Environmental and Genetic Traffic in the Journey from Sperm to Offspring
Abstract
1. Introduction
2. Paternal Influence on Offspring Development: The Underlying Mechanisms
2.1. Non-Adaptive Paternal Effects
2.2. Paternal Effects as Beneficial Adaptive Responses for Offspring
2.3. Paternal Effects to Mediate Sexual Conflict
2.4. Paternal Effects to Control Selfish Genetic Elements
3. Environmental Influences: Relating Paternal Fertility Factors and Offspring Health
3.1. Environmental Factors and Male Fertility: State-of-the-Art Knowledge
3.2. Paternal Exposure to Environmental Factors: Transmission to the Offspring
4. Genetic Causes of Spermatogenesis Disturbances
4.1. Microdeletions of Azoospermic Factor AZF Region
4.2. Paternal Genetic Diseases and Defects
4.2.1. Klinefelter Syndrome
4.2.2. Kallmann Syndrome
5. Epigenetic Markers Transmitted to Offspring
5.1. DNA Methylation and Acetylation of Sperm
5.2. Histone Retention and Modifications
5.3. Sperm-Borne Small RNAs
6. Impact of External Factors on Epigenetics of Sperm: The Connecting Link
7. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Hart, K.; Tadros, N.N. The role of environmental factors and lifestyle on male reproductive health, the epigenome, and resulting offspring. Panminerva Med. 2019, 61, 187–195. [Google Scholar] [CrossRef]
- Mima, M.; Greenwald, D.; Ohlander, S. Environmental Toxins and Male Fertility. Curr. Urol. Rep. 2018, 19, 50. [Google Scholar] [CrossRef]
- Rauh, V.A.; Margolis, A.E. Research Review: Environmental exposures, neurodevelopment, and child mental health—New paradigms for the study of brain and behavioral effects. J. Child. Psychol. Psychiatry 2016, 57, 775–793. [Google Scholar] [CrossRef]
- Jami, E.S.; Hammerschlag, A.R.; Bartels, M.; Middeldorp, C.M. Parental characteristics and offspring mental health and related outcomes: A systematic review of genetically informative literature. Transl. Psychiatry 2021, 11, 197. [Google Scholar] [CrossRef] [PubMed]
- Ralston, A.; Shaw, K. Environment controls gene expression: Sex determination and the onset of genetic disorders. Nat. Educ. 2008, 1, 203. [Google Scholar]
- Bline, A.P.; Le Goff, A.; Allard, P. What Is Lost in the Weismann Barrier? J. Dev. Biol. 2020, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Donkin, I.; Barrès, R. Sperm epigenetics and influence of environmental factors. Mol. Metab. 2018, 14, 1–11. [Google Scholar] [CrossRef]
- Finelli, R.; Mottola, F.; Agarwal, A. Impact of Alcohol Consumption on Male Fertility Potential: A Narrative Review. Int. J. Environ. Res. Public Health 2021, 19, 328. [Google Scholar] [CrossRef]
- Marić, T.; Fučić, A.; Aghayanian, A. Environmental and occupational exposures associated with male infertility. Arh. Hig. Rada Toksikol. 2021, 72, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Xavier, M.J.; Roman, S.D.; Aitken, R.J.; Nixon, B. Transgenerational inheritance: How impacts to the epigenetic and genetic information of parents affect offspring health. Hum. Reprod. Update 2019, 25, 518–540. [Google Scholar] [CrossRef] [PubMed]
- Burton, T.; Metcalfe, N.B. Can environmental conditions experienced in early life influence future generations? Proc. Biol. Sci. 2014, 281, 20140311. [Google Scholar] [CrossRef] [PubMed]
- Day, J.; Savani, S.; Krempley, B.D.; Nguyen, M.; Kitlinska, J.B. Influence of paternal preconception exposures on their offspring: Through epigenetics to phenotype. Am. J. Stem Cells. 2016, 5, 11–18. [Google Scholar] [PubMed]
- Knudsen, M.T.; Rezwan, F.I.; Jiang, Y.; Karmaus, W.; Svanes, C.; Holloway, J.W. Transgenerational and intergenerational epigenetic inheritance in allergic diseases. J. Allergy Clin. Immunol. 2018, 142, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Branje, S.; Geeraerts, S.; de Zeeuw, E.L.; Oerlemans, A.M.; Koopman-Verhoeff, M.E.; Schulz, S.; Nelemans, S.; Meeus, W.; Hartman, C.A.; Hillegers, M.H.J.; et al. Intergenerational transmission: Theoretical and methodological issues and an introduction to four Dutch cohorts. Dev. Cogn. Neurosci. 2020, 45, 100835. [Google Scholar] [CrossRef]
- Magkos, F.; Yannakoulia, M.; Chan, J.L.; Mantzoros, C.S. Management of the metabolic syndrome and type 2 diabetes through lifestyle modification. Annu. Rev. Nutr. 2009, 29, 223–256. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.K.; Hevener, A.L.; Barnard, R.J. Metabolic syndrome and insulin resistance: Underlying causes and modification by exercise training. Compr. Physiol. 2013, 3, 1–58. [Google Scholar] [CrossRef] [PubMed]
- Jung, U.J.; Choi, M.S. Obesity and its metabolic complications: The role of adipokines and the relationship between obesity, inflammation, insulin resistance, dyslipidemia and nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2014, 15, 6184–6223. [Google Scholar] [CrossRef] [PubMed]
- Rando, O.J.; Simmons, R.A. I’m eating for two: Parental dietary effects on offspring metabolism. Cell 2015, 161, 93–105. [Google Scholar] [CrossRef]
- Rosenbusch, B. To What Extent Are Cryopreserved Sperm and Testicular Biopsy Samples Used in Assisted Reproduction? J. Reprod. Infertil. 2018, 19, 115–118. [Google Scholar]
- Sharma, R.; Agarwal, A.; Rohra, V.K.; Assidi, M.; Abu-Elmagd, M.; Turki, R.F. Effects of increased paternal age on sperm quality, reproductive outcome and associated epigenetic risks to offspring. Reprod. Biol. Endocrinol. 2015, 13, 35. [Google Scholar] [CrossRef]
- Kaltsas, A.; Moustakli, E.; Zikopoulos, A.; Georgiou, I.; Dimitriadis, F.; Symeonidis, E.N.; Markou, E.; Michaelidis, T.M.; Tien, D.M.B.; Giannakis, I.; et al. Impact of Advanced Paternal Age on Fertility and Risks of Genetic Disorders in Offspring. Genes 2023, 14, 486. [Google Scholar] [CrossRef] [PubMed]
- Engeland, A.; Bjørge, T.; Daltveit, A.K.; Skurtveit, S.; Vangen, S.; Vollset, S.E.; Furu, K. Effects of preconceptional paternal drug exposure on birth outcomes: Cohort study of 340,000 pregnancies using Norwegian population-based databases. Br. J. Clin. Pharmacol. 2013, 75, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Vallaster, M.P.; Kukreja, S.; Bing, X.Y.; Ngolab, J.; Zhao-Shea, R.; Gardner, P.D.; Tapper, A.R.; Rando, O.J. Paternal nicotine exposure alters hepatic xenobiotic metabolism in offspring. eLife 2017, 6, e24771. [Google Scholar] [CrossRef] [PubMed]
- Rutkowska, J.; Lagisz, M.; Bonduriansky, R.; Nakagawa, S. Mapping the past, present and future research landscape of paternal effects. BMC Biol. 2020, 18, 183. [Google Scholar] [CrossRef] [PubMed]
- Aitken, R.J. Male reproductive ageing: A radical road to ruin. Hum. Reprod. 2023, 38, 1861–1871. [Google Scholar] [CrossRef] [PubMed]
- Fitz-James, M.H.; Cavalli, G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 2022, 23, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Immler, S. The sperm factor: Paternal impact beyond genes. Heredity 2018, 121, 239–247. [Google Scholar] [CrossRef]
- Maklakov, A.A.; Immler, S. The Expensive Germline and the Evolution of Ageing. Curr. Biol. 2016, 26, R577–R586. [Google Scholar] [CrossRef]
- Dhawan, V.; Kumar, M.; Dipika, D.; Malhotra, N.; Singh, N.; Dadhwal, V.; Dada, R. Paternal factors and embryonic development: Role in recurrent pregnancy loss. Andrologia 2019, 51, e13171. [Google Scholar] [CrossRef]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. Piwi-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Ernst, C.; Odom, D.T.; Kutter, C. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat. Commun. 2017, 8, 1411. [Google Scholar] [CrossRef]
- Sharma, U. Paternal Contributions to Offspring Health: Role of Sperm Small RNAs in Intergenerational Transmission of Epigenetic Information. Front. Cell Dev. Biol. 2019, 7, 215. [Google Scholar] [CrossRef]
- Donelan, S.C.; Hellmann, J.K.; Bell, A.M.; Luttbeg, B.; Orrock, J.L.; Sheriff, M.J.; Sih, A. Transgenerational Plasticity in Human-Altered Environments. Trends Ecol. Evol. 2020, 35, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Hales, C.N.; Barker, D.J. The thrifty phenotype hypothesis. Br. Med. Bull. 2001, 60, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Miska, E.A.; Ferguson-Smith, A.C. Transgenerational inheritance: Models and mechanisms of non-DNA sequence-based inheritance. Science 2016, 354, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, S.; Uno, M.; Okabe, E.; Nono, M.; Nishida, E. Environmental stresses induce transgenerationally inheritable survival advantages via germline-to-soma communication in Caenorhabditis elegans. Nat. Commun. 2017, 8, 14031. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.C.; Marshall, D.J. Adaptive parental effects: The importance of estimating environmental predictability and offspring fitness appropriately. Oikos 2014, 123, 769–776. [Google Scholar] [CrossRef]
- Marshall, D.J.; Uller, T. When is a maternal effect adaptive? Oikos 2007, 116, 1957–1963. [Google Scholar] [CrossRef]
- Xue, B.; Leibler, S. Evolutionary learning of adaptation to varying environments through a transgenerational feedback. Proc. Natl. Acad. Sci. USA 2016, 113, 11266–11271. [Google Scholar] [CrossRef]
- Burgess, S.C.; Marshall, D.J. Temperature-induced maternal effects and environmental predictability. J. Exp. Biol. 2011, 214, 2329–2336. [Google Scholar] [CrossRef]
- Crean, A.J.; Dwyer, J.M.; Marshall, D.J. Adaptive paternal effects? Experimental evidence that the paternal environment affects offspring performance. Ecology 2013, 94, 2575–2582. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.T.A. An evolutionary review of human telomere biology: The thrifty telomere hypothesis and notes on potential adaptive paternal effects. Am. J. Hum. Biol. 2011, 23, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Arnqvist, G.; Rowe, L. Sexual Conflict; Princeton University Press: Princeton, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Anvar, Z.; Chakchouk, I.; Demond, H.; Sharif, M.; Kelsey, G.; Van den Veyver, I.B. DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations that Disrupt Genomic Imprinting. Genes 2021, 12, 1214. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.K.; Wolf, J.B. The coadaptation theory for genomic imprinting. Evol. Lett. 2017, 1, 49–59. [Google Scholar] [CrossRef]
- Haig, D. The kinship theory of genomic imprinting. Ann. Rev. Ecol. Syst. 2000, 31, 9–32. [Google Scholar] [CrossRef]
- Haig, D. Genomic imprinting and kinship: How good is the evidence? Ann. Rev. Genet. 2004, 38, 553–585. [Google Scholar] [CrossRef]
- Day, T.; Bonduriansky, R. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics 2004, 167, 1537–1546. [Google Scholar] [CrossRef]
- Wolf, J.B.; Hager, R. A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 2006, 4, 2238–2243. [Google Scholar] [CrossRef] [PubMed]
- Spencer, H.G.; Clark, A.G. Non-conflict theories for the evolution of genomic imprinting. Heredity 2014, 113, 112–118. [Google Scholar] [CrossRef]
- Agren, J.A.; Clark, A.G. Selfish genetic elements. PLoS Genet. 2018, 14, e1007700. [Google Scholar] [CrossRef]
- Wedell, N. Selfish genes and sexual selection: The impact of genomic parasites on host reproduction. J. Zool. 2020, 311, 1–12. [Google Scholar] [CrossRef]
- Verspoor, R.L.; Price, T.A.R.; Wedell, N. Selfish genetic elements and male fertility. Phil. Trans. R. Soc. B 2020, 375, 20200067. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. USA 2011, 108, 10863–10870. [Google Scholar] [CrossRef] [PubMed]
- Wedell, N. The effect of non-self genes on the behaviour of hosts. In Genes and Behaviour: Beyond Nature-Nurture; Hosken, D.J., Hunt, J., Wedell, N., Eds.; John Wiley & Sons: Chichester, UK, 2019; pp. 157–180. [Google Scholar]
- Zanders, S.E.; Unckless, R.L. Fertility costs of meiotic drivers. Curr. Biol. 2019, 29, R512–R520. [Google Scholar] [CrossRef]
- Sutter, A.; Lindholm, A.K. Detrimental effects of an autosomal selfish genetic element on sperm competitiveness in house mice. Proc. Biol. Sci. 2015, 282, 20150974. [Google Scholar] [CrossRef] [PubMed]
- Christie, J.R.; Schaerf, T.M.; Beekman, M. Selection against heteroplasmy explains the evolution of uniparental inheritance of mitochondria. PLoS Genet. 2015, 11, e1005112. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Sobanski, J.; Bock, R. Why are most organelle genomes transmitted maternally? Bioessays 2015, 37, 80–94. [Google Scholar] [CrossRef]
- Cordier, S. Evidence for a role of paternal exposures in developmental toxicity. Basic Clin. Pharmacol. Toxicol. 2008, 102, 176–181. [Google Scholar] [CrossRef]
- Soubry, A.; Hoyo, C.; Jirtle, R.L.; Murphy, S.K. A paternal environmental legacy: Evidence for epigenetic inheritance through the male germ line. Bioessays 2014, 36, 359–371. [Google Scholar] [CrossRef]
- Bonde, J.P.; Tøttenborg, S.S.; Hougaard, K.S. Paternal environmental exposure and offspring health. Curr. Opin. Endocr. Metab. Res. 2019, 7, 14–20. [Google Scholar] [CrossRef]
- Blay, R.M.; Pinamang, A.D.; Sagoe, A.E.; Owusu, E.; Koney, N.K.; Arko-Boham, B. Infuence of lifestyle and environmental factors on semen quality in ghanaian men. Int. J. Reprod. Med. 2020, 2020, 6908458. [Google Scholar] [CrossRef] [PubMed]
- Nateghian, Z.; Aliabadi, E. Aspects of Environmental Pollutants on Male Fertility and Sperm Parameters. J. Environ. Treat. Tech. 2020, 8, 299–309. Available online: http://www.jett.dormaj.com (accessed on 30 August 2023).
- Selvaraju, V.; Baskaran, S.; Agarwal, A.; Henkel, R. Environmental contaminants and male infertility: Effects and mechanisms. Andrologia 2021, 53, e13646. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tian, X.; Ye, B.; Zhang, Y.; Li, C.; Liao, J.; Zou, Y.; Zhang, S.; Zhu, Y.; Yang, J.; et al. Gaseous pollutant exposure affects semen quality in central China: A cross-sectional study. Andrology 2020, 8, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhao, J.; Cao, W.; Lu, W.; Zheng, T.; Zeng, Q. Identifying critical exposure windows for ambient air pollution and semen quality in Chinese men. Environ. Res. 2020, 189, 109894. [Google Scholar] [CrossRef] [PubMed]
- Wdowiak, A.; Wdowiak, E.; Bień, A.; Bojar, I.; Iwanowicz-Palus, G.; Raczkiewicz, D. Air pollution and semen parameters in men seeking fertility treatment for the first time. Int. J. Occup. Med. Environ. Health 2019, 32, 387–399. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, T.; Liu, S.; Cao, Z.; Zhao, Y.; Su, X.; Liao, Z.; Teng, X.; Hua, J. Concentrated ambient PM2.5 exposure affects mice sperm quality and testosterone biosynthesis. PeerJ 2019, 7, e8109. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Sengupta, P.; Bagchi, S.; Chhikara, B.S.; Pavlík, A.; Sláma, P.; Roychoudhury, S. Reproductive toxicity of combined effects of endocrine disruptors on human reproduction. Front. Cell Dev. Biol. 2023, 11, 1162015. [Google Scholar] [CrossRef]
- Mendiola, J.; Moreno, J.M.; Roca, M.; Vergara-Juárez, N.; Martínez-García, M.J.; García-Sánchez, A.; Elvira-Rendueles, B.; Moreno-Grau, S.; LópezEspín, J.J.; Ten, J.; et al. Relationships between heavy metal concentrations in three diferent body fuids and male reproductive parameters: A pilot study. Environ. Health 2011, 10, 6. [Google Scholar] [CrossRef]
- Sukhn, C.; Awwad, J.; Ghantous, A.; Zaatari, G. Associations of semen quality with non-essential heavy metals in blood and seminal fuid: Data from the Environment and Male Infertility (EMI) study in Lebanon. J. Assist. Reprod. Genet. 2018, 35, 1691–1701. [Google Scholar] [CrossRef]
- Manouchehri, A.; Shokri, S.; Pirhadi, M.; Karimi, M.; Abbaszadeh, S.; Mirzaei, G.; Bahmani, M. The Effects of Toxic Heavy Metals Lead, Cadmium and Copper on the Epidemiology of Male and Female Infertility. JBRA Assist. Reprod. 2022, 26, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Santonastaso, M.; Mottola, F.; Iovine, C.; Cesaroni, F.; Colacurci, N.; Rocco, L. In Vitro Effects of Titanium Dioxide Nanoparticles (TiO2NPs) on Cadmium Chloride (CdCl2) Genotoxicity in Human Sperm Cells. Nanomaterials 2020, 10, 1118. [Google Scholar] [CrossRef] [PubMed]
- Calogero, A.E.; Fiore, M.; Giacone, F.; Altomare, M.; Asero, P.; Ledda, C.; Romeo, G.; Mongioì, L.M.; Copat, C.; Giuffrida, M.; et al. Exposure to multiple metals/metalloids and human semen quality: A cross-sectional study. Ecotoxicol. Environ. Saf. 2021, 215, 112165. [Google Scholar] [CrossRef]
- Hardneck, F.; Israel, G.; Pool, E.; Maree, L. Quantitative assessment of heavy metal effects on sperm function using computer-aided sperm analysis and cytotoxicity assays. Andrologia 2018, 50, e13141. [Google Scholar] [CrossRef] [PubMed]
- Mínguez-Alarcón, L.; Hauser, R.; Gaskins, A.J. Effects of bisphenol A on male and couple reproductive health: A review. Fertil. Steril. 2016, 106, 864–870. [Google Scholar] [CrossRef]
- Cariati, F.; D’Uonno, N.; Borrillo, F.; Iervolino, S.; Galdiero, G.; Rb, T. Bisphenol a: An emerging threat to male fertility. Reprod. Biol. Endocrinol. 2019, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Barbonetti, A.; Castellini, C.; Di Giammarco, N.; Santilli, G.; Francavilla, S.; Francavilla, F. In vitro exposure of human spermatozoa to bisphenol A induces pro-oxidative/apoptotic mitochondrial dysfunction. Reprod. Toxicol. 2016, 66, 61–67. [Google Scholar] [CrossRef]
- Bretveld, R.; Brouwers, M.; Ebisch, I.; Roeleveld, N. Infuence of pesticides on male fertility. Scand. J. Work. Environ. Health 2007, 33, 13–28. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering, and Medicine; Division on Earth and Life Studies; Board on Environmental Studies and Toxicology; Committee on Endocrine-Related Low-Dose Toxicity. Application of Systematic Review Methods in an Overall Strategy for Evaluating Low-Dose Toxicity from Endocrine Active Chemicals. National Academies Press (US): Washington, DC, USA, 2017; 3, Phthalates and Male Reproductive-Tract Development. Available online: https://www.ncbi.nlm.nih.gov/books/NBK453249/ (accessed on 30 August 2023).
- Hutson, J.M. Cryptorchidism and Hypospadias. 14 December 2022. In Endotext [Internet]; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Ding, X.; Cao, L.; Zheng, Y.; Zhou, X.; He, X.; Xu, S.; Ren, W. Insights into the Evolution of Spermatogenesis-Related Ubiquitin-Proteasome System Genes in Abdominal Testicular Laurasiatherians. Genes 2021, 12, 1780. [Google Scholar] [CrossRef] [PubMed]
- Al-Otaibi, S.T. Male infertility among bakers associated with exposure to high environmental temperature at the workplace. J. Taibah Univ. Med. Sci. 2018, 13, 103–107. [Google Scholar] [CrossRef]
- Hamerezaee, M.; Dehghan, S.F.; Golbabaei, F.; Fathi, A.; Barzegar, L.; Heidarnejad, N. Assessment of semen quality among workers exposed to heat stress: A cross-sectional study in a steel industry. Saf. Health Work 2018, 9, 232–235. [Google Scholar] [CrossRef]
- Kesari, K.K.; Agarwal, A.; Henkel, R. Radiations and male fertility. Reprod. Biol. Endocrinol. 2018, 16, 118. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Han, D.; Ryu, J.; Kim, K.; Kim, Y.H. Effects of mobile phone usage on sperm quality—No time-dependent relationship on usage: A systematic review and updated meta-analysis. Environ. Res. 2021, 202, 111784. [Google Scholar] [CrossRef] [PubMed]
- Aitken, R.J. Reactive oxygen species as mediators of sperm capacitation and pathological damage. Mol. Reprod. Dev. 2017, 84, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Mottola, F.; Santonastaso, M.; Ronga, V.; Finelli, R.; Rocco, L. Polymorphic Rearrangements of Human Chromosome 9 and Male Infertility: New Evidence and Impact on Spermatogenesis. Biomolecules 2023, 13, 729. [Google Scholar] [CrossRef] [PubMed]
- Leisegang, K.; Roychoudhury, S.; Slama, P.; Finelli, R. The Mechanisms and Management of Age-Related Oxidative Stress in Male Hypogonadism Associated with Non-communicable Chronic Disease. Antioxidants 2021, 10, 1834. [Google Scholar] [CrossRef] [PubMed]
- Durairajanayagam, D. Lifestyle causes of male infertility. Arab. J. Urol. 2018, 16, 10–20. [Google Scholar] [CrossRef]
- Samarasinghe, S.V.A.C.; Krishnan, K.; Naidu, R.; Megharaj, M.; Miller, K.; Fraser, B.; Aitken, R.J. Parabens generate reactive oxygen species in human spermatozoa. Andrology 2018, 6, 532–541. [Google Scholar] [CrossRef]
- Santonastaso, M.; Mottola, F.; Colacurci, N.; Iovine, C.; Pacifico, S.; Cammarota, M.; Cesaroni, F.; Rocco, L. In vitro genotoxic effects of titanium dioxide nanoparticles (n-TiO2) in human sperm cells. Mol. Reprod. Dev. 2019, 86, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Villarreal, J.; Betancourt-Martinex, N.D.; Carranza-Rosales, P.; Valdez, E.V.; Guzman-Delgado, N.E.; Lopez-Marquez, F.C.; Moran-Martinez, J. Formaldehyde induces DNA strand breaks on spermatozoa and lymphocytes of Wistar rats. Tsitol. Genet. 2017, 51, 78–80. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.G.; Valsala Gopalakrishnan, A. The interplay of arsenic, silymarin, and NF-ĸB pathway in male reproductive toxicity: A review. Ecotoxicol. Environ. Saf. 2023, 252, 114614. [Google Scholar] [CrossRef]
- Khan, F.; Niaz, K.; Hassan, F.I.; Abdollahi, M. An evidence-based review of the genotoxic and reproductive effects of sulfur mustard. Arch. Toxicol. 2017, 91, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Perrin, J.; Tassistro, V.; Mandon, M.; Grillo, J.M.; Botta, A.; Sari-Minodier, I. Tobacco consumption and benzo(a)pyrene-diolepoxide-DNA adducts in spermatozoa: In smokers, swim-up procedure selects spermatozoa with decreased DNA damage. Fertil. Steril. 2011, 95, 2013–2017. [Google Scholar] [CrossRef] [PubMed]
- McQueen, D.B.; Zhang, J.; Robins, J.C. Sperm DNA fragmentation and recurrent pregnancy loss: A systematic review and meta-analysis. Fertil. Steril. 2019, 112, 54–60.e3. [Google Scholar] [CrossRef] [PubMed]
- Waber, D.P.; Bryce, C.P.; Fitzmaurice, G.M.; Zichlin, M.L.; McGaughy, J.; Girard, J.M.; Galler, J.R. Neuropsychological outcomes at midlife following moderate to severe malnutrition in infancy. Neuropsychology 2014, 28, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Soneji, S.; Beltrán-Sánchez, H. Association of Maternal Cigarette Smoking and Smoking Cessation With Preterm Birth. JAMA Netw. Open 2019, 2, e192514. [Google Scholar] [CrossRef]
- Meeker, J.D. Exposure to environmental endocrine disruptors and child development. Arch. Pediatr. Adolesc. Med. 2012, 166, E1–E7. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.; Das, M.K.; Duttaroy, A.K. Plastics derived endocrine-disrupting compounds and their effects on early development. Birth Defects Res. 2020, 112, 1308–1325. [Google Scholar] [CrossRef]
- McCanlies, E.C.; Ma, C.C.; Gu, J.K.; Fekedulegn, D.; Sanderson, W.T.; Ludeña-Rodriguez, Y.J.; Hertz-Picciotto, I. The CHARGE study: An assessment of parental occupational exposures and autism spectrum disorder. Occup. Environ. Med. 2019, 76, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shi, J.; Rassoulzadegan, M.; Tuorto, F.; Chen, Q. Sperm RNA code programmes the metabolic health of offspring. Nat. Rev. Endocrinol. 2019, 15, 489–498. [Google Scholar] [CrossRef]
- Gapp, K.; Jawaid, A.; Sarkies, P.; Bohacek, J.; Pelczar, P.; Prados, J.; Farinelli, L.; Miska, E.; Mansuy, I.M. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014, 17, 667–669. [Google Scholar] [CrossRef] [PubMed]
- Zumbrun, E.E.; Sido, J.M.; Nagarkatti, P.S.; Nagarkatti, M. Epigenetic Regulation of Immunological Alterations Following Prenatal Exposure to Marijuana Cannabinoids and its Long Term Consequences in Offspring. J. Neuroimmune Pharmacol. 2015, 10, 245–254. [Google Scholar] [CrossRef]
- McCarthy, D.M.; Morgan, T.J., Jr.; Lowe, S.E.; Williamson, M.J.; Spencer, T.J.; Biederman, J.; Bhide, P.G. Nicotine exposure of male mice produces behavioral impairment in multiple generations of descendants. PLoS Biol. 2018, 16, e2006497. [Google Scholar] [CrossRef] [PubMed]
- Holloway, Z.R.; Hawkey, A.B.; Torres, A.K.; Evans, J.; Pippen, E.; White, H.; Katragadda, V.; Kenou, B.; Wells, C.; Murphy, S.K.; et al. Paternal cannabis extract exposure in rats: Preconception timing effects on neurodevelopmental behavior in offspring. Neurotoxicology 2020, 81, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Ueker, M.E.; Silva, V.M.; Moi, G.P.; Pignati, W.A.; Mattos, I.E.; Silva, A.M.C. Parenteral exposure to pesticides and occurence of congenital malformations: Hospital-based case-control study. BMC Pediatr. 2016, 16, 125. [Google Scholar] [CrossRef]
- Rauh, V.; Arunajadai, S.; Horton, M.; Perera, F.; Hoepner, L.; Barr, D.B.; Whyatt, R. Seven-year neurodevelopmental scores and prenatal exposure to chlorpyrifos, a common agricultural pesticide. Environ. Health Perspect. 2011, 119, 1196–1201. [Google Scholar] [CrossRef]
- Shelton, J.F.; Geraghty, E.M.; Tancredi, D.J.; Delwiche, L.D.; Schmidt, R.J.; Ritz, B.; Hansen, R.L.; Hertz-Picciotto, I. Neurodevelopmental disorders and prenatal residential proximity to agricultural pesticides: The CHARGE study. Environ. Health Perspect. 2014, 122, 1103–1109, Erratum in: Environ Health Perspect. 2014, 122, A266. [Google Scholar] [CrossRef] [PubMed]
- Yorifuji, T.; Tsuda, T.; Kashima, S.; Doi, H. Mercury and autism: Accelerating evidence? Neuroendocrinol. Lett. 2014, 35, 221–226. [Google Scholar]
- Ng, S.-F.; Lin RC, Y.; Laybutt, D.R.; Barres, R.; Owens, J.A.; Morris, M.J. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature 2010, 467, 963–966. [Google Scholar] [CrossRef]
- Wei, Y.; Yang, C.R.; Wei, Y.P.; Ge, Z.J.; Zhao, Z.A.; Zhang, X.H. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc. Natl. Acad. Sci. USA 2015, 112, 5361–5366. [Google Scholar] [CrossRef]
- Wei, Y.; Yang, C.R.; Wei, Y.P.; Zhao, Z.A.; Hou, Y.; Schatten, H.; Sun, Q.Y. Paternally induced transgenerational inheritance of susceptibility to cardiac ischemia-reperfusion injury. Front. Biosci. 2014, 19, 1074–1087. [Google Scholar]
- Janssen, B.G.; Godderis, L.; Pieters, N.; Poels, K.; Kiciński, M.; Cuypers, A.; Fierens, F.; Penders, J.; Plusquin, M.; Gyselaers, W.; et al. Placental DNA hypomethylation in association with particulate air pollution in early life. Part. Fibre Toxicol. 2017, 14, 1–14. [Google Scholar]
- Dolinoy, D.C.; Weidman, J.R.; Waterland, R.A.; Jirtle, R.L. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 2007, 115, 879–885. [Google Scholar]
- Guerrero-Bosagna, C.; Covert, T.R.; Haque, M.M.; Settles, M.; Nilsson, E.E.; Anway, M.D.; Skinner, M.K. Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers. Reprod. Toxicol. 2012, 34, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, M.F.; Bellinger, D.C.; Wright, R.O.; Weisskopf, M.G. Attention-deficit/hyperactivity disorder and urinary metabolites of organophosphate pesticides. Pediatrics 2010, 125, e1270–e1277. [Google Scholar] [CrossRef] [PubMed]
- Rossides, M.; Kampitsi, C.E.; Talbäck, M.; Mogensen, H.; Wiebert, P.; Feychting, M.; Tettamanti, G. Risk of Cancer in Children of Parents Occupationally Exposed to Hydrocarbon Solvents and Engine Exhaust Fumes: A Register-Based Nested Case-Control Study from Sweden (1960–2015). Environ. Health Perspect. 2022, 130, 77002. [Google Scholar] [CrossRef]
- Short, A.K.; Fennell, K.A.; Perreau, V.M.; Fox, A.; O’Bryan, M.K.; Kim, J.H.; Bredy, T.W.; Pang, T.Y.; Hannan, A.J. Elevated paternal glucocorticoid exposure alters the small noncoding RNA profile in sperm and modifies anxiety and depressive phenotypes in the offspring. Transl. Psychiatry 2016, 6, e837. [Google Scholar] [CrossRef]
- Liu, Y.; Zhi, X. Advances in Genetic Diagnosis of Kallmann Syndrome and Genetic Interruption. Reprod. Sci. 2022, 29, 1697–1709. [Google Scholar] [CrossRef]
- Cox, C.M.; Thoma, M.E.; Tchangalova, N.; Mburu, G.; Bornstein, M.J.; Johnson, C.L.; Kiarie, J. Infertility prevalence and the methods of estimation from 1990 to 2021: A systematic review and meta-analysis. Hum. Reprod. Open 2022, 2022, hoac051. [Google Scholar] [CrossRef]
- Mazzilli, R.; Rucci, C.; Vaiarelli, A.; Cimadomo, D.; Ubaldi, F.M.; Foresta, C.; Ferlin, A. Male factor infertility and assisted reproductive technologies: Indications, minimum access criteria and outcomes. J. Endocrinol. Investig. 2023, 46, 1079–1085. [Google Scholar] [CrossRef]
- Xie, C.; Chen, X.; Liu, Y.; Wu, Z.; Ping, P. Multicenter study of genetic abnormalities associated with severe oligospermia and non-obstructive azoospermia. J. Int. Med. Res. 2018, 46, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Colaco, S.; Modi, D. Genetics of the human Y chromosome and its association with male infertility. Reprod. Biol. Endocrinol. 2018, 16, 14. [Google Scholar] [CrossRef] [PubMed]
- Bardoni, B.; Zuffardi, O.; Guioli, S.; Ballabio, A.; Simi, P.; Cavalli, P.; Grimoldi, M.G.; Fraccaro, M.; Camerino, G. A deletion map of the human Yq11 region: Implications for the evolution of the Y chromosome and tentative mapping of a locus involved in spermatogenesis. Genomics 1991, 11, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, M.J.; Huffman, P.J.; Haney, N.M.; Kohn, T.P. Y-Chromosome Microdeletions: A Review of Prevalence, Screening, and Clinical Considerations. Appl. Clin. Genet. 2021, 14, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Foresta, C.; Moro, E.; Ferlin, A. Y chromosome microdeletions and alterations of spermatogenesis. Endocr. Rev. 2001, 22, 226–239. [Google Scholar] [PubMed]
- Vogt, P.H.; Edelmann, A.; Kirsch, S.; Henegariu, O.; Hirschmann, P.; Kiesewetter, F.; Kohn, F.M.; Schill, W.B.; Farah, S.; Ramos, C.; et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum. Mol. Genet. 1996, 5, 933–943. [Google Scholar] [CrossRef] [PubMed]
- SAl-Ouqaili, M.T.; Al-Ani, S.K.; Alaany, R.; Al-Qaisi, M.N. Detection of partial and/or complete Y chromosome microdeletions of azoospermia factor a (AZFa) sub-region in infertile Iraqi patients with azoospermia and severe oligozoospermia. J. Clin. Lab. Anal. 2022, 36, e24272. [Google Scholar] [CrossRef]
- Repping, S.; Skaletsky, H.; Lange, J.; Silber, S.; Van Der Veen, F.; Oates, R.D.; Page, D.C.; Rozen, S. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am. J. Hum. Genet. 2002, 71, 906–922. [Google Scholar] [CrossRef]
- Suganthi, R.; Vijesh, V.V.; Vandana, N.; Fathima Ali Benazir, J. Y choromosomal microdeletion screening in the workup of male infertility and its current status in India. Int. J. Fertil. Steril. 2014, 7, 253–266. [Google Scholar]
- Navarro-Costa, P.; Plancha, C.E.; Goncalves, J. Genetic dissection of the AZF regions of the human Y chromosome: Thriller or filler for male (in) fertility? J. Biomed. Biotechnol. 2010, 2010, 936569. [Google Scholar] [CrossRef] [PubMed]
- Yuen, W.; Golin, A.P.; Flannigan, R.; Schlegel, P.N. Histology and sperm retrieval among men with Y chromosome microdeletions. Transl. Androl. Urol. 2021, 10, 1442–1456. [Google Scholar] [CrossRef] [PubMed]
- Kuroda-Kawaguchi, T.; Skaletsky, H.; Brown, L.G.; Minx, P.J.; Cordum, H.S.; Waterston, R.H.; Wilson, R.K.; Silber, S.; Oates, R.; Rozen, S.; et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat. Genet. 2001, 29, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Vogt, P.H.; Bender, U.; Deibel, B.; Kiesewetter, F.; Zimmer, J.; Strowitzki, T. Human AZFb deletions cause distinct testicular pathologies depending on their extensions in Yq11 and the Y haplogroup: New cases and review of literature. Cell Biosci. 2021, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Witherspoon, L.; Dergham, A.; Flannigan, R. Y-microdeletions: A review of the genetic basis for this common cause of male infertility. Transl. Androl. Urol. 2021, 10, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Klinefelter, H.F., Jr.; Reifenstein, E.C., Jr.; Albright, F., Jr. Syndrome characterized by gynecomastia, aspermatogenesis without A-Leydigism, and increased excretion of follicle-stimulating hormone. J. Clin. Endocrinol. 1942, 2, 615–627. [Google Scholar] [CrossRef]
- Groth, K.A.; Skakkebæk, A.; Høst, C.; Gravholt, C.H.; Bojesen, A. Klinefelter syndrome—A clinical update. J. Clin. Endocrinol. Metab. 2013, 98, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, V.O.; Polisseni, F.; Pannain, G.D.; Carvalho, M.A.G. Genetics in human reproduction. JBRA Assist. Reprod. 2020, 24, 480–491. [Google Scholar] [CrossRef]
- Thomas, N.; Hassold, T. Aberrant recombination and the origin of Klinefelter syndrome. Hum. Reprod. Update 2003, 9, 309–317. [Google Scholar] [CrossRef]
- Jo, D.G.; Seo, J.T.; Lee, J.S.; Park, S.Y.; Kim, J.W. Klinefelter syndrome diagnosed by prenatal screening tests in high-risk groups. Korean J. Urol. 2013, 54, 263. [Google Scholar] [CrossRef]
- Bonomi, M.; Rochira, V.; Pasquali, D.; Balercia, G.; Jannini, E.; Ferlin, A. Klinefelter syndrome (KS): Genetics, clinical phenotype and hypogonadism. J. Endocrinol. Investig. 2017, 40, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Lanfranco, F.; Kamischke, A.; Zitzmann, M.; Nieschlag, E. Klinefelter’s syndrome. Lancet 2004, 364, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Turriff, A.; Macnamara, E.; Levy, H.P.; Biesecker, B. The impact of living with Klinefelter syndrome: A qualitative exploration of adolescents and adults. J. Genet. Couns. 2017, 26, 728–737. [Google Scholar] [CrossRef] [PubMed]
- Høst, C.; Skakkebæk, A.; Groth, K.A.; Bojesen, A. The role of hypogonadism in Klinefelter syndrome. Asian J. Androl. 2014, 16, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yuan, T.; Song, S.; Chen, S.; Wang, L.; Fu, Y.; Dong, Y.; Tang, Y.; Zhao, W. Glucose metabolic disorder in Klinefelter syndrome: A retrospective analysis in a single Chinese hospital and literature review. BMC Endocr. Disord. 2021, 21, 239. [Google Scholar] [CrossRef]
- O’Connor, M.J.; Snyder, E.A.; Hayes, F.J. Klinefelter syndrome and diabetes. Curr. Diabetes Rep. 2019, 19, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bojesen, A.; Juul, S.; Birkebæk, N.H.; Gravholt, C.H. Morbidity in Klinefelter syndrome: A Danish register study based on hospital discharge diagnoses. J. Clin. Endocrinol. Metab. 2006, 91, 1254–1260. [Google Scholar] [CrossRef]
- Davis, S.M.; DeKlotz, S.; Nadeau, K.J.; Kelsey, M.M.; Zeitler, P.S.; Tartaglia, N.R. High prevalence of cardiometabolic risk features in adolescents with 47,XXY/Klinefelter syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 327–333. [Google Scholar] [CrossRef]
- Agarwal, S.; Dekam, M. Multiple cardiac anomalies in an elderly man with Klinefelter’s syndrome. Singap. Med. J. 2011, 52, e15–e17. [Google Scholar]
- Jørgensen, I.N.; Skakkebaek, A.; Andersen, N.H.; Pedersen, L.N.; Hougaard, D.M.; Bojesen, A.; Trolle, C.; Gravholt, C.H. Short QTc interval in males with klinefelter syndrome—Influence of CAG repeat length, body composition, and testosterone replacement therapy. Pacing Clin. Electrophysiol. 2015, 38, 472–482. [Google Scholar] [CrossRef]
- Zitzmann, M.; Bongers, R.; Werler, S.; Bogdanova, N.; Wistuba, J.; Kliesch, S.; Gromoll, J.; Tüttelmann, F. Gene expression patterns in relation to the clinical phenotype in Klinefelter syndrome. J. Clin. Endocrinol. Metab. 2015, 100, E518–E523. [Google Scholar] [CrossRef]
- Fricke, G.; Mattern, H.; Schweikert, H.; Schwanitz, G. Klinefelter’s syndrome and mitral valve prolapse. An echocardiographic study in twenty-two patients. Biomed. Pharmacother. 1984, 38, 88–97. [Google Scholar] [PubMed]
- Pasquali, D.; Arcopinto, M.; Renzullo, A.; Rotondi, M.; Accardo, G.; Salzano, A.; Esposito, D.; Saldamarco, L.; Isidori, A.M.; Marra, A.M.; et al. Cardiovascular abnormalities in Klinefelter syndrome. Int. J. Cardiol. 2013, 168, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, K.; Matsuyama, H. Klinefelter syndrome: From pediatrics to geriatrics. Reprod. Med. Biol. 2019, 18, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Turriff, A.; Levy, H.P.; Biesecker, B. Prevalence and psychosocial correlates of depressive symptoms among adolescents and adults with Klinefelter syndrome. Genet. Med. 2011, 13, 966–972. [Google Scholar] [CrossRef] [PubMed]
- Sasco, A.J.; Lowenfels, A.B.; Jong, P.P.D. epidemiology of male breast cancer. A meta-analysis of published case-control studies and discussion of selected aetiological factors. Int. J. Cancer 1993, 53, 538–549. [Google Scholar] [CrossRef]
- Völkl, T.M.; Langer, T.; Aigner, T.; Greess, H.; Beck, J.D.; Rauch, A.M.; Dörr, H.G. Klinefelter syndrome and mediastinal germ cell tumors. Am. J. Med. Genet. Part A 2006, 140, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Raposo, C.; Tévar, F.Z.; Moyano, M.S.; Gómez, M.L.; Casado, E. Male breast cancer. Cancer Treat. Rev. 2010, 36, 451–457. [Google Scholar] [CrossRef]
- Brinton, L.A. Breast cancer risk among patients with Klinefelter syndrome. Acta Paediatr. 2011, 100, 814–818. [Google Scholar] [CrossRef]
- De Sanctis, V.; Fiscina, B.; Soliman, A.; Giovannini, M.; Yassin, M. Klinefelter syndrome and cancer: From childhood to adulthood. Pediatr. Endocrinol. Rev. 2013, 11, 44–50. [Google Scholar]
- Ji, J.; Zöller, B.; Sundquist, J.; Sundquist, K. Risk of solid tumors and hematological malignancy in persons with Turner and Klinefelter syndromes: A national cohort study. Int. J. Cancer 2016, 139, 754–758. [Google Scholar] [CrossRef]
- Lazúrová, I.; Rovenský, J.; Imrich, R.; Blažíčková, S.; Lazúrová, Z.; Payer, J. Autoimmune rheumatic diseases and Klinefelter syndrome Autoimunitné reumatické choroby a Klinefelterov syndróm. Eur. Pharm. J. 2016, 63, 18–22. [Google Scholar] [CrossRef]
- Dode, C.; Hardelin, J.P. Kallmann syndrome. Eur. J. Hum. Genet. 2009, 17, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Shima, H.; Tokuhiro, E.; Okamoto, S.; Nagamori, M.; Ogata, T.; Narumi, S.; Nakamura, A.; Izumi, Y.; Jinno, T.; Suzuki, E.; et al. SOX10 Mutation Screening for 117 Patients with Kallmann Syndrome. J. Endocr. Soc. 2021, 5, bvab056. [Google Scholar] [CrossRef] [PubMed]
- Marhari, H.; Chahdi Ouazzani, F.Z.; Ouahabi, H.E.; Bouguenouch, L. Le syndrome de Kallmann-de Morsier: À propos de trois cas [Kallmann-de Morsier syndrome: About 3 cases]. Pan Afr. Med. J. 2019, 33, 221. [Google Scholar] [CrossRef] [PubMed]
- Maestre de San Juan, A. Teratolagia: Falta total de los nervios olfactorios con anosmia en un individuo en quien existia una atrofifia congenita de los testiculos y miembro viril. El Siglo Me’dico 1856, 3, 211–221. [Google Scholar]
- Kallmann, F.J.; Schoenfeld, W.A.; Barrera, S.E. The genetic aspects of primary eunuchoidism. Am. J. Ment. Defificiency 1944, 158, 203–236. [Google Scholar]
- Stamou, M.I.; Georgopoulos, N.A. Kallmann syndrome: Phenotype and genotype of hypogonadotropic hypogonadism. Metabolism 2018, 86, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Tanriverdi, F.; MacColl, G.S.; Bouloux, P.M. Kallmann syndrome: Molecular pathogenesis. Int. J. Biochem. Cell. Biol. 2003, 35, 1157–1162. [Google Scholar] [CrossRef]
- Dode, C.; Teixetra, L.; Levillers Fouveaut, C.; Bouchard, P.; Kottler, M.L.; Lespinasse, J.; Lienhardt-Roussie, A.; Mathieu, M.; Moerman, A.; Morgan, G. Kallmann syndrome: Mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006, 2, 1648–1652. [Google Scholar] [CrossRef]
- Ogata, T.; Fujiwara, I.; Ogawa, E.; Sato, N.; Udaka, T.; Kosaki, K. Kallman syndrome phenotype in a female patient with CHARGE syndrome and CHD7 mutation. Endocr J. 2006, 53, 741–743. [Google Scholar] [CrossRef]
- Young, J.; Xu, C.; Papadakis, G.E.; Acierno, J.S.; Maione, L.; Hietamäki, J.; Raivio, T.; Pitteloud, N. Clinical Management of Congenital Hypogonadotropic Hypogonadism. Endocr. Rev. 2019, 40, 669–710. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.J.; Zhang, Q.; Wang, Y.Q.; Yang, G.Q.; Hong, T.P.; Zhu, D.L.; Yang, J.K.; Ning, G.; Jin, N.; Chen, K.; et al. Mutation analyses in pedigrees and sporadic cases of ethnic Han Chinese Kallmann syndrome patients. Exp. Biol. Med. 2015, 240, 1480–1489. [Google Scholar] [CrossRef] [PubMed]
- Soussi-Yanicostas, N.; Faivre-Sarrailh, C.; Hardelin, J.P.; Levilliers, J.; Rougon, G.; Petit, C. Anosmin-1 underlying the X chromosomelinked Kallmann syndrome is an adhesion molecule that can modulate neurite growth in a cell-type specifific manner. J. Cell Sci. 1998, 111, 2953–2965. [Google Scholar] [CrossRef] [PubMed]
- Soussi-Yanicostas, N.; de Castro, F.; Julliard, A.K.; Perfettini, I.; Chedotal, A.; Petit, C. Anosmin1, defective in the X-linked form of Kallmann syndrome, promotes axonal branch formation from olfactory bulb output neurons. Cell 2002, 109, 217–228. [Google Scholar] [CrossRef]
- Gonzalez-Martinez, D.; Kim, S.H.; Hu, Y.; Guimond, S.; Schofifield, J.; Winyard, P.; Vannelli, G.B.; Turnbull, J.; Bouloux, P.M. Anosmin-1 modulates fifibroblast growth factor receptor 1 signaling in human gonadotropinreleasing hormone olfactory neuroblasts through a heparan sulfatedependent mechanism. J. Neurosci. 2004, 24, 10384–10392. [Google Scholar] [CrossRef]
- Hardelin, J.P.; Levilliers, J.; Blanchard, S.; Carel, J.C.; Leutenegger, M.; Pinard-ertelletto, J.P.; Bouloux, P.; Petit, C. Heterogeneity in the mutations responsible for X chromosome-linked Kallmann syndrome. Hum. Mol. Genet. 1993, 2, 373–377. [Google Scholar] [CrossRef]
- Quinton, R.; Duke, V.M.; de Zoysa, P.A.R.; Platts, A.D.; Valentine, A.; Kendall, B.; Pickman, S.; Kirk, J.M.; Besser, G.M.; Jacobs, H.S.; et al. The neuroradiology of Kallmann’s syndrome: A genotypic and phenotypic analysis. J. Clin. Endocrinol. Metab. 1996, 81, 3010–3017. [Google Scholar] [CrossRef][Green Version]
- Albuisson, J.; Pecheux, C.; Carel, J.C.; Lacombe, D.; Leheup, B.; Lapuzina, P.; Bouchard, P.; Legius, E.; Matthijs, G.; Wasniewska, M.; et al. Kallmann syndrome: 14 novel mutations in KAL1 and FGFR1 (KAL2). Hum. Mutat. 2005, 25, 98–99. [Google Scholar] [CrossRef]
- Dode, C.; Levilliers, J.; Dupont, J.M.; De Paepe, A.; Le Du, N.; SoussiYanicostas, N.; Coimbra, R.S.; Delmaghani, S.; Compain-Nouaille, S.; Baverel, F.; et al. Loss of function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat. Genet. 2003, 33, 463–465. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Katsumata, N.; Kagami, M.; Hasegawa, T.; Hori, X.; Kawakita, S.; Minowada, S.; Shimotsuka, A.; Shishiba, Y.; Yokozawa, M.; et al. Clinical assessment and mutation analysis of Kallmann syndrome 1 (KAL1) and fifibroblast growth factor receptor 1 (FGFR1, or KAL2) in five families and 18 sporadic patients. J. Clin. Endocrinol. Metab. 2004, 89, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Stamou, M.I.; Cox, K.H.; Crowley, W.F., Jr. Discovering genes essential to the hypothalamic regulation of human reproduction using a human disease model: Adjusting to life in the “-omics” era. Endocr. Rev. 2015, 36, 603–621. [Google Scholar] [CrossRef] [PubMed]
- Topaloglu, A.K.; Kotan, L.D. Genetics of hypogonadotropic hypogonadism. Endocr. Dev. 2016, 29, 36–49. [Google Scholar] [PubMed]
- Cariboni, A.; Balasubramanian, R. Kallmann syndrome and idiopathic hypogonadotropic hypogonadism: The role of semaphorin signaling on GnRH neurons. Handb. Clin. Neurol. 2021, 182, 307–315. [Google Scholar] [PubMed]
- Lacal, I.; Ventura, R. Epigenetic Inheritance: Concepts, Mechanisms and Perspectives. Front. Mol. Neurosci. 2018, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.E.; Ben Maamar, M.; Skinner, M.K. Role of epigenetic transgenerational inheritance in generational toxicology. Environ. Epigenet. 2022, 8, dvac001. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Li, Z.; Wang, G.; Wang, H.; Zhou, Y.; Zhao, X.; Cheng, C.Y.; Qiao, Y.; Sun, F. Sperm epigenetic alterations contribute to inter- and transgenerational effects of paternal exposure to long-term psychological stress via evading offspring embryonic reprogramming. Cell Discov. 2021, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Allegrucci, C.; Thurston, A.; Lucas, E.; Young, L. Epigenetics and the germline. Reproduction 2005, 129, 137–149. [Google Scholar] [CrossRef]
- Kiselev, I.S.; Kulakova, O.G.; Boyko, A.N.; Favorova, O.O. DNA Methylation As an Epigenetic Mechanism in the Development of Multiple Sclerosis. Acta Naturae 2021, 13, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Shanthikumar, S.; Neeland, M.R.; Maksimovic, J.; Ranganathan, S.C.; Saffery, R. DNA methylation biomarkers of future health outcomes in children. Mol. Cell Pediatr. 2020, 7, 7. [Google Scholar] [CrossRef]
- Luján, S.; Caroppo, E.; Niederberger, C.; Arce, J.-C.; Sadler-Riggleman, I.; Beck, D.; Nilsson, E.; Skinner, M.K. Sperm DNA methylation epimutation biomarkers for male infertility and FSH therapeutic responsiveness. Sci. Rep. 2019, 9, 16786. [Google Scholar] [CrossRef] [PubMed]
- Ichiyanagi, T.; Ichiyanagi, K.; Miyake, M.; Sasaki, H. Accumulation and loss of asymmetric non-CpG methylation during male germ-cell development. Nucleic Acids Res. 2013, 41, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Gkountela, S.; Zhang, K.X.; Shafiq, T.A.; Liao, W.-W.; Hargan-Calvopiña, J.; Chen, P.-Y.; Clark, A.T. DNA demethylation dynamics in the human prenatal germline. Cell 2015, 161, 1425–1436. [Google Scholar] [CrossRef]
- Tang, W.W.; Dietmann, S.; Irie, N.; Leitch, H.G.; Floros, V.I.; Bradshaw, C.R.; Hackett, J.A.; Chinnery, P.F.; Surani, M.A. A unique gene regulatory network resets the human germline epigenome for development. Cell 2015, 161, 1453–1467. [Google Scholar] [CrossRef] [PubMed]
- Rousseaux, S.; Khochbin, S. Histone acylation beyond acetylation: Terra incognita in chromatin biology. Cell J. 2015, 17, 1. [Google Scholar]
- Henikoff, S.; Smith, M.M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 2015, 7, a019364. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, J.; Reynoird, N.; Montellier, E.; Boussouar, F.; Rousseaux, S.; Khochbin, S. From meiosis to postmeiotic events: The secrets of histone disappearance. FEBS J. 2010, 277, 599–604. [Google Scholar] [CrossRef]
- Goudarzi, A.; Zhang, D.; Huang, H.; Barral, S.; Kwon, O.K.; Qi, S.; Tang, Z.; Buchou, T.; Vitte, A.-L.; He, T.; et al. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Mol. Cell 2016, 62, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Barral, S.; Morozumi, Y.; Tanaka, H.; Montellier, E.; Govin, J.; de Dieuleveult, M.; Charbonnier, G.; Coute, Y.; Puthier, D.; Buchou, T.; et al. Histone variant H2A. L. 2 guides transition protein-dependent protamine assembly in male germ cells. Mol. Cell 2017, 66, 89–101.e8. [Google Scholar] [CrossRef]
- Skinner, M.K.; Maamar, M.B.; Sadler-Riggleman, I.; Beck, D.; Nilsson, E.; McBirney, M.; Klukovich, R.; Xie, Y.; Tang, C.; Yan, W. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin 2018, 11, 1–24. [Google Scholar] [CrossRef]
- Carrell, D.T.; Emery, B.R.; Hammoud, S. The aetiology of sperm protamine abnormalities and their potential impact on the sperm epigenome. Int. J. Androl. 2008, 31, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Maamar, M.B.; Beck, D.; Nilsson, E.; McCarrey, J.R.; Skinner, M.K. Developmental origins of transgenerational sperm histone retention following ancestral exposures. Dev. Biol. 2020, 465, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, S.S.; Nix, D.A.; Hammoud, A.O.; Gibson, M.; Cairns, B.R.; Carrell, D.T. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum. Reprod. 2011, 26, 2558–2569. [Google Scholar] [CrossRef] [PubMed]
- Ihara, M.; Meyer-Ficca, M.L.; Leu, N.A.; Rao, S.; Li, F.; Gregory, B.D.; Zalenskaya, I.A.; Schultz, R.M.; Meyer, R.G. Paternal poly (ADP-ribose) metabolism modulates retention of inheritable sperm histones and early embryonic gene expression. PLoS Genet. 2014, 10, e1004317. [Google Scholar] [CrossRef] [PubMed]
- Luense, L.J.; Wang, X.; Schon, S.B.; Weller, A.H.; Shiao, E.L.; Bryant, J.M.; Bartolomei, M.S.; Coutifaris, C.; Garcia, B.A.; Berger, S.L. Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenetics Chromatin 2016, 9, 24. [Google Scholar] [CrossRef]
- Van der Heijden, G.W.; Ramos, L.; Baart, E.B.; van den Berg, I.M.; Derijck, A.A.; van der Vlag, J.; Martini, E.; de Boer, P. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev. Biol. 2008, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Carone, B.R.; Hung, J.-H.; Hainer, S.J.; Chou, M.-T.; Carone, D.M.; Weng, Z.; Fazzio, T.G.; Rando, O.J. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev. Cell 2014, 30, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Samans, B.; Yang, Y.; Krebs, S.; Sarode, G.V.; Blum, H.; Reichenbach, M.; Wolf, E.; Steger, K.; Dansranjavin, T.; Schagdarsurengin, U. Uniformity of nucleosome preservation pattern in Mammalian sperm and its connection to repetitive DNA elements. Dev. Cell 2014, 30, 23–35. [Google Scholar] [CrossRef]
- Kurimoto, K.; Saitou, M. (Eds.) Mechanism and reconstitution in vitro of germ cell development in mammals. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2015. [Google Scholar]
- Teperek, M.; Simeone, A.; Gaggioli, V.; Miyamoto, K.; Allen, G.E.; Erkek, S.; Kwon, T.; Marcotte, E.M.; Zegerman, P.; Bradshaw, C.R.; et al. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 2016, 26, 1034–1046. [Google Scholar] [CrossRef]
- Bao, J.; Bedford, M.T. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction 2016, 151, R55. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.; Brinkworth, M.; Iles, D. Paternal DNA packaging in spermatozoa: More than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 2010, 139, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Rivera, C.; Gurard-Levin, Z.A.; Almouzni, G.; Loyola, A. Histone lysine methylation and chromatin replication. Biochim. Biophys. Acta-Gene Regul. Mech. 2014, 1839, 1433–1439. [Google Scholar] [CrossRef]
- Verma, A.; Rajput, S.; Kumar, S.; De, S.; Chakravarty, A.K.; Kumar, R.; Datta, T.K. Differential histone modification status of spermatozoa in relation to fertility of buffalo bulls. J. Cell. Biochem. 2015, 116, 743–753. [Google Scholar] [CrossRef]
- Kutchy, N.; Menezes, E.; Chiappetta, A.; Tan, W.; Wills, R.W.; Kaya, A.; Topper, E.; Moura, A.A.; Perkins, A.D.; Memili, E. Acetylation and methylation of sperm histone 3 lysine 27 (H3K27ac and H3K27me3) are associated with bull fertility. Andrologia 2018, 50, e12915. [Google Scholar] [CrossRef] [PubMed]
- Barratt, C.L.; Aitken, R.J.; Björndahl, L.; Carrell, D.T.; de Boer, P.; Kvist, U.; Lewis, S.E.; Perreault, S.D.; Perry, M.J.; Ramos, L.; et al. Sperm DNA: Organization, protection and vulnerability: From basic science to clinical applications—A position report. Hum. Reprod. 2010, 25, 824–838. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Shaman, J.A.; Boaz, S.M.; Ward, W.S. Paternal pronuclear DNA degradation is functionally linked to DNA replication in mouse oocytes. Biol. Reprod. 2007, 77, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Shaman, J.A.; Yamauchi, Y.; Steven Ward, W. Function of the sperm nuclear matrix. Arch. Androl. 2007, 53, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Barbu, M.G.; Thompson, D.C.; Suciu, N.; Voinea, S.C.; Cretoiu, D.; Predescu, D.V. The Roles of MicroRNAs in Male Infertility. Int. J. Mol. Sci. 2021, 22, 2910. [Google Scholar] [CrossRef]
- Cecere, G. Small RNAs in epigenetic inheritance: From mechanisms to trait transmission. FEBS Lett. 2021, 595, 2953–2977. [Google Scholar] [CrossRef] [PubMed]
- Suh, N.; Blelloch, R. Small RNAs in early mammalian development: From gametes to gastrulation. Development 2011, 138, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Yang, W.; Xuan, J.; Gupta, S.; Krylov, S.N.; Ma, X.; Yang, Q.; Yang, B.Z. Reciprocal regulation of miRNAs and piRNAs in embryonic development. Cell Death Differ. 2016, 23, 1458–1470. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Schuster, A.; Tang, C.; Yu, T.; Ortogero, N.; Bao, J.; Zheng, H.; Yan, W. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Dev. Camb. Engl. 2016, 143, 635e47. [Google Scholar] [CrossRef] [PubMed]
- Champroux, A.; Cocquet, J.; Henry-Berger, J.; Drevet, J.R.; Kocer, A. A decade of exploring the mammalian sperm epigenome: Paternal epigenetic and transgenerational inheritance. Front. Cell Dev. Biol. 2018, 6, 50. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Grandjean, V.; Fourré, S.; De Abreu, D.A.; Derieppe, M.A.; Remy, J.J.; Rassoulzadegan, M. RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci. Rep. 2015, 5, 18193. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Schuster, A.; Tang, C.; Yu, T.; Ortogero, N.; Bao, J.; Zheng, H.; Yan, W.; Wang, Z. Sperm-borne small RNA profiling reveals piRNAs in human seminal plasma. Oncotarget 2020, 11, 55–70. [Google Scholar]
- Sendler, E.; Johnson, G.D.; Mao, S.; Goodrich, R.D.; Diamond, M.P.; Hauser, R.; Krawetz, S.A. The effect of smoking on the small non-coding RNAome in human sperm. Hum. Reprod. 2016, 31, 2525–2537. [Google Scholar]
- Rodgers, A.B.; Morgan, C.P.; Leu, N.A.; Bale, T.L. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc. Natl. Acad. Sci. USA 2015, 112, 13699–13704. [Google Scholar] [CrossRef]
- Fullston, T.; Ohlsson Teague, E.M.; Palmer, N.O.; DeBlasio, M.J.; Mitchell, M.; Corbett, M.; Print, C.G.; Owens, J.A.; Lane, M. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J. 2013, 27, 4226–4243. [Google Scholar] [CrossRef]
- Benchaib, M.; Braun, V.; Ressnikof, D.; Lornage, J.; Durand, P.; Niveleau, A. Influence of global sperm DNA methylation on IVF results. Hum. Reprod. 2010, 25, 2158–2168. [Google Scholar] [CrossRef] [PubMed]
- Skakkebæk, A.; Viuff, M.; Nielsen, M.M.; Gravholt, C.H. (Eds.) Epigenetics and genomics in Klinefelter syndrome. In American Journal of Medical Genetics Part C: Seminars in Medical Genetics; Wiley Online Library: Hoboken, NJ, USA, 2020. [Google Scholar]
- Passerini, V.; Ozeri-Galai, E.; De Pagter, M.S.; Donnelly, N.; Schmalbrock, S.; Kloosterman, W.P.; Kerem, B.; Storchová, Z. The presence of extra chromosomes leads to genomic instability. Nat. Commun. 2016, 7, 10754. [Google Scholar] [CrossRef]
- Jowhar, Z.; Shachar, S.; Gudla, P.R.; Wangsa, D.; Torres, E.; Russ, J.L.; Pegoraro, G.; Ried, T.; Raznahan, A.; Misteli, T. Effects of human sex chromosome dosage on spatial chromosome organization. Mol. Biol. Cell 2018, 29, 2458–2469. [Google Scholar] [CrossRef] [PubMed]
- Migicovsky, Z.; Kovalchuk, I. Epigenetic memory in mammals. Front. Genet. 2011, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Tiffon, C. The Impact of Nutrition and Environmental Epigenetics on Human Health and Disease. Int. J. Mol. Sci. 2018, 19, 3425. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.; Estanyol, J.M.; Ballescá, J.L.; Oliva, R. Human sperm chromatin epigenetic potential: Genomics, proteomics, and male infertility. Asian J. Androl. 2015, 17, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Barrachina, F.; Battistone, M.A.; Castillo, J.; Mallofré, C.; Jodar, M.; Breton, S.; Oliva, R. Sperm acquire epididymis-derived proteins through epididymosomes. Hum. Reprod. 2022, 37, 651–668. [Google Scholar] [CrossRef]
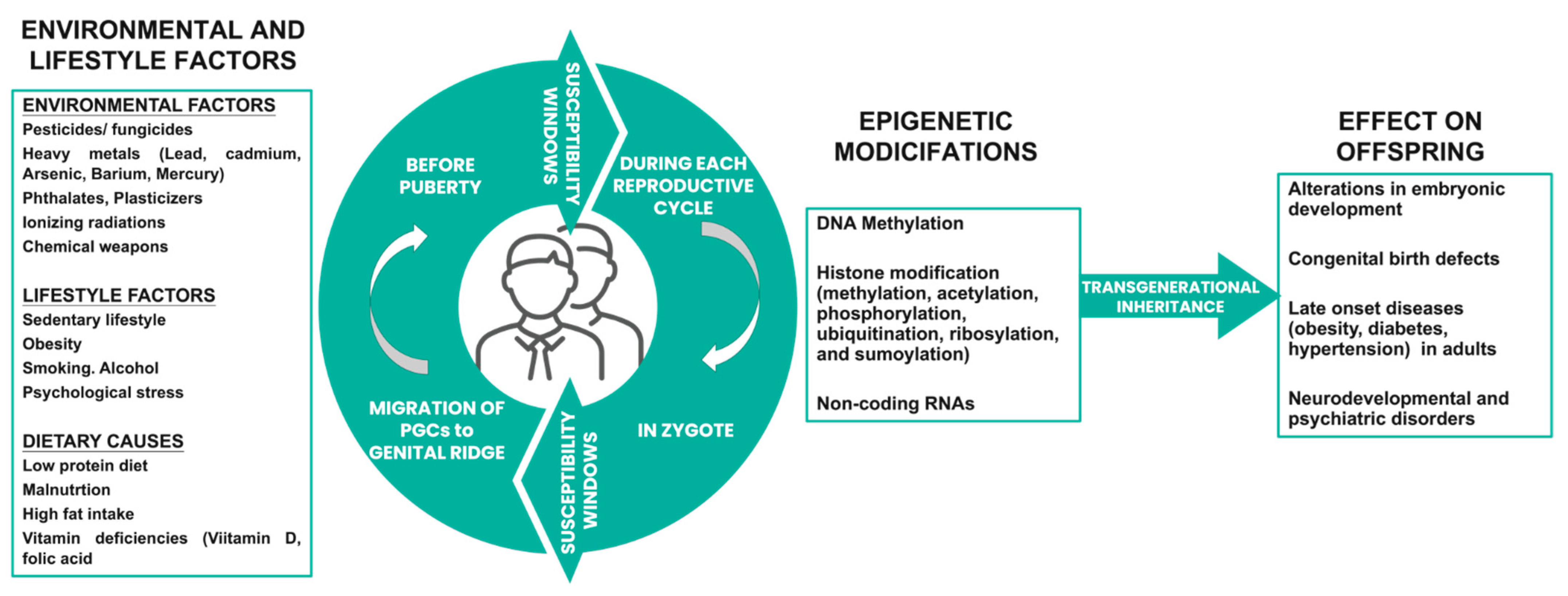
| Environmental Factors | Evidence of Paternal Transmission | Health Outcomes in Offspring | References |
|---|---|---|---|
| Heavy metals (mercury, lead, etc.) | Epigenetic changes in sperm DNA | Autism, attention-deficit hyperactivity disorder (ADHD) | [111,112,113,114] |
| High-fat diet/obesity | Changes in sperm DNA methylation | Obesity, type 2 diabetes | |
| Psychological stress/drugs | Changes in sperm DNA methylation and RNA expression | Anxiety, depression, behavioral abnormalities | [105,106] |
| Air pollution/cigarette smoking | Epigenetic changes in sperm DNA | Cardiovascular diseases, respiratory diseases, cognitive impairment, reduced growth of fetus | [115,116] |
| Radiation | Epigenetic changes in sperm DNA | Cancer | [117,118] |
| Pesticides | Epigenetic changes in sperm DNA | Birth defects, behavioral abnormalities, cognitive impairment | [110,119] |
| PATHOGENETIC MECHANISMS AND CLINICAL CHARACTERISTICS | |||
|---|---|---|---|
| Supernumerary X Chromosome | Testosterone Deficiency | Supernumerary X Chromosome and Testosterone Deficiency | |
| ONSET | Before puberty | At puberty/adulthood | Before puberty, with progressive worsening after puberty |
| SIGNS | Longer legs Small testes Congenital malformations (cleft lip, cleft palate, hernia)—rare | Sparse body and facial hair; decreased muscle mass; female pubic escutcheon; bilateral gynecomastia; eunuchoid skeleton; longer legs (due to testosterone deficiency in fetal life); impaired estradiol/testosterone ratio | Tall stature; eunuchoid skeleton; gynecoid pelvis; elevated FSH/LH; increased BMI (overweight/obese); metabolic abnormalities; reduced bone mineral density; genital abnormalities at birth (rare) |
| SYMPTOMS | Disability in speech and language; azoospermia | Erectile dysfunction; reduced libido; weakness; impaired well-being | Mood disturbances |
| Epigenetic Marker(s) | Target Gene(s)/Pathway(s) | Findings | References |
|---|---|---|---|
| miRNAs | Metabolic genes | Paternal transfer of miRNAs contributed to the inheritance of diet-induced obesity and metabolic disorders in offspring. | [229] |
| tRNA fragments | Gene expression | tRNA fragments in sperm played a role in gene expression regulation during fertilization and embryonic development. | [228] |
| tsRNAs | Metabolic genes | Sperm tsRNAs contributed to the transgenerational inheritance of acquired metabolic disorders in offspring. | [230] |
| Sperm-borne RNAs | Neurodevelopmental genes | Sperm-borne RNAs were implicated in the transgenerational inheritance of the effects of early trauma on neurodevelopmental outcomes in offspring. | [105] |
| piRNAs | Epigenetic regulation | piRNAs were identified in human seminal plasma, suggesting a potential role in transgenerational epigenetic inheritance and the regulation of gene expression in offspring. | [231] |
| Small non-coding RNAs | Epigenetic modifications | Smoking influenced the small non-coding RNAome in human sperm, suggesting a potential role in transgenerational effects on epigenetic modifications and gene regulation in offspring. | [232] |
| microRNAs | Behavior-related genes | Paternal stress led to alterations in sperm microRNA content and transgenerational effects on offspring behavior. | [233] |
| microRNAs | Metabolic genes | Sperm microRNAs were altered by diet and paternal obesity, influencing offspring metabolic health and susceptibility to metabolic disorders. | [234] |
| miRNAs | Fertility-related genes | Abnormal sperm miRNA profiles were associated with male infertility and could potentially be transmitted to offspring, impacting fertility and reproductive health. | [235] |
| piRNAs | Reproductive genes | Aberrant piRNA expression in sperm was linked to male infertility and may contribute to transgenerational effects on reproductive health and fertility. | [230] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sengupta, P.; Dutta, S.; Liew, F.F.; Dhawan, V.; Das, B.; Mottola, F.; Slama, P.; Rocco, L.; Roychoudhury, S. Environmental and Genetic Traffic in the Journey from Sperm to Offspring. Biomolecules 2023, 13, 1759. https://doi.org/10.3390/biom13121759
Sengupta P, Dutta S, Liew FF, Dhawan V, Das B, Mottola F, Slama P, Rocco L, Roychoudhury S. Environmental and Genetic Traffic in the Journey from Sperm to Offspring. Biomolecules. 2023; 13(12):1759. https://doi.org/10.3390/biom13121759
Chicago/Turabian StyleSengupta, Pallav, Sulagna Dutta, Fong Fong Liew, Vidhu Dhawan, Biprojit Das, Filomena Mottola, Petr Slama, Lucia Rocco, and Shubhadeep Roychoudhury. 2023. "Environmental and Genetic Traffic in the Journey from Sperm to Offspring" Biomolecules 13, no. 12: 1759. https://doi.org/10.3390/biom13121759
APA StyleSengupta, P., Dutta, S., Liew, F. F., Dhawan, V., Das, B., Mottola, F., Slama, P., Rocco, L., & Roychoudhury, S. (2023). Environmental and Genetic Traffic in the Journey from Sperm to Offspring. Biomolecules, 13(12), 1759. https://doi.org/10.3390/biom13121759










