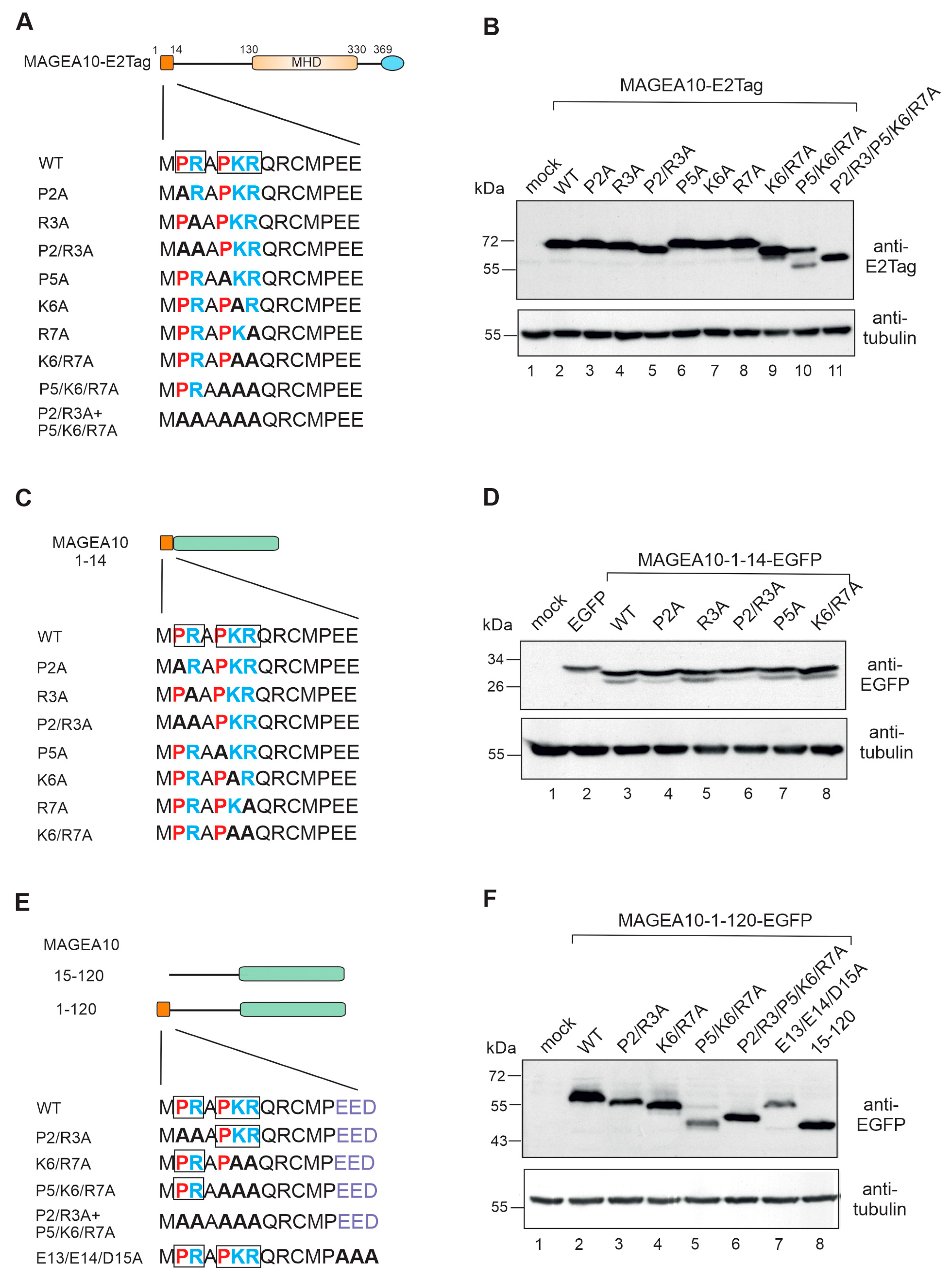
3.3. The First Seven Amino Acids in the N-Terminus Are Responsible for Aberrant Migration
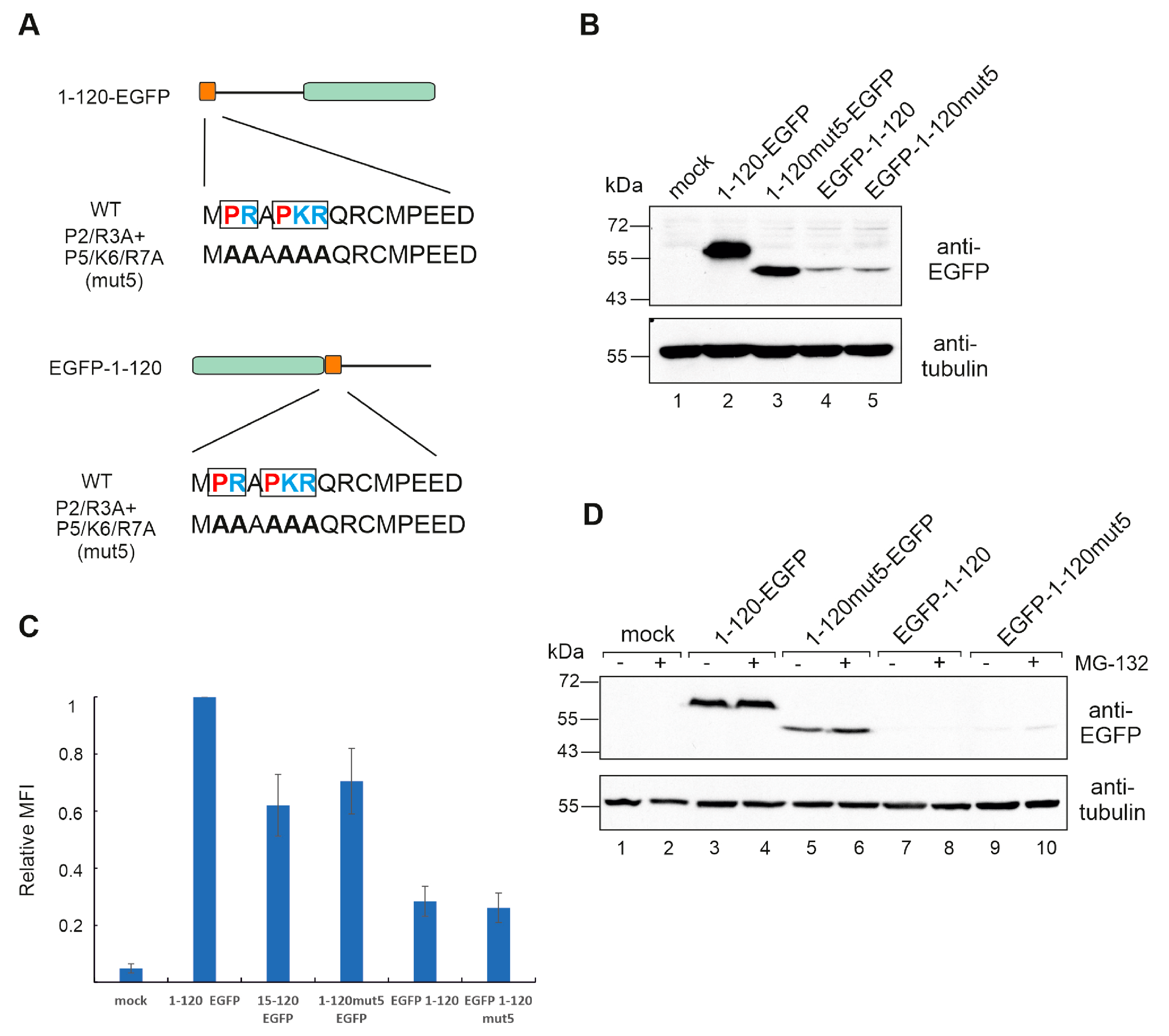
3.4. Hiding the N-Terminus Affects MAGEA10 Expression and Mobility in Gel
3.5. Mutations within the N-Terminus Does Not Affect MAGEA10 Incorporation into EVs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simpson, A.J.G.; Caballero, O.L.; Jungbluth, A.; Chen, Y.-T.; Old, L.J. Cancer/Testis Antigens, Gametogenesis and Cancer. Nat. Rev. Cancer 2005, 5, 615–625. [Google Scholar] [CrossRef]
- Jäger, E.; Chen, Y.T.; Drijfhout, J.W.; Karbach, J.; Ringhoffer, M.; Jäger, D.; Arand, M.; Wada, H.; Noguchi, Y.; Stockert, E.; et al. Simultaneous Humoral and Cellular Immune Response against Cancer-Testis Antigen NY-ESO-1: Definition of Human Histocompatibility Leukocyte Antigen (HLA)-A2-Binding Peptide Epitopes. J. Exp. Med. 1998, 187, 265–270. [Google Scholar] [CrossRef]
- Tanzarella, S.; Russo, V.; Lionello, I.; Dalerba, P.; Rigatti, D.; Bordignon, C.; Traversari, C. Identification of A Promiscuous T-Cell Epitope Encoded by Multiple Members of the MAGE Family. Cancer Res. 1999, 59, 2668–2674. [Google Scholar]
- Gjerstorff, M.F.; Andersen, M.H.; Ditzel, H.J. Oncogenic Cancer/Testis Antigens: Prime Candidates for Immunotherapy. Oncotarget 2015, 6, 15772–15787. [Google Scholar] [CrossRef]
- Van der Bruggen, P.; Traversari, C.; Chomez, P.; Lurquin, C.; Plaen, E.D.; den Eynde, B.V.; Knuth, A.; Boon, T. A Gene Encoding an Antigen Recognized by Cytolytic T Lymphocytes on a Human Melanoma. Science 1991, 254, 1643–1647. [Google Scholar] [CrossRef]
- Plaen, E.D.; Traversari, C.; Gaforio, J.J.; Szikora, J.-P.; Smet, C.D.; Brasseur, F.; van der Bruggen, P.; Lethé, B.; Lurquin, C.; Chomez, P.; et al. Structure, Chromosomal Localization, and Expression of 12 Genes of the MAGE Family. Immunogenetics 1994, 40, 360–369. [Google Scholar] [CrossRef]
- Gjerstorff, M.F.; Harkness, L.; Kassem, M.; Frandsen, U.; Nielsen, O.; Lutterodt, M.; Møllgård, K.; Ditzel, H.J. Distinct GAGE and MAGE-A Expression during Early Human Development indicate Specific Roles in Lineage Differentiation. Hum. Reprod. 2008, 23, 2194–2201. [Google Scholar] [CrossRef]
- De Smet, C.; Lurquin, C.; Lethé, B.; Martelange, V.; Boon, T. DNA Methylation Is the Primary Silencing Mechanism for a Set of Germ Line- and Tumor-Specific Genes with a Cpg-Rich Promoter. Mol. Cell. Biol. 1999, 19, 7327–7335. [Google Scholar] [CrossRef]
- Lian, Y.; Meng, L.; Ding, P.; Sang, M. Epigenetic Regulation of MAGE Family in Human Cancer Progression-DNA Methylation, Histone Modification, and Non-Coding RNAs. Clin. Epigenetics 2018, 10, 115. [Google Scholar] [CrossRef]
- Pineda, C.T.; Ramanathan, S.; Fon Tacer, K.; Weon, J.L.; Potts, M.B.; Ou, Y.-H.; White, M.A.; Potts, P.R. Degradation of AMPK by a Cancer-Specific Ubiquitin Ligase. Cell 2015, 160, 715–728. [Google Scholar] [CrossRef]
- Liu, W.; Cheng, S.; Asa, S.L.; Ezzat, S. The Melanoma-Associated Antigen A3 Mediates Fibronectin-Controlled Cancer Progression and Metastasis. Cancer Res. 2008, 68, 8104–8112. [Google Scholar] [CrossRef]
- Hartmann, S.; Brisam, M.; Rauthe, S.; Driemel, O.; Brands, R.C.; Rosenwald, A.; Kübler, A.C.; Müller-Richter, U.D.A. Contrary Melanoma-Associated Antigen-A Expression at the Tumor Front and Center: A Comparative Analysis of Stage I and IV Head and Neck Squamous Cell Carcinoma. Oncol. Lett. 2016, 12, 2942–2947. [Google Scholar] [CrossRef]
- Gure, A.O.; Chua, R.; Williamson, B.; Gonen, M.; Ferrera, C.A.; Gnjatic, S.; Ritter, G.; Simpson, A.J.G.; Chen, Y.-T.; Old, L.J.; et al. Cancer-Testis Genes Are Coordinately Expressed and Are Markers of Poor Outcome in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2005, 11, 8055–8062. [Google Scholar] [CrossRef]
- Gordeeva, O.F. Cancer-Testis Antigens: Unique Cancer Stem Cell Biomarkers and Targets for Cancer therapy. Semin. Cancer Biol. 2018, 53, 75–89. [Google Scholar] [CrossRef]
- Doyle, J.M.; Gao, J.; Wang, J.; Yang, M.; Potts, P.R. MAGE-RING Protein Complexes Comprise a Family of E3 Ubiquitin Ligases. Mol. Cell 2010, 39, 963–974. [Google Scholar] [CrossRef]
- Hagiwara, Y.; Sieverling, L.; Hanif, F.; Anton, J.; Dickinson, E.R.; Bui, T.T.T.; Andreeva, A.; Barran, P.E.; Cota, E.; Nikolova, P.V. Consequences of Point Mutations in Melanoma-Associated Antigen 4 (MAGE-A4) Protein: Insights from Structural and Biophysical Studies. Sci. Rep. 2016, 6, 25182. [Google Scholar] [CrossRef]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A Majority of the Cancer/Testis Antigens Are Intrinsically Disordered Proteins. J. Cell. Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef]
- Marcar, L.; MacLaine, N.J.; Hupp, T.R.; Meek, D.W. Mage-A Cancer/Testis Antigens inhibit P53 Function by Blocking Its interaction with Chromatin. Cancer Res. 2010, 70, 10362–10370. [Google Scholar] [CrossRef]
- Peikert, T.; Specks, U.; Farver, C.; Erzurum, S.C.; Comhair, S.A.A. Melanoma Antigen A4 Is Expressed in Non–Small Cell Lung Cancers and Promotes Apoptosis. Cancer Res. 2006, 66, 4693–4700. [Google Scholar] [CrossRef]
- Bhan, S.; Chuang, A.; Negi, S.; Glazer, C.; Califano, J. MAGEA4 induces Growth in Normal Oral Keratinocytes by inhibiting Growth Arrest and Apoptosis. Oncol. Rep. 2012, 28, 1498–1502. [Google Scholar] [CrossRef]
- Laduron, S.; Deplus, R.; Zhou, S.; Kholmanskikh, O.; Godelaine, D.; De Smet, C.; Hayward, S.D.; Fuks, F.; Boon, T.; De Plaen, E. MAGE-A1 interacts with Adaptor SKIP and the Deacetylase HDAC1 to Repress Transcription. Nucleic Acids Res. 2004, 32, 4340–4350. [Google Scholar] [CrossRef]
- Schultz-Thater, E.; Iezzi, G.; Le Magnen, C.; Zajac, P.; Spagnoli, G.C.; Piscuoglio, S.; Carafa, V.; Terracciano, L.; Tornillo, L. MAGE-A10 Is a Nuclear Protein Frequently Expressed in High Percentages of Tumor Cells in Lung, Skin and Urothelial Malignancies. Int. J. Cancer 2011, 129, 1137–1148. [Google Scholar] [CrossRef]
- Õunap, K.; Kurg, K.; Võsa, L.; Maiväli, Ü.; Teras, M.; Planken, A.; Ustav, M.; Kurg, R. Antibody Response against Cancer-Testis Antigens MAGEA4 and MAGEA10 in Patients with Melanoma. Oncol. Lett. 2018, 16, 211–218. [Google Scholar] [CrossRef]
- Rimoldi, D.; Salvi, S.; Reed, D.; Coulie, P.; Jongeneel, V.C.; De Plaen, E.; Brasseur, F.; Rodriguez, A.-M.; Boon, T.; Cerottini, J.-C. CDNA and Protein Characterization of Human MAGE-10. Int. J. Cancer 1999, 82, 901–907. [Google Scholar] [CrossRef]
- Kuldkepp, A.; Karakai, M.; Toomsoo, E.; Reinsalu, O.; Kurg, R. Cancer-Testis Antigens MAGEA Proteins Are incorporated into Extracellular Vesicles Released by Cells. Oncotarget 2019, 10, 3694–3708. [Google Scholar] [CrossRef]
- Kurg, R.; Reinsalu, O.; Jagur, S.; Õunap, K.; Võsa, L.; Kasvandik, S.; Padari, K.; Gildemann, K.; Ustav, M. Biochemical and Proteomic Characterization of Retrovirus Gag Based Microparticles Carrying Melanoma Antigens. Sci. Rep. 2016, 6, 29425. [Google Scholar] [CrossRef]
- Brūmele, B.; Mutso, M.; Telanne, L.; Õunap, K.; Spunde, K.; Abroi, A.; Kurg, R. Human TRMT112-Methyltransferase Network Consists of Seven Partners interacting with a Common Co-Factor. Int. J. Mol. Sci. 2021, 22, 13593. [Google Scholar] [CrossRef]
- Klock, H.E.; Lesley, S.A. The Polymerase incomplete Primer Extension (PIPE) Method Applied to High-Throughput Cloning and Site-Directed Mutagenesis. Methods Mol. Biol. 2009, 498, 91–103. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Reinsalu, O.; Samel, A.; Niemeister, E.; Kurg, R. MAGEA4 Coated Extracellular Vesicles Are Stable and Can Be Assembled in Vitro. Int. J. Mol. Sci. 2021, 22, 5208. [Google Scholar] [CrossRef]
- Van der Lee, R.; Lang, B.; Kruse, K.; Gsponer, J.; Sánchez de Groot, N.; Huynen, M.A.; Matouschek, A.; Fuxreiter, M.; Babu, M.M. Intrinsically Disordered Segments Affect Protein Half-Life in the Cell and during Evolution. Cell Rep. 2014, 8, 1832–1844. [Google Scholar] [CrossRef]
- Guo, L.-W.; Ruoho, A.E. N-Terminal Half of the CGMP Phosphodiesterase Γ-Subunit Contributes to Stabilization of the Gtpase-Accelerating Protein Complex. J. Biol. Chem. 2011, 286, 15260–15267. [Google Scholar] [CrossRef]
- Hegde, M.L.; Tsutakawa, S.E.; Hegde, P.M.; Holthauzen, L.M.F.; Li, J.; Oezguen, N.; Hilser, V.J.; Tainer, J.A.; Mitra, S. The Disordered C-Terminal Domain of Human DNA Glycosylase NEIL1 Contributes to Its Stability via intramolecular interactions. J. Mol. Biol. 2013, 425, 2359–2371. [Google Scholar] [CrossRef]
- Fishbain, S.; Inobe, T.; Israeli, E.; Chavali, S.; Yu, H.; Kago, G.; Babu, M.M.; Matouschek, A. Sequence Composition of Disordered Regions Fine-Tunes Protein Half-Life. Nat. Struct. Mol. Biol. 2015, 22, 214–221. [Google Scholar] [CrossRef]
- Armstrong, D.J.; Roman, A. The Anomalous Electrophoretic Behavior of the Human Papillomavirus Type 16 E7 Protein Is Due to the High Content of Acidic Amino Acid Residues. Biochem. Biophys. Res. Commun. 1993, 192, 1380–1387. [Google Scholar] [CrossRef]
- Klenova, E.M.; Nicolas, R.H.; Sally, U.; Carne, A.F.; Lee, R.E.; Lobanenkov, V.V.; Goodwin, G.H. Molecular Weight Abnormalities of the CTCF Transcription Factor: CTCF Migrates Aberrantly in SDS-PAGE and the Size of the Expressed Protein Is Affected by the UTRs and Sequences within the Coding Region of the CTCF Gene. Nucleic Acids Res. 1997, 25, 466–473. [Google Scholar] [CrossRef]
- Popelka, H.; Uversky, V.N.; Klionsky, D.J. Identification of atg3 as an intrinsically Disordered Polypeptide Yields insights into the Molecular Dynamics of Autophagy-Related Proteins in Yeast. Autophagy 2014, 10, 1093–1104. [Google Scholar] [CrossRef]
- Tiwari, P.; Kaila, P.; Guptasarma, P. Understanding Anomalous Mobility of Proteins on SDS-PAGE with Special Reference to the Highly Acidic Extracellular Domains of Human E- and N-Cadherins. Electrophoresis 2019, 40, 1273–1281. [Google Scholar] [CrossRef]
- Ramazi, S.; Zahiri, J. Post-Translational Modifications in Proteins: Resources, Tools and Prediction Methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef]
- Colemon, A.; Harris, T.M.; Ramanathan, S. DNA Hypomethylation Drives Changes in MAGE-A Gene Expression Resulting in Alteration of Proliferative Status of Cells. Genes Environ. 2020, 42, 24. [Google Scholar] [CrossRef]
- Gee, R.R.F.; Chen, H.; Lee, A.K.; Daly, C.A.; Wilander, B.A.; Fon Tacer, K.; Potts, P.R. Emerging Roles of the MAGE Protein Family in Stress Response Pathways. J. Biol. Chem. 2020, 295, 16121–16155. [Google Scholar] [CrossRef]
- Yang, S.W.; Li, L.; Connelly, J.P.; Porter, S.N.; Kodali, K.; Gan, H.; Park, J.M.; Fon Tacer, K.; Tillman, H.; Peng, J.; et al. A Cancer-Specific Ubiquitin Ligase Drives mRNA Alternative Polyadenylation by Ubiquitinating the mRNA 3′ End Processing Complex. Mol. Cell 2020, 77, 1206–1221.e7. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Redl, I.; Fisicaro, C.; Dutton, O.; Hoffmann, F.; Henderson, L.; Owens, B.M.J.; Heberling, M.; Paci, E.; Tamiola, K. ADOPT: Intrinsic protein disorder prediction through deep bidirectional transformers. NAR Genom. Bioiform. 2023, 5, lqad041. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef]
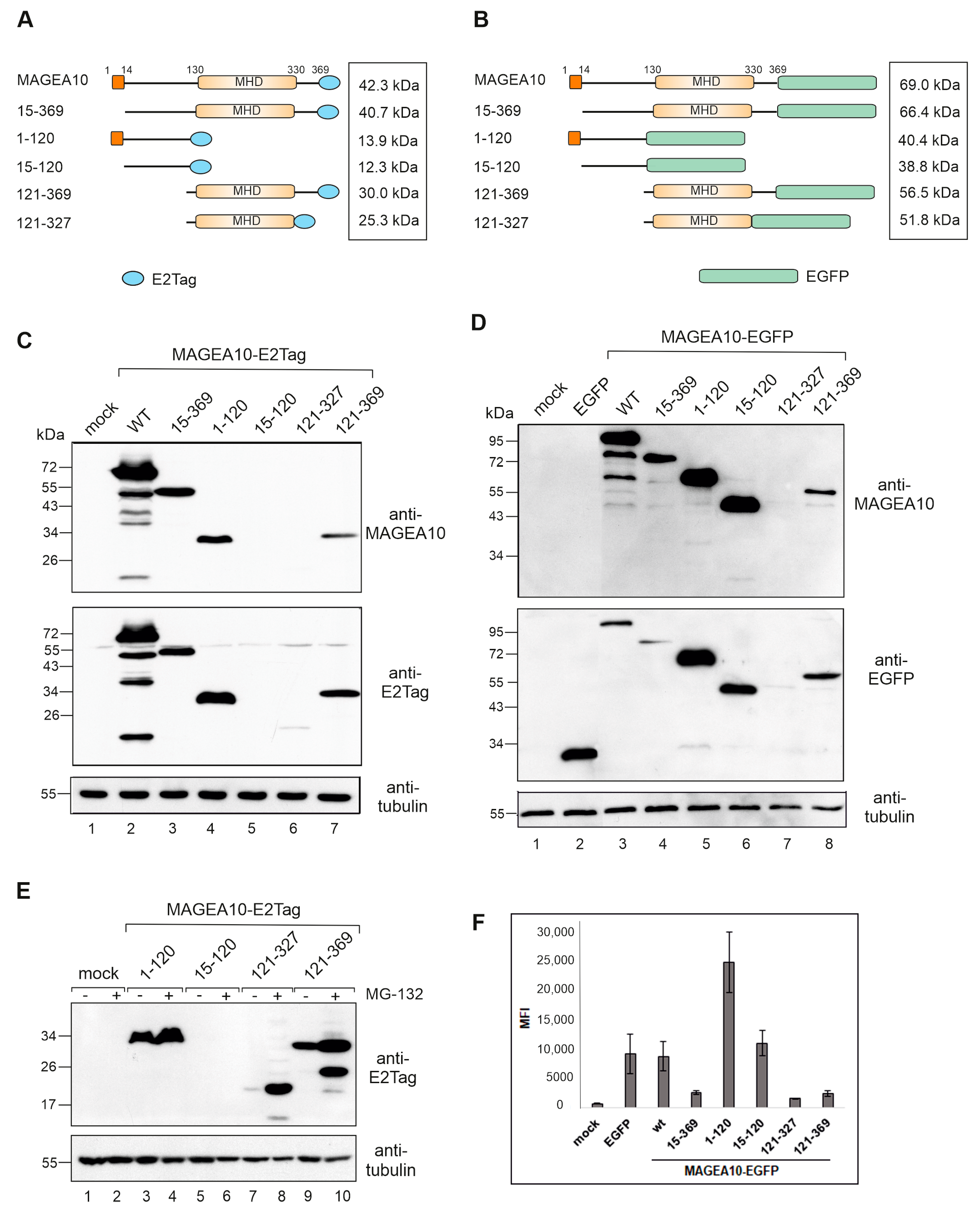


| Protein Length (aa) | Calculated MW (kDa) | Mobility in SDS-PAGE (kDa) | Times Higher in SDS-PAGE than Calculated | |
|---|---|---|---|---|
| MAGEA10-E2Tag | 383 | 42.3 | 68 | 1.6 |
| 15-369-E2Tag | 370 | 40.7 | 46 | 1.1 |
| 1-120-E2Tag | 133 | 13.9 | 30 | 2.2 |
| 121-327-E2Tag | 221 | 25.3 | 25 | 1.0 |
| 121-369-E2Tag | 264 | 30 | 32 | 1.1 |
| MAGEA10-EGFP | 620 | 69 | 98 | 1.4 |
| 15-369-EGFP | 598 | 66.4 | 74 | 1.1 |
| 1-120-EGFP | 369 | 40.4 | 60 | 1.5 |
| 15-120-EGFP | 356 | 38.8 | 48 | 1.2 |
| 121-327-EGFP | 457 | 51.8 | 52 | 1.0 |
| 121-369-EGFP | 500 | 56.5 | 56 | 1.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samel, A.; Väärtnõu, F.; Verk, L.; Kurg, K.; Mutso, M.; Kurg, R. How the Intrinsically Disordered N-Terminus of Cancer/Testis Antigen MAGEA10 Is Responsible for Its Expression, Nuclear Localisation and Aberrant Migration. Biomolecules 2023, 13, 1704. https://doi.org/10.3390/biom13121704
Samel A, Väärtnõu F, Verk L, Kurg K, Mutso M, Kurg R. How the Intrinsically Disordered N-Terminus of Cancer/Testis Antigen MAGEA10 Is Responsible for Its Expression, Nuclear Localisation and Aberrant Migration. Biomolecules. 2023; 13(12):1704. https://doi.org/10.3390/biom13121704
Chicago/Turabian StyleSamel, Anneli, Fred Väärtnõu, Lisbeth Verk, Kristiina Kurg, Margit Mutso, and Reet Kurg. 2023. "How the Intrinsically Disordered N-Terminus of Cancer/Testis Antigen MAGEA10 Is Responsible for Its Expression, Nuclear Localisation and Aberrant Migration" Biomolecules 13, no. 12: 1704. https://doi.org/10.3390/biom13121704






