The Role of Glutamine Synthetase (GS) and Glutamate Synthase (GOGAT) in the Improvement of Nitrogen Use Efficiency in Cereals
Abstract
1. Introduction
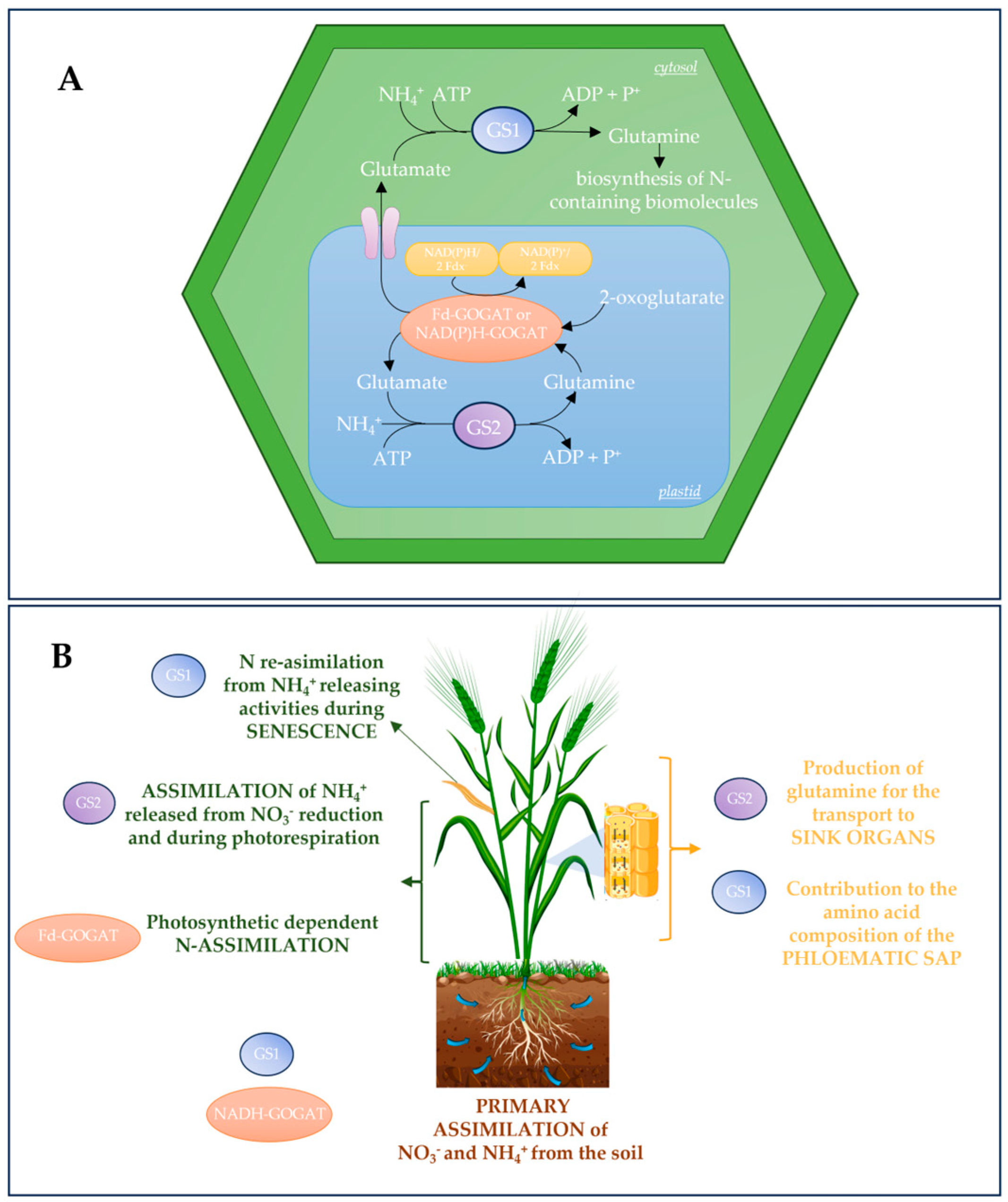
2. GS-GOGAT Cycle
3. GS and GOGAT Isoforms in NUE of Cereals
4. Correlation of NUE Quantitative Trait Loci with GS and GOGAT
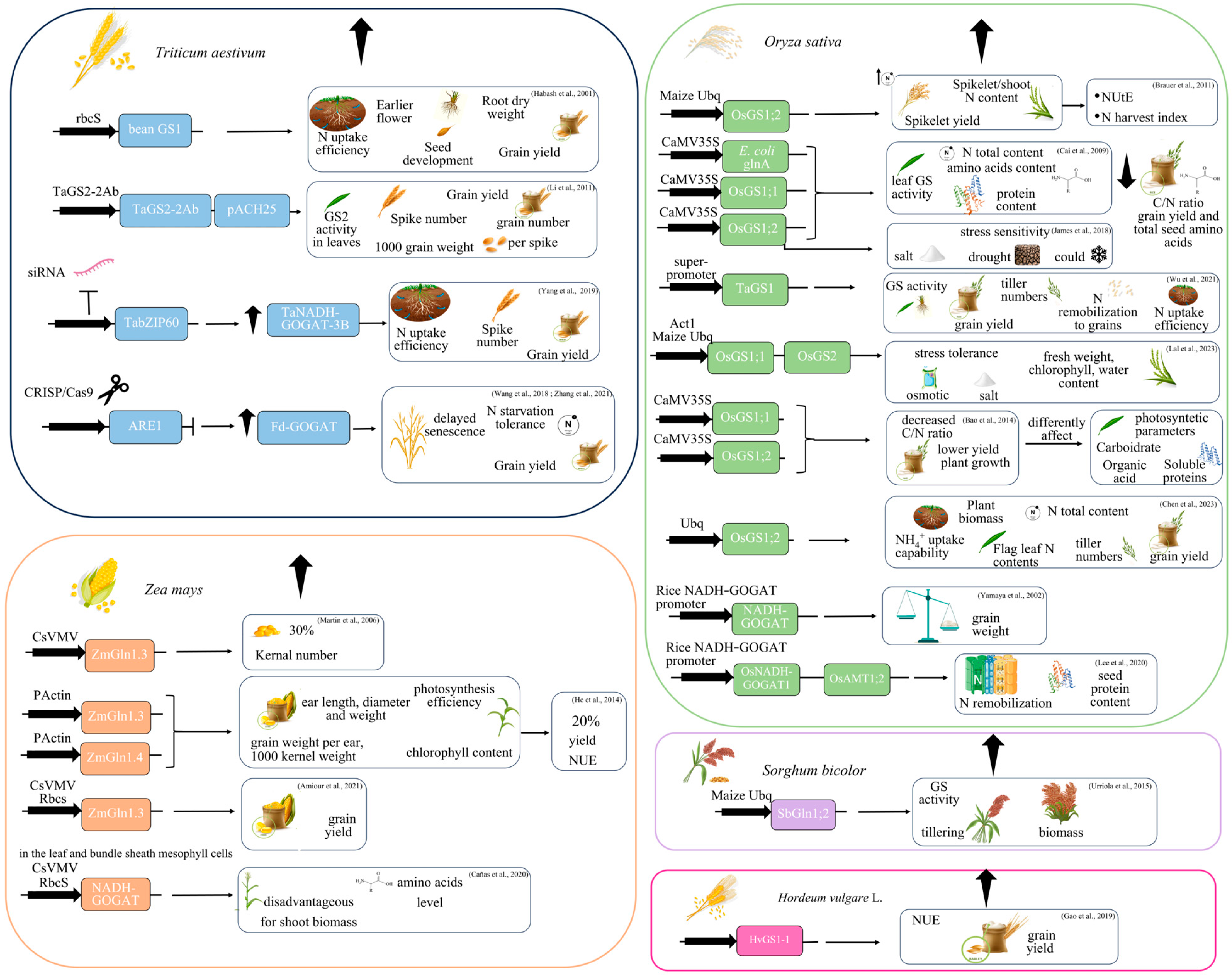
5. Transgenic Cereals Overexpressing GS or GOGAT
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. World Food and Agriculture—Statistical Yearbook 2021; FAO: Rome, Italy, 2021; ISBN 978-92-5-134332-6. [Google Scholar] [CrossRef]
- Ladha, J.K.; Tirol-Padre, A.; Reddy, C.K.; Cassman, K.G.; Verma, S.; Powlson, D.S.; van Kessel, C.; de Richter, D.B.; Chakraborty, D.; Pathak, H. Global nitrogen budgets in cereals: A 50-year assessment for maize, rice, and wheat production systems. Sci. Rep. 2016, 6, 19355. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Ormeno-Orrillo, E.; Lopez-Lopez, A.; Rogel, M.A.; Reyes-Hernandez, B.J.; Martinez-Romero, J.C.; Reddy, P.M.; Martinez-Romero, E. Nitrogen fixation in cereals. Front. Microbiol. 2018, 9, 1794. [Google Scholar] [CrossRef] [PubMed]
- Muitire, C.; Kamutando, C.; Moyo, M. Building Stress Resilience of Cereals under Future Climatic Scenarios: ‘The Case of Maize, Wheat, Rice and Sorghum’. In Cereal Grains-Volume 1; IntechOpen: London, UK, 2021. [Google Scholar]
- Singer, S.D.; Foroud, N.A.; Laurie, J.D. Molecular Improvement of Grain: Target Traits for a Changing World. In Encyclopedia of Food Security and Sustainability; Elsevier: Oxford, UK, 2019; pp. 545–555. [Google Scholar]
- OECD/FAO. OECD-FAO Agricultural Outlook 2020–2029. Available online: https://www.oecd-ilibrary.org/agriculture-and-food/oecd-fao-agricultural-outlook-2020-2029_1112c23b-en (accessed on 11 September 2023).
- Chen, L.Y.; Liao, H. Engineering crop nutrient efficiency for sustainable agriculture. J. Integr. Plant Biol. 2017, 59, 710–735. [Google Scholar] [CrossRef] [PubMed]
- Mulvaney, R.L.; Khan, S.A.; Ellsworth, T.R. Synthetic nitrogen fertilizers deplete soil nitrogen: A global dilemma for sustainable cereal production. J. Environ. Qual. 2009, 38, 2295–2314. [Google Scholar] [CrossRef] [PubMed]
- Marschner, H. Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: New York, NY, USA, 1995. [Google Scholar]
- Fowler, D.; Pyle, J.A.; Raven, J.A.; Sutton, M.A. The global nitrogen cycle in the twenty-first century: Introduction. Philos. Trans. R. Soc. B 2013, 368, 20130165. [Google Scholar] [CrossRef] [PubMed]
- Kopittke, P.M.; Menzies, N.W.; Wang, P.; McKenna, B.A.; Lombi, E. Soil and the intensification of agriculture for global food security. Environ. Int. 2019, 132, 105078. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.N.; Sharma, M.; Aneja, V.P.; Balasubramanian, R. Ammonia in the atmosphere: A review on emission sources, atmospheric chemistry and deposition on terrestrial bodies. Environ. Sci. Pollut. Res. 2013, 20, 8092–8131. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.H.; deKok, T.M.; Levallois, P.; Brender, J.; Gulis, G.; Nolan, B.T.; VanDerslice, J. Workgroup report: Drinking-water nitrate and health-recent findings and research needs. Environ. Health Perspect. 2005, 113, 1607–1614. [Google Scholar] [CrossRef]
- Ma, G.; Liu, W.X.; Li, S.S.; Zhang, P.P.; Wang, C.Y.; Lu, H.F.; Wang, L.F.; Xie, Y.X.; Ma, D.Y.; Kang, G.Z. Determining the optimal N input to improve grain yield and quality in winter wheat with reduced apparent N loss in the North China plain. Front. Plant Sci. 2019, 1, 181. [Google Scholar] [CrossRef]
- Adjesiwor, A.T.; Islam, M.A. Rising nitrogen fertilizer prices and projected increase in maize ethanol production: The future of forage production and the potential of legumes in forage production systems. Grassl. Sci. 2016, 62, 203–212. [Google Scholar] [CrossRef]
- Mueller, N.D.; West, P.C.; Gerber, J.S.; MacDonald, G.K.; Polasky, S.; Foley, J.A. A tradeoff frontier for global nitrogen use and cereal production. Environ. Res. Lett. 2014, 9, 054002. [Google Scholar] [CrossRef]
- Zhu, X.C.; Zhang, J.; Zhang, Z.P.; Deng, A.X.; Zhang, W.J. Dense planting with less basal nitrogen fertilization might benefit rice cropping for high yield with less environmental impacts. Eur. J. Agron. 2016, 75, 50–59. [Google Scholar] [CrossRef]
- Fischer, R.A.; Byerlee, D.; Edmeades, G.O. Can technology deliver on the yield challenge to 2050? In Proceedings of the FAO Expert Meeting on How to Feed the World in 2050, Rome, Italy, 24–26 June 2009; pp. 1–48. [Google Scholar]
- Hawkesford, M.J. Genetic variation in traits for nitrogen use efficiency in wheat. J. Exp. Bot. 2017, 68, 2627–2632. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef]
- Garnett, T.; Conn, V.; Kaiser, B.N. Root based approaches to improving nitrogen use efficiency in plants. Plant Cell Environ. 2009, 32, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Avice, J.C.; Etienne, P. Leaf senescence and nitrogen remobilization efficiency in oilseed rape (Brassica napus L.). J. Exp. Bot. 2014, 65, 3813–3824. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Bi, Y.M.; Rothstein, S.J. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 2011, 62, 1499–1509. [Google Scholar] [CrossRef]
- Li, H.; Hu, B.; Chu, C. Nitrogen use efficiency in crops: Lessons from Arabidopsis and rice. J. Exp. Bot. 2017, 68, 2477–2488. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.J.; Hu, B.; Chu, C.C. Nitrogen assimilation in plants: Current status and future prospects. J. Genet. Genom. 2022, 49, 394–404. [Google Scholar] [CrossRef]
- Zhang, X.; Davidson, E.A.; Mauzerall, D.L.; Searchinger, T.D.; Dumas, P.; Shen, Y. Managing nitrogen for sustainable development. Nature 2015, 528, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Anas, M.; Liao, F.; Verma, K.K.; Sarwar, M.A.; Mahmood, A.; Chen, Z.L.; Li, Q.; Zeng, X.P.; Liu, Y.; Li, Y.R. Fate of nitrogen in agriculture and environment: Agronomic, eco-physiological and molecular approaches to improve nitrogen use efficiency. Biol. Res. 2020, 53, 47. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, K.; Song, W.; Zhong, N.; Wu, Y.; Fu, X. Improving crop nitrogen use efficiency toward sustainable green revolution. Annu. Rev. Plant Biol. 2022, 73, 523–551. [Google Scholar] [CrossRef] [PubMed]
- Melino, V.J.; Tester, M.A.; Okamoto, M. Strategies for engineering improved nitrogen use efficiency in crop plants via redistribution and recycling of organic nitrogen. Curr. Opin. Biotechnol. 2022, 73, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Okamoto, M.; Beatty, P.H.; Rothstein, S.J.; Good, A.G. The genetics of nitrogen use efficiency in crop plants. Ann. Rev. Genet. 2015, 49, 269–289. [Google Scholar] [CrossRef]
- Bandyopadhyay, T.; Prasad, M. A precise method for analyzing nitrogen use in foxtail millet. Methods Mol. Biol. 2020, 2057, 113–118. [Google Scholar] [PubMed]
- Hirel, B.; Le Gouis, J.; Ney, B.; Gallais, A. The challenge of improving nitrogen use efficiency in crop plants: Towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 2007, 58, 2369–2387. [Google Scholar] [CrossRef]
- Garnett, T.; Plett, D.; Heuer, S.; Okamoto, M. Genetic approaches to enhancing nitrogen-use efficiency (NUE) in cereals: Challenges and future directions. Funct. Plant Biol. 2015, 42, 921–941. [Google Scholar] [CrossRef]
- Chichkova, S.; Arellano, J.; Vance, C.P.; Hernandez, G. Transgenic tobacco plants that overexpress alfalfa NADH-glutamate synthase have higher carbon and nitrogen content. J. Exp. Bot. 2001, 52, 2079–2087. [Google Scholar] [CrossRef] [PubMed]
- Brauer, E.K.; Rochon, A.; Bi, Y.M.; Bozzo, G.G.; Rothstein, S.J.; Shelp, B.J. Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase1. Physiol. Plant. 2011, 141, 361–372. [Google Scholar] [CrossRef]
- Miflin, B.J.; Habash, D.Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 2002, 53, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Bernard, S.M.; Habash, D.Z. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 2009, 182, 608–620. [Google Scholar] [CrossRef] [PubMed]
- McNally, S.F.; Hirel, B.; Gadal, P.; Mann, A.F.; Stewart, G.R. Glutamine synthetases of higher plants: Evidence for a specific isoform content related to their possible physiological role and their compartmentation within the Leaf. Plant Physiol. 1983, 72, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Zozaya-Hinchliffe, M.; Potenza, C.; Ortega, J.L.; Sengupta-Gopalan, C. Nitrogen and metabolic regulation of the expression of plastidic glutamine synthetase in alfalfa (Medicago sativa). Plant Sci. 2005, 168, 1041–1052. [Google Scholar] [CrossRef]
- Bernard, S.M.; Moller, A.L.B.; Dionisio, G.; Kichey, T.; Jahn, T.P.; Dubois, F.; Baudo, M.; Lopes, M.S.; Terce-Laforgue, T.; Foyer, C.H.; et al. Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.). Plant Mol. Biol. 2008, 67, 89–105. [Google Scholar] [CrossRef] [PubMed]
- Swarbreck, S.M.; Defoin-Platel, M.; Hindle, M.; Saqi, M.; Habash, D.Z. New perspectives on glutamine synthetase in grasses. J. Exp. Bot. 2011, 62, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Guan, M.; de Bang, T.C.; Pedersen, C.; Schjoerring, J.K. Cytosolic glutamine synthetase Gln1;2 is the main isozyme contributing to GS1 activity and can be up-regulated to relieve ammonium toxicity. Plant Physiol. 2016, 171, 1921–1933. [Google Scholar] [CrossRef]
- Konishi, N.; Saito, M.; Imagawa, F.; Kanno, K.; Yamaya, T.; Kojima, S. Cytosolic glutamine synthetase isozymes play redundant roles in ammonium assimilation under low-ammonium conditions in roots of Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 601–613. [Google Scholar] [CrossRef]
- Kusano, M.; Fukushima, A.; Tabuchi-Kobayashi, M.; Funayama, K.; Kojima, S.; Maruyama, K.; Yamamoto, Y.Y.; Nishizawa, T.; Kobayashi, M.; Wakazaki, M.; et al. Cytosolic GLUTAMINE SYNTHETASE1;1 modulates metabolism and chloroplast development in roots. Plant Physiol. 2020, 182, 1894–1909. [Google Scholar] [CrossRef]
- Canton, F.R.; Suarez, M.F.; Canovas, F.M. Molecular aspects of nitrogen mobilization and recycling in trees. Photosynth. Res. 2005, 83, 265–278. [Google Scholar] [CrossRef]
- Tabuchi, M.; Sugiyama, K.; Ishiyama, K.; Inoue, E.; Sato, T.; Takahashi, H.; Yamaya, T. Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J. 2005, 42, 641–651. [Google Scholar] [CrossRef]
- Avila-Ospina, L.; Moison, M.; Yoshimoto, K.; Masclaux-Daubresse, C. Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 2014, 65, 3799–3811. [Google Scholar] [CrossRef] [PubMed]
- Krapp, A. Plant nitrogen assimilation and its regulation: A complex puzzle with missing pieces. Curr. Opin. Plant Biol. 2015, 25, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Caputo, C.; Criado, M.V.; Roberts, I.N.; Gelso, M.A.; Barneix, A.J. Regulation of glutamine synthetase 1 and amino acids transport in the phloem of young wheat plants. Plant Physiol. Biochem. 2009, 47, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Wallsgrove, R.M.; Turner, J.C.; Hall, N.P.; Kendall, A.C.; Bright, S.W.J. Barley Mutants Lacking Chloroplast Glutamine-Synthetase—Biochemical and Genetic-Analysis. Plant Physiol. 1987, 83, 155–158. [Google Scholar] [PubMed]
- Brestic, M.; Zivcak, M.; Olsovska, K.; Shao, H.B.; Kalaji, H.M.; Allakhverdiev, S.I. Reduced glutamine synthetase activity plays a role in control of photosynthetic responses to high light in barley leaves. Plant Physiol. Biochem. 2014, 81, 74–83. [Google Scholar] [PubMed]
- Perez-Delgado, C.M.; Garcia-Calderon, M.; Marquez, A.J.; Betti, M. Reassimilation of photorespiratory ammonium in lotus japonicus leplants deficient in plastidic glutamine synthetase. PLoS ONE 2015, 10, e0130438. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weber, A.; Flugge, U.I. Interaction of cytosolic and plastidic nitrogen metabolism in plants. J. Exp. Bot. 2002, 53, 865–874. [Google Scholar] [CrossRef]
- Brugiere, N.; Dubois, F.; Limami, A.M.; Lelandais, M.; Roux, Y.; Sangwan, R.S.; Hirel, B. Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 1999, 11, 1995–2011. [Google Scholar] [CrossRef]
- Ortega, J.L.; Temple, S.J.; Sengupta-Gopalan, C. Constitutive overexpression of cytosolic glutamine synthetase (GS1) gene in transgenic alfalfa demonstrates that GS1 may be regulated at the level of RNA stability and protein turnover. Plant Physiol. 2001, 126, 109–121. [Google Scholar] [CrossRef]
- Larios, B.; Aguera, E.; Cabello, P.; Maldonado, J.M.; de la Haba, P. The rate of CO2 assimilation controls the expression and activity of glutamine synthetase through sugar formation in sunflower (Helianthus annuus L.) leaves. J. Exp. Bot. 2004, 55, 69–75. [Google Scholar] [CrossRef]
- Finnemann, J.; Schjoerring, J.K. Post-translational regulation of cytosolic glutamine synthetase by reversible phosphorylation and 14-3-3 protein interaction. Plant J. 2000, 24, 171–181. [Google Scholar] [CrossRef]
- Riedel, J.; Tischner, R.; Mack, G. The chloroplastic glutamine synthetase (GS-2) of tobacco is phosphorylated and associated with 14-3-3 proteins inside the chloroplast. Planta 2001, 213, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.; Seabra, A.; Melo, P.; Cullimore, J.; Carvalho, H. Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula. J. Exp. Bot. 2006, 57, 2751–2761. [Google Scholar] [CrossRef] [PubMed]
- Ortega, J.L.; Roche, D.; Sengupta-Gopalan, C. Oxidative turnover of soybean root glutamine synthetase. In vitro and in vivo studies. Plant Physiol. 1999, 119, 1483–1495. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Carrillo, N.; Valle, E.M. The role of photosynthetic electron transport in the oxidative degradation of chloroplastic glutamine synthetase. Plant Physiol. 1999, 121, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Anzawa, D.; Kokubun, N.; Makino, A.; Mae, T. Direct evidence for non-enzymatic fragmentation of chloroplastic glutamine synthetase by a reactive oxygen species. Plant Cell Environ. 2002, 25, 625–631. [Google Scholar] [CrossRef]
- Melo, P.M.; Silva, L.S.; Ribeiro, I.; Seabra, A.R.; Carvalho, H.G. Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 2011, 157, 1505–1517. [Google Scholar] [CrossRef]
- Suzuki, A. Glutamate synthase and amino acid synthesis in higher plants. In Advances in Botanical Research; Jacquot, J.-P., Ed.; Academic Press: Cambridge, MA, USA, 2021; Volume 100, pp. 129–144. [Google Scholar]
- van den Heuvel, R.H.H.; Ferrari, D.; Bossi, R.T.; Ravasio, S.; Curti, B.; Vanoni, M.A.; Florencio, F.J.; Mattevi, A. Structural studies on the synchronization of catalytic centers in glutamate synthase. J. Biol. Chem. 2002, 277, 24579–24583. [Google Scholar] [CrossRef] [PubMed]
- Lea, P.J.; Miflin, B.J. Glutamate synthase and the synthesis of glutamate in plants. Plant Physiol. Biochem. 2003, 41, 555–564. [Google Scholar] [CrossRef]
- Suzuki, A.; Knaff, D.B. Glutamate synthase: Structural, mechanistic and regulatory properties, and role in the amino acid metabolism. Photosynth. Res. 2005, 83, 191–217. [Google Scholar] [CrossRef] [PubMed]
- Vanoni, M.A.; Dossena, L.; van den Heuvel, R.H.H.; Curti, B. Structure-function studies on the complex iron-sulfur flavoprotein glutamate synthase: The key enzyme of ammonia assimilation. Photosynth. Res. 2005, 83, 219–238. [Google Scholar] [CrossRef] [PubMed]
- Valadier, M.H.; Yoshida, A.; Grandjean, O.; Morin, H.; Kronenberger, J.; Boutet, S.; Raballand, A.; Hase, T.; Yoneyama, T.; Suzuki, A. Implication of the glutamine synthetase/glutamate synthase pathway in conditioning the amino acid metabolism in bundle sheath and mesophyll cells of maize leaves. FEBS J. 2008, 275, 3193–3206. [Google Scholar] [CrossRef]
- Potel, F.; Valadier, M.H.; Ferrario-Mery, S.; Grandjean, O.; Morin, H.; Gaufichon, L.; Boutet-Mercey, S.; Lothier, J.; Rothstein, S.J.; Hirose, N.; et al. Assimilation of excess ammonium into amino acids and nitrogen translocation in Arabidopsis thaliana—Roles of glutamate synthases and carbamoylphosphate synthetase in leaves. FEBS J. 2009, 276, 4061–4076. [Google Scholar] [CrossRef] [PubMed]
- Bowsher, C.G.; Lacey, A.E.; Hanke, G.T.; Clarkson, D.T.; Saker, L.R.; Stulen, I.; Emes, M.J. The effect of G1c6P uptake and its subsequent oxidation within pea root plastids on nitrite reduction and glutamate synthesis. J. Exp. Bot. 2007, 58, 1109–1118. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hudson, D.; Guevara, D.; Yaish, M.W.; Hannam, C.; Long, N.; Clarke, J.D.; Bi, Y.M.; Rothstein, S.J. GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 2011, 6, e26765. [Google Scholar] [CrossRef] [PubMed]
- Bi, Z.Z.; Zhang, Y.X.; Wu, W.X.; Zhan, X.D.; Yu, N.; Xu, T.T.; Liu, Q.E.; Li, Z.; Shen, X.H.; Chen, D.B.; et al. ES7, encoding a ferredoxin-dependent glutamate synthase, functions in nitrogen metabolism and impacts leaf senescence in rice. Plant Sci. 2017, 259, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Jamai, A.; Salome, P.A.; Schilling, S.H.; Weber, A.P.M.; McClung, C.R. Arabidopsis photorespiratory serine hydroxymethyltransferase activity requires the mitochondrial accumulation of ferredoxin-dependent glutamate synthase. Plant Cell 2009, 21, 595–606. [Google Scholar] [CrossRef]
- Tabuchi, M.; Abiko, T.; Yamaya, T. Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J. Exp. Bot. 2007, 58, 2319–2327. [Google Scholar] [CrossRef]
- Thomsen, H.C.; Eriksson, D.; Moller, I.S.; Schjoerring, J.K. Cytosolic glutamine synthetase: A target for improvement of crop nitrogen use efficiency? Trends Plant Sci. 2014, 19, 656–663. [Google Scholar] [CrossRef]
- Sakurai, N.; Hayakawa, T.; Nakamura, T.; Yamaya, T. Changes in the cellular localization of cytosolic glutamine synthetase protein in vascular bundles of rice leaves at various stages of development. Planta 1996, 200, 306–311. [Google Scholar] [CrossRef]
- Funayama, K.; Kojima, S.; Tabuchi-Kobayashi, M.; Sawa, Y.; Nakayama, Y.; Hayakawa, T.; Yamaya, T. Cytosolic glutamine synthetase1;2 is responsible for the primary assimilation of ammonium in rice roots. Plant Cell Physiol. 2013, 54, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, M.; Ishiyama, K.; Kusano, M.; Fukushima, A.; Kojima, S.; Hanada, A.; Kanno, K.; Hayakawa, T.; Seto, Y.; Kyozuka, J.; et al. Lack of cytosolic glutamine synthetase1;2 in vascular tissues of axillary buds causes severe reduction in their outgrowth and disorder of metabolic balance in rice seedlings. Plant J. 2015, 81, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Beier, M.P.; Tabuchi-Kobayashi, M.; Hayatsu, Y.; Nakamura, H.; Umetsu-Ohashi, T.; Sasaki, K.; Ishiyama, K.; Murozuka, E.; Kojima, M.; et al. Cytosolic Glutamine Synthetase GS1;3 Is Involved in Rice Grain Ripening and Germination. Front. Plant Sci. 2022, 13, 835835. [Google Scholar]
- Xiong, Y.F.; Ren, Y.; Li, W.; Wu, F.S.; Yang, W.J.; Huang, X.L.; Yao, J.L. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J. Exp. Bot. 2019, 70, 3765–3780. [Google Scholar] [CrossRef]
- Singh, K.K.; Ghosh, S. Regulation of glutamine synthetase isoforms in two differentially drought-tolerant rice (Oryza sativa L.) cultivars under water deficit conditions. Plant Cell Rep. 2013, 32, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Muhitch, M.J.; Liang, H.; Rastogi, R.; Sollenberger, K.G. Isolation of a promoter sequence from the glutamine synthetase(1-2) gene capable of conferring tissue-specific gene expression in transgenic maize. Plant Sci. 2002, 163, 865–872. [Google Scholar] [CrossRef]
- Muhitch, M.J. Distribution of the glutamine synthetase isozyme GS(p1) in maize (Zea mays). J. Plant Physiol. 2003, 160, 601–605. [Google Scholar] [CrossRef][Green Version]
- Li, M.G.; Villemur, R.; Hussey, P.J.; Silflow, C.D.; Gantt, J.S.; Snustad, D.P. differential expression of 6 glutamine-synthetase genes in Zea mays. Plant Mol. Biol. 1993, 23, 401–407. [Google Scholar] [CrossRef]
- Martin, A.; Lee, J.; Kichey, T.; Gerentes, D.; Zivy, M.; Tatout, C.; Dubois, F.; Balliau, T.; Valot, B.; Davanture, M.; et al. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 2006, 18, 3252–3274. [Google Scholar] [CrossRef]
- Hirel, B.; Andrieu, B.; Valadier, M.H.; Renard, S.; Quillere, I.; Chelle, M.; Pommel, B.; Fournier, C.; Drouet, J.L. Physiology of maize II: Identification of physiological markers representative of the nitrogen status of maize (Zea mays) leaves during grain filling. Physiol. Plant. 2005, 124, 178–188. [Google Scholar] [CrossRef]
- Goodall, A.J.; Kumar, P.; Tobin, A.K. Identification and expression analyses of cytosolic glutamine synthetase genes in barley (Hordeum vulgare L.). Plant Cell Physiol. 2013, 54, 492–505. [Google Scholar] [CrossRef]
- Wei, Y.; Xiong, S.; Zhang, Z.; Meng, X.; Wang, L.; Zhang, X.; Yu, M.; Yu, H.; Wang, X.; Ma, X. Localization, gene expression, and functions of glutamine synthetase isozymes in wheat grain (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 580405. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, X.; Zhang, Z.; Xiong, S.; Meng, X.; Zhang, J.; Wang, L.; Zhang, X.; Yu, M.; Ma, X. Nitrogen regulating the expression and localization of four glutamine synthetase isoforms in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2020, 21, 6299. [Google Scholar] [CrossRef]
- Li, X.G.; Liu, H.Y.; Jin, Z.X.; Liu, H.L.; Huang, X.; Xu, M.L.; Zhang, F.Z. Changes in activities of key enzymes for starch synthesis and glutamine synthetase in grains of progenies from a rice cross during grain filling. Rice Sci. 2010, 17, 243–246. [Google Scholar] [CrossRef]
- Habash, D.Z.; Massiah, A.J.; Rong, H.L.; Wallsgrove, R.M.; Leigh, R.A. The role of cytosolic glutamine synthetase in wheat. Ann. Appl. Biol. 2001, 138, 83–89. [Google Scholar] [CrossRef]
- Le, T.N.N.; Lee, B.; Back, K.; Kim, Y.S.; Cheong, H. Coordinated expression of cytosolic and chloroplastic glutamine synthetase during reproductive stage and its impact in GS1 RNAi transgenic rice. Rice Sci 2018, 25, 250–260. [Google Scholar] [CrossRef]
- Liu, X.L.; Tian, Y.L.; Chi, W.C.; Zhang, H.Z.; Yu, J.; Chen, G.M.; Wu, W.; Jiang, X.Z.; Wang, S.S.; Lin, Z.X.; et al. Alternative splicing of OsGS1;1 affects nitrogen-use efficiency, grain development, and amylose content in rice. Plant J. 2022, 110, 1751–1762. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.; Tabuchi, M.; Fukushima, A.; Funayama, K.; Diaz, C.; Kobayashi, M.; Hayashi, N.; Tsuchiya, Y.N.; Takahashi, H.; Kamata, A.; et al. Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice. Plant J. 2011, 66, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Q.; Shi, W.M. Expression analysis of the glutamine synthetase and glutamate synthase gene families in young rice (Oryza sativa) seedlings. Plant Sci. 2006, 170, 748–754. [Google Scholar] [CrossRef]
- Ishiyama, K.; Kojima, S.; Takahashi, H.; Hayakawa, T.; Yamaya, T. Cell type distinct accumulations of mRNA and protein for NADH-dependent glutamate synthase in rice roots in response to the supply of NH4+. Plant Physiol. Biochem. 2003, 41, 643–647. [Google Scholar] [CrossRef]
- Kojima, S.; Kimura, M.; Nozaki, Y.; Yamaya, T. Analysis of a promoter for the NADH-glutamate synthase gene in rice (Oryza sativa): Cell type-specific expression in developing organs of transgenic rice plants. Aust. J. Plant Physiol. 2000, 27, 787–793. [Google Scholar] [CrossRef]
- Tamura, W.; Hidaka, Y.; Tabuchi, M.; Kojima, S.; Hayakawa, T.; Sato, T.; Obara, M.; Kojima, M.; Sakakibara, H.; Yamaya, T. Reverse genetics approach to characterize a function of NADH-glutamate synthase1 in rice plants. Amino Acids 2010, 39, 1003–1012. [Google Scholar] [CrossRef]
- Tamura, W.; Kojima, S.; Toyokawa, A.; Watanabe, H.; Tabuchi-Kobayashi, M.; Hayakawa, T.; Yamaya, T. Disruption of a novel NADH-glutamate synthase2 gene caused marked reduction in spikelet number of rice. Front. Plant Sci. 2011, 2, 57. [Google Scholar] [CrossRef]
- Yamaya, T.; Kusano, M. Evidence supporting distinct functions of three cytosolic glutamine synthetases and two NADH-glutamate synthases in rice. J. Exp. Bot. 2014, 65, 5519–5525. [Google Scholar] [CrossRef] [PubMed]
- Imagawa, F.; Minagawa, H.; Nakayama, Y.; Kanno, K.; Hayakawa, T.; Kojima, S. Tos17 insertion in NADH-dependent glutamate synthase genes leads to an increase in grain protein content in rice. J. Cereal Sci. 2018, 84, 38–43. [Google Scholar] [CrossRef]
- Coschigano, K.T.; Melo-Oliveira, R.; Lim, J.; Coruzzi, G.M. Arabidopsis gls mutants and distinct Fd-GOGAT genes. Implications for photorespiration and primary nitrogen assimilation. Plant Cell 1998, 10, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Ishiyama, K.; Hayakawa, T.; Yamaya, T. Expression of NADH-dependent glutamate synthase protein in the epidermis and exodermis of rice roots in response to the supply of ammonium ions. Planta 1998, 204, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.D.; Qin, R.; Li, M.; Alamin, M.; Jin, X.L.; Liu, Y.; Shi, C.H. The ferredoxin-dependent glutamate synthase (OsFd-GOGAT) participates in leaf senescence and the nitrogen remobilization in rice. Mol. Genet. Genomics 2017, 292, 385–395. [Google Scholar] [CrossRef]
- Yang, X.L.; Nian, J.Q.; Xie, Q.J.; Feng, J.; Zhang, F.X.; Jing, H.W.; Zhang, J.; Dong, G.J.; Liang, Y.; Peng, J.L.; et al. Rice ferredoxin-dependent glutamate synthase regulates nitrogen-carbon metabolomes and is genetically differentiated between japonica and indica subspecies. Mol. Plant 2016, 9, 1520–1534. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.W.; Carrayol, E.; Hirel, B. Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: Localization, relative proportion and their role in ammonium assimilation or nitrogen transport. Planta 2000, 211, 800–806. [Google Scholar] [CrossRef]
- Sakakibara, H.; Kawabata, S.; Takahashi, H.; Hase, T.; Sugiyama, T. Molecular-cloning of the family of glutamine-synthetase genes from maize—Expression of genes for glutamine-synthetase and ferredoxin-dependent glutamate synthase in photosynthetic and nonphotosynthetic tissues. Plant Cell Physiol. 1992, 33, 49–58. [Google Scholar]
- Cañas, R.A.; Quillere, I.; Lea, P.J.; Hirel, B. Analysis of amino acid metabolism in the ear of maize mutants deficient in two cytosolic glutamine synthetase isoenzymes highlights the importance of asparagine for nitrogen translocation within sink organs. Plant Biotechnol. J. 2010, 8, 966–978. [Google Scholar] [CrossRef] [PubMed]
- Broyart, C.; Fontaine, J.X.; Molinie, R.; Cailleu, D.; Terce-Laforgue, T.; Dubois, F.; Hirel, B.; Mesnard, F. Metabolic profiling of maize mutants deficient for two glutamine synthetase isoenzymes using 1H-NMR-based metabolomics. Phytochem. Anal. 2010, 21, 102–109. [Google Scholar] [CrossRef]
- Avila-Ospina, L.; Marmagne, A.; Talbotec, J.; Krupinska, K.; Masclaux-Daubresse, C. The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence. J. Exp. Bot. 2015, 66, 2013–2026. [Google Scholar] [CrossRef]
- Shah, J.M.; Bukhari, S.A.H.; Zeng, J.B.; Quan, X.Y.; Ali, E.; Muhammad, N.; Zhang, G.P. Nitrogen (N) metabolism related enzyme activities, cell ultrastructure and nutrient contents as affected by N level and barley genotype. J. Integr. Agric. 2017, 16, 190–198. [Google Scholar] [CrossRef]
- Wang, X.C.; Wei, Y.H.; Shi, L.X.; Ma, X.M.; Theg, S.M. New isoforms and assembly of glutamine synthetase in the leaf of wheat (Triticum aestivum L.). J. Exp. Bot. 2015, 66, 6827–6834. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhao, X.; Liu, Q.; Hong, X.; Zhang, W.; Zhang, Y.; Sun, L.; Li, H.; Tong, Y. Transgenic expression of plastidic glutamine synthetase increases nitrogen uptake and yield in wheat. Plant Biotechnol. J. 2018, 16, 1858–1867. [Google Scholar] [CrossRef]
- Nemeth, E.; Nagy, Z.; Pecsvaradi, A. Chloroplast glutamine synthetase, the key regulator of nitrogen metabolism in wheat, performs its role by fine regulation of enzyme activity via negative cooperativity of its subunits. Front. Plant Sci. 2018, 9, 191. [Google Scholar] [CrossRef]
- Kichey, T.; Heumez, E.; Pocholle, D.; Pageau, K.; Vanacker, H.; Dubois, F.; Le Gouis, J.; Hirel, B. Combined agronomic and physiological aspects of nitrogen management in wheat highlight a central role for glutamine synthetase. New Phytol. 2006, 169, 265–278. [Google Scholar] [CrossRef]
- Kichey, T.; Hirel, B.; Heumez, E.; Dubois, F.; Le Gouis, J. In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crop. Res. 2007, 102, 22–32. [Google Scholar] [CrossRef]
- Habash, D.Z.; Bernard, S.; Schondelmaier, J.; Weyen, J.; Quarrie, S.A. The genetics of nitrogen use in hexaploid wheat: N utilisation, development and yield. TAG Theor. Appl. Genet. 2007, 114, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Nigro, D.; Fortunato, S.; Giove, S.L.; Paradiso, A.; Gu, Y.Q.; Blanco, A.; de Pinto, M.C.; Gadaleta, A. Glutamine synthetase in durum wheat: Genotypic variation and relationship with grain protein content. Front. Plant Sci. 2016, 7, 971. [Google Scholar] [CrossRef]
- Gaju, O.; Allard, V.; Martre, P.; Snape, J.W.; Heumez, E.; Le Gouis, J.; Moreau, D.; Bogard, M.; Griffiths, S.; Orford, S.; et al. Identification of traits to improve the nitrogen-use efficiency of wheat genotypes. Field Crops Res. 2011, 123, 139–152. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiong, S.; Wei, Y.; Meng, X.; Wang, X.; Ma, X. The role of glutamine synthetase isozymes in enhancing nitrogen use efficiency of N-efficient winter wheat. Sci. Rep. 2017, 7, 1000. [Google Scholar] [CrossRef]
- Nigro, D.; Blanco, A.; Anderson, O.D.; Gadaleta, A. Characterization of ferredoxin-dependent glutamine-oxoglutarate amidotransferase (Fd-GOGAT) genes and their relationship with grain protein content QTL in wheat. PLoS ONE 2014, 9, e103869. [Google Scholar] [CrossRef] [PubMed]
- Nigro, D.; Gu, Y.Q.; Huo, N.X.; Marcotuli, I.; Blanco, A.; Gadaleta, A.; Anderson, O.D. Structural analysis of the wheat genes encoding NADH-dependent glutamine-2-oxoglutarate amidotransferases and correlation with grain protein content. PLoS ONE 2013, 8, e73751. [Google Scholar] [CrossRef]
- Nigro, D.; Fortunato, S.; Giove, S.L.; Mangini, G.; Yacoubi, I.; Simeone, R.; Blanco, A.; Gadaleta, A. Allelic variants of glutamine synthetase and glutamate synthase genes in a collection of durum wheat and association with grain protein content. Diversity 2017, 9, 52. [Google Scholar] [CrossRef]
- Quraishi, U.M.; Abrouk, M.; Murat, F.; Pont, C.; Foucrier, S.; Desmaizieres, G.; Confolent, C.; Riviere, N.; Charmet, G.; Paux, E.; et al. Cross-genome map based dissection of a nitrogen use efficiency ortho-metaQTL in bread wheat unravels concerted cereal genome evolution. Plant J. 2011, 65, 745–756. [Google Scholar] [CrossRef]
- Obara, M.; Kashiba, K.; Nagano, A.; Tateshita, N.; Ebitani, T.; Yano, M.; Sato, T.; Yamaya, T. Linkage analysis for QTL associated with panicle number on chromosome 2 in rice (Oryza sativa L.); Toward identification of the regulatory gene. Plant Cell Physiol. 2004, 45, S126. [Google Scholar]
- Obara, M.; Kajiura, M.; Fukuta, Y.; Yano, M.; Hayashi, M.; Yamaya, T.; Sato, T. Mapping of QTLs associated with cytosolic glutamine synthetase and NADH-glutamate synthase in rice (Oryza sativa L.). J. Exp. Bot. 2001, 52, 1209–1217. [Google Scholar]
- Hirel, B.; Bertin, P.; Quillere, I.; Bourdoncle, W.; Attagnant, C.; Dellay, C.; Gouy, A.; Cadiou, S.; Retailliau, C.; Falque, M.; et al. Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 2001, 125, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Gallais, A.; Hirel, B. An approach to the genetics of nitrogen use efficiency in maize. J. Exp. Bot. 2004, 55, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Masclaux, C.; Quillere, I.; Gallais, A.; Hirel, B. The challenge of remobilisation in plant nitrogen economy. A survey of physio-agronomic and molecular approaches. Ann. Appl. Biol. 2001, 138, 69–81. [Google Scholar] [CrossRef]
- Fontaine, J.X.; Ravel, C.; Pageau, K.; Heumez, E.; Dubois, F.; Hirel, B.; Le Gouis, J. A quantitative genetic study for elucidating the contribution of glutamine synthetase, glutamate dehydrogenase and other nitrogen-related physiological traits to the agronomic performance of common wheat. Theor. Appl. Genet. 2009, 119, 645–662. [Google Scholar] [CrossRef] [PubMed]
- Bordes, J.; Ravel, C.; Jaubertie, J.P.; Duperrier, B.; Gardet, O.; Heumez, E.; Pissavy, A.L.; Charmet, G.; Le Gouis, J.; Balfourier, F. Genomic regions associated with the nitrogen limitation response revealed in a global wheat core collection. Theor. Appl. Genet. 2013, 126, 805–822. [Google Scholar] [CrossRef]
- Gadaleta, A.; Nigro, D.; Giancaspro, A.; Blanco, A. The glutamine synthetase (GS2) genes in relation to grain protein content of durum wheat. Funct. Integr. Genom. 2011, 11, 665–670. [Google Scholar] [CrossRef]
- Nigro, D.; Gadaleta, A.; Mangini, G.; Colasuonno, P.; Marcotuli, I.; Giancaspro, A.; Giove, S.L.; Simeone, R.; Blanco, A. Candidate genes and genome-wide association study of grain protein content and protein deviation in durum wheat. Planta 2019, 249, 1157–1175. [Google Scholar] [CrossRef]
- Nigro, D.; Fortunato, S.; Giove, S.L.; Mazzucotelli, E.; Gadaleta, A. Functional validation of glutamine synthetase and glutamate synthase genes in durum wheat near isogenic lines with QTL for high GPC. Int. J. Mol. Sci. 2020, 21, 9253. [Google Scholar] [CrossRef]
- Gadaleta, A.; Nigro, D.; Marcotuli, I.; Giancaspro, A.; Giove, S.L.; Blanco, A. Isolation and characterisation of cytosolic glutamine synthetase (GSe) genes and association with grain protein content in durum wheat. Crop Pasture Sci. 2014, 65, 38–45. [Google Scholar] [CrossRef]
- Guo, Y.; Sun, J.J.; Zhang, G.Z.; Wang, Y.Y.; Kong, F.M.; Zhao, Y.; Li, S.S. Haplotype, molecular marker and phenotype effects associated with mineral nutrient and grain size traits of TaGS1 a in wheat. Field Crops Res. 2013, 154, 119–125. [Google Scholar] [CrossRef]
- Laperche, A.; Brancourt-Hulmel, M.; Heumez, E.; Gardet, O.; Hanocq, E.; Devienne-Barret, F.; Le Gouis, J. Using genotype x nitrogen interaction variables to evaluate the QTL involved in wheat tolerance to nitrogen constraints. Theor. Appl. Genet. 2007, 115, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Li, P.C.; Chen, F.J.; Cai, H.G.; Liu, J.C.; Pan, Q.C.; Liu, Z.G.; Gu, R.L.; Mi, G.H.; Zhang, F.S.; Yuan, L.X. A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J. Exp. Bot. 2015, 66, 3175–3188. [Google Scholar] [CrossRef] [PubMed]
- Cormier, F.; Le Gouis, J.; Dubreuil, P.; Lafarge, S.; Praud, S. A genome-wide identification of chromosomal regions determining nitrogen use efficiency components in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2014, 127, 2679–2693. [Google Scholar] [CrossRef]
- Li, X.P.; Zhao, X.Q.; He, X.; Zhao, G.Y.; Li, B.; Liu, D.C.; Zhang, A.M.; Zhang, X.Y.; Tong, Y.P.; Li, Z.S. Haplotype analysis of the genes encoding glutamine synthetase plastic isoforms and their association with nitrogen-use- and yield-related traits in bread wheat. New Phytol. 2011, 189, 449–458. [Google Scholar] [CrossRef] [PubMed]
- He, C.M.; Liu, C.X.; Liu, Q.; Gao, X.X.; Li, N.; Zhang, J.R.; Wang, L.M.; Liu, T.S. Over-expression of glutamine synthetase genes Gln1-3/Gln1-4 improved nitrogen assimilation and maize yields. Maydica 2014, 59, 250–256. [Google Scholar]
- Amiour, N.; Decousset, L.; Rouster, J.; Quenard, N.; Buet, C.; Dubreuil, P.; Quillere, I.; Brule, L.; Cukier, C.; Dinant, S.; et al. Impacts of environmental conditions, and allelic variation of cytosolic glutamine synthetase on maize hybrid kernel production. Commun. Biol. 2021, 4, 1095. [Google Scholar] [CrossRef]
- Cai, H.M.; Zhou, Y.; Xiao, J.H.; Li, X.H.; Zhang, Q.F.; Lian, X.M. Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep. 2009, 28, 527–537. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Fernie, A.R.; Stitt, M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 2010, 3, 973–996. [Google Scholar] [CrossRef]
- Bao, A.L.; Zhao, Z.Q.; Ding, G.D.; Shi, L.; Xu, F.S.; Cai, H.M. Accumulated expression level of cytosolic glutamine synthetase 1 gene (OsGS1; 1 or OsGS1; 2) alter plant development and the carbon-nitrogen metabolic status in rice. PLoS ONE 2014, 9, e95581. [Google Scholar] [CrossRef]
- Chen, T.; Yang, S.; Wei, T.; Li, Y.; Wang, S.; Su, Y. Overexpression of OsGS1;2 for improved nitrogen use efficiency and grain yield of rice: A field test. Field Crops Res. 2023, 303, 109146. [Google Scholar]
- Wu, D.; Li, Y.; Cao, Y.; Hu, R.; Wu, X.; Zhang, W.; Tao, W.; Xu, G.; Wang, X.; Zhang, Y. Increased glutamine synthetase by overexpression of TaGS1 improves grain yield and nitrogen use efficiency in rice. Plant Physiol. Biochem. 2021, 169, 259–268. [Google Scholar] [CrossRef]
- Sun, H.; Huang, Q.-M.; Su, J. Highly effective expression of glutamine synthetase genes GS1 and GS2 in transgenic rice plants increases nitrogen-deficiency tolerance. J. Plant Physiol. Mol. Biol. 2005, 31, 492–498. [Google Scholar]
- James, D.; Borphukan, B.; Fartyal, D.; Ram, B.; Singho, J.; Manna, M.; Sheri, V.; Panditi, V.; Yadav, R.; Achary, V.M. Met al. Concurrent overexpression of OsGS1;1 and OsGS2 genes in transgenic rice (Oryza sativa L.): Impact on tolerance to abiotic stresses. Front. Plant Sci. 2018, 9, 786. [Google Scholar] [CrossRef] [PubMed]
- Lal, S.K.; Mehta, S.; Raju, D.; Achary, V.M.M.; Venkatapuram, A.K.; Yadav, S.K.; Parmar, H.; Pandey, R.; Panditi, V.; Sheri, V.; et al. Concurrent overexpression of rice GS1;1 and GS2 genes to enhance the nitrogen use efficiency (NUE) in transgenic rice. J. Plant Growth Regul. 2023, 42, 6699–6720. [Google Scholar] [CrossRef]
- Urriola, J.; Rathore, K.S. Overexpression of a glutamine synthetase gene affects growth and development in sorghum. Transgenic Res. 2015, 24, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; de Bang, T.C.; Schjoerring, J.K. Cisgenic overexpression of cytosolic glutamine synthetase improves nitrogen utilization efficiency in barley and prevents grain protein decline under elevated CO(2). Plant Biotechnol. J. 2019, 17, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, regulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef] [PubMed]
- Yamaya, T.; Obara, M.; Nakajima, H.; Sasaki, S.; Hayakawa, T.; Sato, T. Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. J. Exp. Bot. 2002, 53, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Cañas, R.A.; Yesbergenova-Cuny, Z.; Belanger, L.; Rouster, J.; Brule, L.; Gilard, F.; Quillere, I.; Sallaud, C.; Hirel, B. NADH-GOGAT overexpression does not improve maize (Zea mays L.) performance even when pyramiding with NAD-IDH, GDH and GS. Plants 2020, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Marmagne, A.; Park, J.; Fabien, C.; Yim, Y.; Kim, S.J.; Kim, T.H.; Lim, P.O.; Masclaux-Daubresse, C.; Nam, H.G. Concurrent activation of OsAMT1;2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J. 2020, 103, 7–20. [Google Scholar] [CrossRef]
- Yang, J.B.; Wang, M.Y.; Li, W.J.; He, X.; Teng, W.; Ma, W.Y.; Zhao, X.Q.; Hu, M.Y.; Li, H.; Zhang, Y.J.; et al. Reducing expression of a nitrate-responsive bZIP transcription factor increases grain yield and N use in wheat. Plant Biotechnol. J. 2019, 17, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Nian, J.Q.; Xie, X.Z.; Yu, H.; Zhang, J.; Bai, J.T.; Dong, G.J.; Hu, J.; Bai, B.; Chen, L.C.; et al. Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice. Nat. Commun. 2018, 9, 735. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Zhang, H.T.; Li, S.Y.; Li, J.Y.; Yan, L.; Xia, L.Q. Increasing yield potential through manipulating of an ARE1 ortholog related to nitrogen use efficiency in wheat by CRISPR/Cas9. J. Integr. Plant Biol. 2021, 63, 1649–1663. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Y.; Liu, L.L.; Wang, C.; Gan, T.; Zhang, Z.; Wang, D.; Niu, M.; Long, W.; Li, X.; et al. Isolation and characterization of a spotted leaf 32 mutant with early leaf senescence and enhanced defense response in rice. Sci. Rep. 2017, 7, 41846. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Yang, F.; He, X.; Du, X.; Mu, P.; Ma, W. Advances in the functional study of glutamine synthetase in plant abiotic stress tolerance response. Crop J. 2022, 10, 917–923. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Y.; Li, K.; Yan, M.; Zhang, J.; Yu, M.; Tang, S.; Wang, L.; Qu, H.; Luo, L.; et al. Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice. Curr. Biol. 2021, 31, 671–683.e5. [Google Scholar] [CrossRef]
- Gutierrez, R.A.; Stokes, T.L.; Thum, K.; Xu, X.; Obertello, M.; Katari, M.S.; Tanurdzic, M.; Dean, A.; Nero, D.C.; McClung, C.R.; et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl. Acad. Sci. USA 2008, 105, 4939–4944. [Google Scholar] [CrossRef] [PubMed]
- Baslam, M.; Mitsui, T.; Sueyoshi, K.; Ohyama, T. Recent advances in carbon and nitrogen metabolism in C3 plants. Int. J. Mol. Sci. 2020, 22, 318. [Google Scholar] [CrossRef] [PubMed]
| Species | Genes | Principal Localization of Gene Products | Function | References |
|---|---|---|---|---|
| Oryza sativa | OsGS1;1 | vascular tissue of mature leaves | grain filling | [75] |
| OsGS1;2 | leaves, stems, and roots | primary NH4+ assimilation | [78,79,82] | |
| OsGS1;3 | spikelet | grain ripening and germination | [46,75] | |
| Zea mays | ZmGln1;2 | pedicel and pericarp | development of kernel | [83,84] |
| ZmGln1;3 | leaves, roots, and stems | development of cobs in relation to kernel number | [85] | |
| ZmGln1;4 | leaves, roots, stems, and older leaves; bundle sheath cells | NH4+ reassimilation during protein degradation in senescing leaves; development of cobs in relation to kernel size | [86,87] | |
| ZmGln1;5 | cortical tissue of seedling roots, vasculature of roots; seedling shoots and in stems | not reported | [85] | |
| Hordeum vulgare | HvGS1;1 | vascular tissues | N transport and remobilization | [88] |
| HvGS1;2 | mesophyll cells of leaves; cortex and pericycle of roots | primary N assimilation | [88] | |
| HvGS1;3 | specifically expressed in grains; expressed in roots under high NH4+ fertilization | defense against NH4+ toxicity | [88] | |
| Triticum aestivum | TaGS1;1 | perifascicular sheath; mesophyll cells; chalaza and placentochalaza | cytoplasmic NH4+ assimilation | [40,89] |
| TaGS1;2/GSr | vascular cells of leaves and roots; vascular bundle; chalaza and placentochalaza | N transport | [49,89] | |
| TaGS1;3/GSe | mesophyll cells; endosperm transfer cells; aleurone layer | cytoplasmic NH4+ assimilation; alleviating NH4+ toxicity; gluten synthesis | [89,90] |
| Species | Gene | Chromosome Localization | Colocalizing QTLs | References |
|---|---|---|---|---|
| Oryza sativa | GS1 | 2 | Soluble protein content | [126,127] |
| 2 | SPN, PNW | |||
| 11 | PNW, SPW, and RFD | |||
| NADH-GOGAT | 1 | Soluble protein content | [126,127] | |
| 2 | SPN, PNW | |||
| 2 | Soluble protein content, SPN, RFD, and RHD | |||
| Zea mays | Gln2 (cytosolic) | 1 | TKW, KN | [128,129] |
| Gln4 (cytosolic) | 5 | TKW, KN | ||
| Gln1-3 | 5 | GY, TKW, leaf GS activity, NR activity, and leaf nitrate content | [130] | |
| Gln1-3 | 5 | kernel yield and GS activity | [129] | |
| Triticum aestivum | GS2 | 2 | GS activity, soluble protein content/leaf | [118] |
| GS1 | 6 | TGW, grain N | ||
| GSr | 4 | GS activity, grain %N | ||
| GSe | 4 | GS activity | [40] | |
| GSe | 4 | GS activity | [131] | |
| Fd-GOGAT | 2 | GY, GN, and GPC | [132] | |
| NADH-GOGAT | 3 | NUE | [125] | |
| Triticum turgidum | GS2 | 2 | GPC | [124,133,134,135] |
| GPD | [134] | |||
| Gse | 4 | GPC | [133,136] | |
| GPD | [134] | |||
| GS1 | 6 | GPC | ||
| GSr | 4 | GPC | ||
| Fd-GOGAT | 2 | GPC | ||
| NADH-GOGAT | 3 | GPD | [134,135] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fortunato, S.; Nigro, D.; Lasorella, C.; Marcotuli, I.; Gadaleta, A.; de Pinto, M.C. The Role of Glutamine Synthetase (GS) and Glutamate Synthase (GOGAT) in the Improvement of Nitrogen Use Efficiency in Cereals. Biomolecules 2023, 13, 1771. https://doi.org/10.3390/biom13121771
Fortunato S, Nigro D, Lasorella C, Marcotuli I, Gadaleta A, de Pinto MC. The Role of Glutamine Synthetase (GS) and Glutamate Synthase (GOGAT) in the Improvement of Nitrogen Use Efficiency in Cereals. Biomolecules. 2023; 13(12):1771. https://doi.org/10.3390/biom13121771
Chicago/Turabian StyleFortunato, Stefania, Domenica Nigro, Cecilia Lasorella, Ilaria Marcotuli, Agata Gadaleta, and Maria Concetta de Pinto. 2023. "The Role of Glutamine Synthetase (GS) and Glutamate Synthase (GOGAT) in the Improvement of Nitrogen Use Efficiency in Cereals" Biomolecules 13, no. 12: 1771. https://doi.org/10.3390/biom13121771
APA StyleFortunato, S., Nigro, D., Lasorella, C., Marcotuli, I., Gadaleta, A., & de Pinto, M. C. (2023). The Role of Glutamine Synthetase (GS) and Glutamate Synthase (GOGAT) in the Improvement of Nitrogen Use Efficiency in Cereals. Biomolecules, 13(12), 1771. https://doi.org/10.3390/biom13121771








