Recent Advances in Bioimage Analysis Methods for Detecting Skeletal Deformities in Biomedical and Aquaculture Fish Species
Abstract
:1. Introduction
2. Imaging Techniques Used in Fish Bioimages
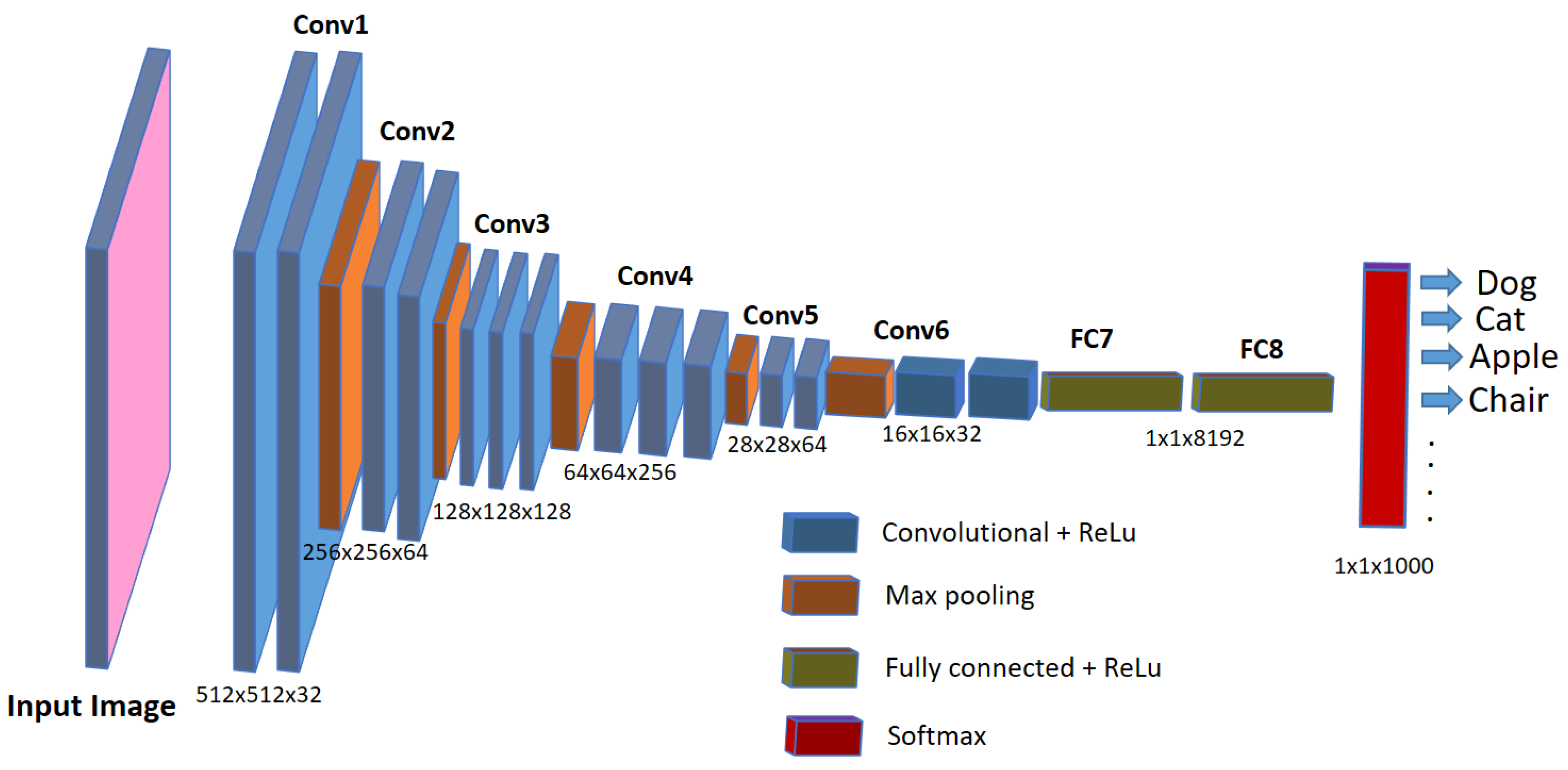
3. Bioimage Analysis in Biomedical Research
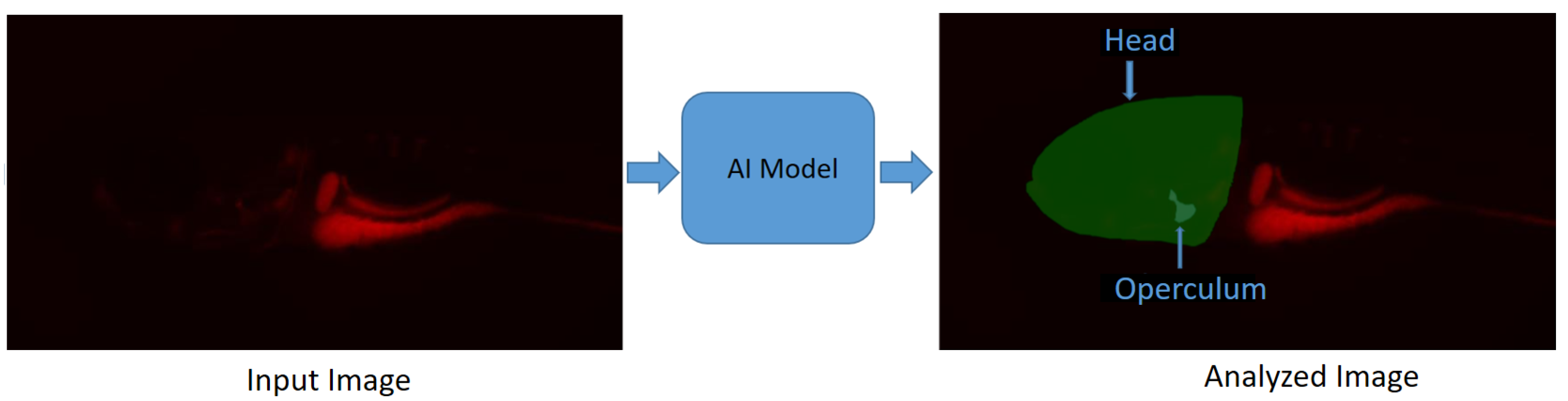
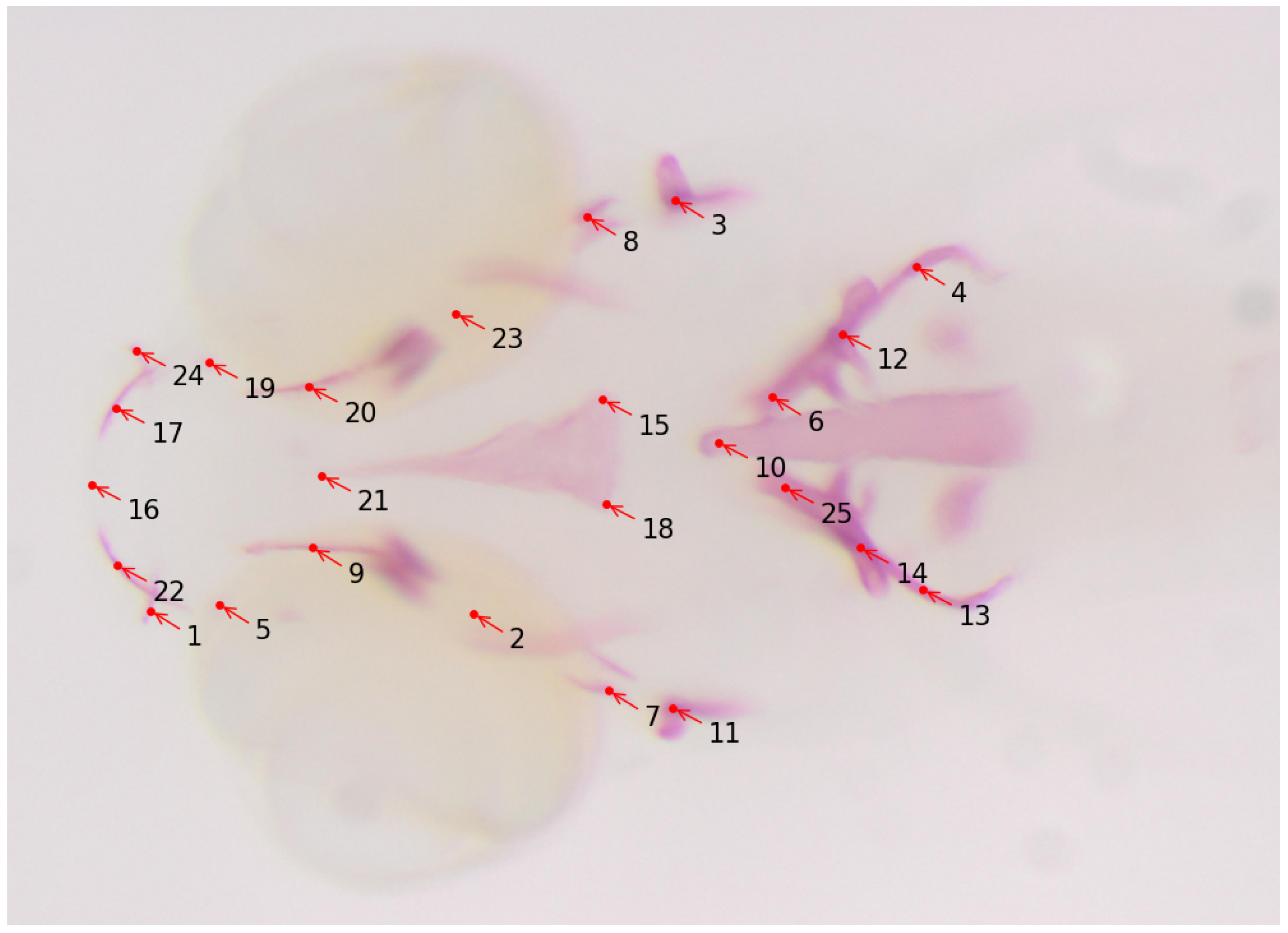
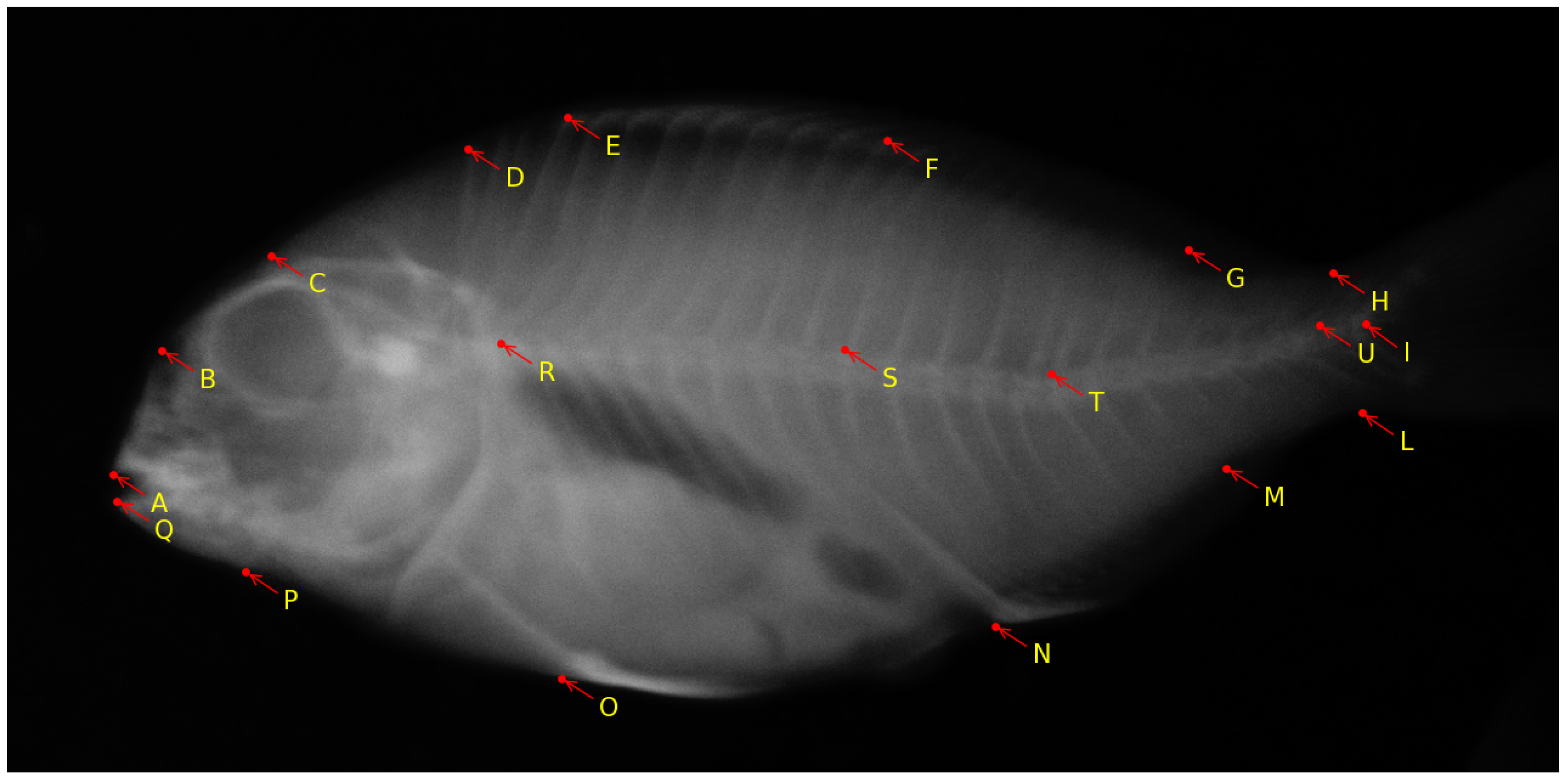
4. Bioimage Analysis in Aquaculture
5. Challenges in Bioimage Analysis Tasks
5.1. High-Content, High-Throughput Imaging
5.2. Choice of Image Analysis Methods/Protocols
5.3. Lack of Annotated Bioimages
5.4. Miscellaneous
6. Conclusions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Name | Ref. Paper | Image Modality | Fish Type | Application | Open Source? | Data Availability | Research Area |
|---|---|---|---|---|---|---|---|
| EmbryoNet | [47] D. Capek et al. | Light microscopy | Zebrafish embryos (1–26 hpf ), Medaka embryos (0–48 hpf), Stickleback larvae (0–140 hpf) | Phenotype classification | Yes | Yes | Biomedical |
| ZF-AutoML | [90] R. Sawaki et al. | Light microscopy | Zebrafish larvae (0–96 hpf) | Phenotype classification | Yes | No | Biomedical |
| ZFTool | [91] M. J. Carreira et al. | Fluorescent Green Channel Microscopy | Zebrafish larvae (0–72 hpf) | Toxicity screening | Yes | Yes | Biomedical |
| FishInspector | [50] E. Teixido et al. | Light microscopy | Zebrafish embryos (0–96 hpf) | Phenotype screening | Yes | Yes | Biomedical |
| ZebrafishMiner | [92] M. Reischl et al. | Fluorescent microscopy | Zebrafish embryos and larvae (32 hpf) | Fluorescent quantification in body parts | No | No | Biomedical |
| ZFBone | [54] M. Tarasco et al. | Fluorescent microscopy | Zebrafish larvae (6 dpf ) | Morphometric analysis | Yes | Yes | Biomedical |
| IMAFISH_ML | [93] A. Navarro et al. | Microscopy, RGB, radiography | Adult gilthead seabream, meagre, red porgy | Morphometric analysis | Yes | No | Aquaculture |
| Cytomine | [58] R. Marée et al. | All types | All types | General image analysis | Yes | Yes | Biomedical, Aquaculture |
References
- Selderslaghs, I.W.; Hooyberghs, J.; Blust, R.; Witters, H.E. Assessment of the developmental neurotoxicity of compounds by measuring locomotor activity in zebrafish embryos and larvae. Neurotoxicology Teratol. 2013, 37, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Rihel, J.; Prober, D.A.; Arvanites, A.; Lam, K.; Zimmerman, S.; Jang, S.; Haggarty, S.J.; Kokel, D.; Rubin, L.L.; Peterson, R.T.; et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 2010, 327, 348–351. [Google Scholar] [CrossRef] [PubMed]
- Bugel, S.M.; Tanguay, R.L. Multidimensional chemobehavior analysis of flavonoids and neuroactive compounds in zebrafish. Toxicol. Appl. Pharmacol. 2018, 344, 23–34. [Google Scholar] [CrossRef]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721–731. [Google Scholar] [CrossRef]
- Love, D.R.; Pichler, F.B.; Dodd, A.; Copp, B.R.; Greenwood, D.R. Technology for high-throughput screens: The present and future using zebrafish. Curr. Opin. Biotechnol. 2004, 15, 564–571. [Google Scholar] [CrossRef]
- Scholz, S.; Fischer, S.; Gündel, U.; Küster, E.; Luckenbach, T.; Voelker, D. The zebrafish embryo model in environmental risk assessment—Applications beyond acute toxicity testing. Environ. Sci. Pollut. Res. 2008, 15, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.T.; MacRae, C.A. Systematic approaches to toxicology in the zebrafish. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 433–453. [Google Scholar] [CrossRef]
- Braunbeck, T.; Kais, B.; Lammer, E.; Otte, J.; Schneider, K.; Stengel, D.; Strecker, R. The fish embryo test (FET): Origin, applications, and future. Environ. Sci. Pollut. Res. 2015, 22, 16247–16261. [Google Scholar] [CrossRef]
- Hill, A.J.; Teraoka, H.; Heideman, W.; Peterson, R.E. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol. Sci. 2005, 86, 6–19. [Google Scholar] [CrossRef]
- Ali, S.; Champagne, D.L.; Spaink, H.P.; Richardson, M.K. Zebrafish embryos and larvae: A new generation of disease models and drug screens. Birth Defects Res. Part Embryo Today Rev. 2011, 93, 115–133. [Google Scholar] [CrossRef]
- Strähle, U.; Scholz, S.; Geisler, R.; Greiner, P.; Hollert, H.; Rastegar, S.; Schumacher, A.; Selderslaghs, I.; Weiss, C.; Witters, H.; et al. Zebrafish embryos as an alternative to animal experiments—A commentary on the definition of the onset of protected life stages in animal welfare regulations. Reprod. Toxicol. 2012, 33, 128–132. [Google Scholar] [CrossRef]
- Pruvot, B.; Curé, Y.; Djiotsa, J.; Voncken, A.; Muller, M. Developmental defects in zebrafish for classification of EGF pathway inhibitors. Toxicol. Appl. Pharmacol. 2014, 274, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Spoorendonk, K.M.; Hammond, C.L.; Huitema, L.F.; Vanoevelen, J.; Schulte-Merker, S. Zebrafish as a unique model system in bone research: The power of genetics and in vivo imaging. J. Appl. Ichthyol. 2010, 26, 219–224. [Google Scholar] [CrossRef]
- Zon, L.I.; Peterson, R.T. In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 2005, 4, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Driever, W.; Solnica-Krezel, L.; Schier, A.; Neuhauss, S.; Malicki, J.; Stemple, D.; Stainier, D.; Zwartkruis, F.; Abdelilah, S.; Rangini, Z.; et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 1996, 123, 37–46. [Google Scholar] [CrossRef]
- Martos-Sitcha, J.A.; Mancera, J.M.; Prunet, P.; Magnoni, L.J. Welfare and stressors in fish: Challenges facing aquaculture. Front. Physiol. 2020, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Llorente, I.; Fernández-Polanco, J.; Baraibar-Diez, E.; Odriozola, M.D.; Bjørndal, T.; Asche, F.; Guillen, J.; Avdelas, L.; Nielsen, R.; Cozzolino, M.; et al. Assessment of the economic performance of the seabream and seabass aquaculture industry in the European Union. Mar. Policy 2020, 117, 103876. [Google Scholar] [CrossRef]
- Verhaegen, Y.; Adriaens, D.; De Wolf, T.; Dhert, P.; Sorgeloos, P. Deformities in larval gilthead sea bream (Sparus aurata): A qualitative and quantitative analysis using geometric morphometrics. Aquaculture 2007, 268, 156–168. [Google Scholar] [CrossRef]
- Hu, W.C.; Chen, L.B.; Huang, B.K.; Lin, H.M. A computer vision-based intelligent fish feeding system using deep learning techniques for aquaculture. IEEE Sens. J. 2022, 22, 7185–7194. [Google Scholar] [CrossRef]
- Santosh, K.; Das, N.; Ghosh, S. Deep Learning Models for Medical Imaging; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Mostafa, S.; Wu, F.X. Diagnosis of autism spectrum disorder with convolutional autoencoder and structural MRI images. In Neural Engineering Techniques for Autism Spectrum Disorder; Elsevier: Amsterdam, The Netherlands, 2021; pp. 23–38. [Google Scholar]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, Y.; Oerlemans, A.; Lao, S.; Wu, S.; Lew, M.S. Deep learning for visual understanding: A review. Neurocomputing 2016, 187, 27–48. [Google Scholar] [CrossRef]
- Fan, Y.L.; Hsu, F.R.; Wang, Y.; Liao, L.D. Unlocking the potential of zebrafish research with artificial intelligence: Advancements in tracking, processing, and visualization. Med. Biol. Eng. Comput. 2023, 61, 2797–2814. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Aponte-Rivera, R.; Barghout, R.M.; Trapani, J.G.; Kindt, K.S. In Vivo Analysis of Hair Cell Sensory Organs in Zebrafish: From Morphology to Function. In Developmental, Physiological, and Functional Neurobiology of the Inner Ear; Humana: New York, NY, USA, 2022; pp. 175–220. [Google Scholar]
- Bauer, B.; Mally, A.; Liedtke, D. Zebrafish embryos and larvae as alternative animal models for toxicity testing. Int. J. Mol. Sci. 2021, 22, 13417. [Google Scholar] [CrossRef] [PubMed]
- Mikut, R.; Dickmeis, T.; Driever, W.; Geurts, P.; Hamprecht, F.A.; Kausler, B.X.; Ledesma-Carbayo, M.J.; Marée, R.; Mikula, K.; Pantazis, P.; et al. Automated processing of zebrafish imaging data: A survey. Zebrafish 2013, 10, 401–421. [Google Scholar] [CrossRef] [PubMed]
- Shamir, L.; Delaney, J.D.; Orlov, N.; Eckley, D.M.; Goldberg, I.G. Pattern recognition software and techniques for biological image analysis. PLoS Comput. Biol. 2010, 6, e1000974. [Google Scholar] [CrossRef] [PubMed]
- Jeanray, N.; Marée, R.; Pruvot, B.; Stern, O.; Geurts, P.; Wehenkel, L.; Muller, M. Phenotype classification of zebrafish embryos by supervised learning. PLoS ONE 2015, 10, e0116989. [Google Scholar] [CrossRef]
- Kuchmiy, A.; Efimov, G.; Nedospasov, S. Methods for in vivo molecular imaging. Biochemistry 2012, 77, 1339–1353. [Google Scholar] [CrossRef]
- Abu-Siniyeh, A.; Al-Zyoud, W. Highlights on selected microscopy techniques to study zebrafish developmental biology. Lab. Anim. Res. 2020, 36, 12. [Google Scholar] [CrossRef]
- Bruneel, B.; Witten, P.E. Power and challenges of using zebrafish as a model for skeletal tissue imaging. Connect. Tissue Res. 2015, 56, 161–173. [Google Scholar] [CrossRef]
- Høgset, H.; Horgan, C.C.; Armstrong, J.P.; Bergholt, M.S.; Torraca, V.; Chen, Q.; Keane, T.J.; Bugeon, L.; Dallman, M.J.; Mostowy, S.; et al. In vivo biomolecular imaging of zebrafish embryos using confocal Raman spectroscopy. Nat. Commun. 2020, 11, 6172. [Google Scholar] [CrossRef]
- Bennet, M.; Akiva, A.; Faivre, D.; Malkinson, G.; Yaniv, K.; Abdelilah-Seyfried, S.; Fratzl, P.; Masic, A. Simultaneous Raman microspectroscopy and fluorescence imaging of bone mineralization in living zebrafish larvae. Biophys. J. 2014, 106, L17–L19. [Google Scholar] [CrossRef]
- Fiedler, I.A.; Schmidt, F.N.; Wölfel, E.M.; Plumeyer, C.; Milovanovic, P.; Gioia, R.; Tonelli, F.; Bale, H.A.; Jähn, K.; Besio, R.; et al. Severely impaired bone material quality in chihuahua zebrafish resembles classical dominant human osteogenesis imperfecta. J. Bone Miner. Res. 2018, 33, 1489–1499. [Google Scholar] [CrossRef]
- da Silva, K.M.; Iturrospe, E.; Bars, C.; Knapen, D.; Van Cruchten, S.; Covaci, A.; van Nuijs, A.L. Mass spectrometry-based zebrafish toxicometabolomics: A review of analytical and data quality challenges. Metabolites 2021, 11, 635. [Google Scholar] [CrossRef]
- Ding, Y.; Vanselow, D.J.; Yakovlev, M.A.; Katz, S.R.; Lin, A.Y.; Clark, D.P.; Vargas, P.; Xin, X.; Copper, J.E.; Canfield, V.A.; et al. Computational 3D histological phenotyping of whole zebrafish by X-ray histotomography. Elife 2019, 8, e44898. [Google Scholar] [CrossRef] [PubMed]
- Merrifield, G.D.; Mullin, J.; Gallagher, L.; Tucker, C.; Jansen, M.A.; Denvir, M.; Holmes, W.M. Rapid and recoverable in vivo magnetic resonance imaging of the adult zebrafish at 7T. Magn. Reson. Imaging 2017, 37, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Babaei, F.; Hong, T.L.C.; Yeung, K.; Cheng, S.H.; Lam, Y.W. Contrast-enhanced X-ray micro-computed tomography as a versatile method for anatomical studies of adult zebrafish. Zebrafish 2016, 13, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Carnovali, M.; Banfi, G.; Mariotti, M. Zebrafish models of human skeletal disorders: Embryo and adult swimming together. BioMed Res. Int. 2019, 2019, 1253710. [Google Scholar] [CrossRef] [PubMed]
- Dellacqua, Z.; Di Biagio, C.; Costa, C.; Pousão-Ferreira, P.; Ribeiro, L.; Barata, M.; Gavaia, P.J.; Mattei, F.; Fabris, A.; Izquierdo, M.; et al. Distinguishing the Effects of Water Volumes versus Stocking Densities on the Skeletal Quality during the Pre-Ongrowing Phase of Gilthead Seabream (Sparus aurata). Animals 2023, 13, 557. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, M.C.; Gilliam, J.F.; Langerhans, R.B. X-ray imaging as a time-saving, non-invasive technique for diet analysis. Fish. Res. 2015, 161, 1–7. [Google Scholar] [CrossRef]
- Suganyadevi, S.; Seethalakshmi, V.; Balasamy, K. A review on deep learning in medical image analysis. Int. J. Multimed. Inf. Retr. 2022, 11, 19–38. [Google Scholar] [CrossRef]
- Liu, X.; Gao, K.; Liu, B.; Pan, C.; Liang, K.; Yan, L.; Ma, J.; He, F.; Zhang, S.; Pan, S.; et al. Advances in deep learning-based medical image analysis. Health Data Sci. 2021, 2021, 8786793. [Google Scholar] [CrossRef]
- Dürr, O.; Sick, B. Single-cell phenotype classification using deep convolutional neural networks. J. Biomol. Screen. 2016, 21, 998–1003. [Google Scholar] [CrossRef]
- Dong, B.; Shao, L.; Da Costa, M.; Bandmann, O.; Frangi, A.F. Deep learning for automatic cell detection in wide-field microscopy zebrafish images. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; pp. 772–776. [Google Scholar]
- Čapek, D.; Safroshkin, M.; Morales-Navarrete, H.; Toulany, N.; Arutyunov, G.; Kurzbach, A.; Bihler, J.; Hagauer, J.; Kick, S.; Jones, F.; et al. EmbryoNet: Using deep learning to link embryonic phenotypes to signaling pathways. Nat. Methods 2023, 20, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Ishaq, O.; Sadanandan, S.K.; Wählby, C. Deep fish: Deep learning–based classification of zebrafish deformation for high-throughput screening. SLAS Discov. Adv. Life Sci. R&D 2017, 22, 102–107. [Google Scholar]
- Allen, M.R.; Krohn, K. Skeletal imaging. In Basic and Applied Bone Biology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 93–113. [Google Scholar]
- Teixidó, E.; Kießling, T.R.; Krupp, E.; Quevedo, C.; Muriana, A.; Scholz, S. Automated morphological feature assessment for zebrafish embryo developmental toxicity screens. Toxicol. Sci. 2019, 167, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Genest, D.; Puybareau, É.; Léonard, M.; Cousty, J.; De Crozé, N.; Talbot, H. High throughput automated detection of axial malformations in Medaka embryo. Comput. Biol. Med. 2019, 105, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Rosa, J.T.; Tarasco, M.; Gavaia, P.J.; Cancela, M.L.; Laizé, V. Screening of Mineralogenic and Osteogenic Compounds in Zebrafish—Tools to Improve Assay Throughput and Data Accuracy. Pharmaceuticals 2022, 15, 983. [Google Scholar] [CrossRef] [PubMed]
- Tarasco, M.; Laizé, V.; Cardeira, J.; Cancela, M.L.; Gavaia, P.J. The zebrafish operculum: A powerful system to assess osteogenic bioactivities of molecules with pharmacological and toxicological relevance. Comp. Biochem. Physiol. Part Toxicol. Pharmacol. 2017, 197, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Tarasco, M.; Cordelieres, F.P.; Cancela, M.L.; Laizé, V. ZFBONE: An ImageJ toolset for semi-automatic analysis of zebrafish bone structures. Bone 2020, 138, 115480. [Google Scholar] [CrossRef] [PubMed]
- Westhoff, J.H.; Steenbergen, P.J.; Thomas, L.S.; Heigwer, J.; Bruckner, T.; Cooper, L.; Tönshoff, B.; Hoffmann, G.F.; Gehrig, J. In vivo high-content screening in zebrafish for developmental nephrotoxicity of approved drugs. Front. Cell Dev. Biol. 2020, 8, 583. [Google Scholar] [CrossRef]
- Di Biagio, C.; Dellacqua, Z.; Martini, A.; Huysseune, A.; Scardi, M.; Witten, P.E.; Boglione, C. A baseline for skeletal investigations in Medaka (Oryzias latipes): The effects of rearing density on the postcranial phenotype. Front. Endocrinol. 2022, 13, 893699. [Google Scholar] [CrossRef]
- Kumar, N.; Carletti, A.; Gavaia, P.J.; Muller, M.; Cancela, M.L.; Geurts, P.; Marée, R. Deep Learning Approaches for Head and Operculum Segmentation in Zebrafish Microscopy Images. In Proceedings of the Computer Analysis of Images and Patterns: 19th International Conference, CAIP 2021, Virtual Event, 28–30 September 2021; pp. 154–164. [Google Scholar]
- Marée, R.; Rollus, L.; Stévens, B.; Hoyoux, R.; Louppe, G.; Vandaele, R.; Begon, J.M.; Kainz, P.; Geurts, P.; Wehenkel, L. Collaborative analysis of multi-gigapixel imaging data using Cytomine. Bioinformatics 2016, 32, 1395–1401. [Google Scholar] [CrossRef]
- Kumar, N.; Biagio, C.D.; Dellacqua, Z.; Raman, R.; Martini, A.; Boglione, C.; Muller, M.; Geurts, P.; Marée, R. Empirical Evaluation of Deep Learning Approaches for Landmark Detection in Fish Bioimages. In Proceedings of the European Conference on Computer Vision; Springer: Cham, Switzerland, 2022; pp. 470–486. [Google Scholar]
- Bergen, D.J.; Kague, E.; Hammond, C.L. Zebrafish as an emerging model for osteoporosis: A primary testing platform for screening new osteo-active compounds. Front. Endocrinol. 2019, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Caetano da Silva, C.; Ostertag, A.; Raman, R.; Muller, M.; Cohen-Solal, M.; Collet, C. wnt11f2 Zebrafish, an Animal Model for Development and New Insights in Bone Formation. Zebrafish 2023, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Foessl, I.; Bassett, J.; Bjørnerem, Å.; Busse, B.; Calado, Â.; Chavassieux, P.; Christou, M.; Douni, E.; Fiedler, I.A.; Fonseca, J.E.; et al. Bone phenotyping approaches in human, mice and zebrafish–Expert overview of the EU cost action GEMSTONE (“GEnomics of MusculoSkeletal traits TranslatiOnal NEtwork”). Front. Endocrinol. 2021, 12, 1476. [Google Scholar] [CrossRef]
- Dietrich, K.; Fiedler, I.A.; Kurzyukova, A.; López-Delgado, A.C.; McGowan, L.M.; Geurtzen, K.; Hammond, C.L.; Busse, B.; Knopf, F. Skeletal biology and disease modeling in zebrafish. J. Bone Miner. Res. 2021, 36, 436–458. [Google Scholar] [CrossRef]
- Caetano da Silva, C.; Edouard, T.; Fradin, M.; Aubert-Mucca, M.; Ricquebourg, M.; Raman, R.; Salles, J.P.; Charon, V.; Guggenbuhl, P.; Muller, M.; et al. WNT11, a new gene associated with early onset osteoporosis, is required for osteoblastogenesis. Hum. Mol. Genet. 2022, 31, 1622–1634. [Google Scholar] [CrossRef] [PubMed]
- Cotti, S.; Huysseune, A.; Larionova, D.; Koppe, W.; Forlino, A.; Witten, P.E. Compression fractures and partial phenotype rescue with a low phosphorus diet in the Chihuahua zebrafish osteogenesis imperfecta model. Front. Endocrinol. 2022, 13, 851879. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, F.; Bek, J.W.; Besio, R.; De Clercq, A.; Leoni, L.; Salmon, P.; Coucke, P.J.; Willaert, A.; Forlino, A. Zebrafish: A resourceful vertebrate model to investigate skeletal disorders. Front. Endocrinol. 2020, 11, 489. [Google Scholar] [CrossRef]
- Tacon, A.G.; Metian, M. Fish matters: Importance of aquatic foods in human nutrition and global food supply. Rev. Fish. Sci. 2013, 21, 22–38. [Google Scholar] [CrossRef]
- Toni, M.; Manciocco, A.; Angiulli, E.; Alleva, E.; Cioni, C.; Malavasi, S. Review: Assessing fish welfare in research and aquaculture, with a focus on European directives. Animal 2019, 13, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, S.; Liu, J.; Gao, Q.; Dong, S.; Zhou, C. Deep learning for smart fish farming: Applications, opportunities and challenges. Rev. Aquac. 2021, 13, 66–90. [Google Scholar] [CrossRef]
- Li, D.; Du, L. Recent advances of deep learning algorithms for aquacultural machine vision systems with emphasis on fish. Artif. Intell. Rev. 2022, 55, 4077–4116. [Google Scholar] [CrossRef]
- Tonachella, N.; Martini, A.; Martinoli, M.; Pulcini, D.; Romano, A.; Capoccioni, F. An affordable and easy-to-use tool for automatic fish length and weight estimation in mariculture. Sci. Rep. 2022, 12, 15642. [Google Scholar] [CrossRef] [PubMed]
- Hough, C. Manual of control of malformations in fish aquaculture. Science and Practice. In Federation of European Aquaculture Producers; RapidPRess: Luxembourg, 2009; Volume 150. [Google Scholar]
- Dara, M.; Carbonara, P.; La Corte, C.; Parrinello, D.; Cammarata, M.; Parisi, M.G. Fish Welfare in Aquaculture: Physiological and Immunological Activities for Diets, Social and Spatial Stress on Mediterranean Aqua Cultured Species. Fishes 2023, 8, 414. [Google Scholar] [CrossRef]
- Boglione, C.; Costa, C. Skeletal deformities and juvenile quality. In Sparidae; Wiley: Hoboken, NJ, USA, 2011; pp. 233–294. [Google Scholar]
- Lubin, A.; Otterstrom, J.; Hoade, Y.; Bjedov, I.; Stead, E.; Whelan, M.; Gestri, G.; Paran, Y.; Payne, E. A versatile, automated and high-throughput drug screening platform for zebrafish embryos. Biol. Open 2021, 10, bio058513. [Google Scholar] [CrossRef] [PubMed]
- Kithcart, A.; MacRae, C.A. Using zebrafish for high-throughput screening of novel cardiovascular drugs. Basic Transl. Sci. 2017, 2, 1–12. [Google Scholar] [CrossRef]
- Laleh, N.G.; Muti, H.S.; Loeffler, C.M.L.; Echle, A.; Saldanha, O.L.; Mahmood, F.; Lu, M.Y.; Trautwein, C.; Langer, R.; Dislich, B.; et al. Benchmarking weakly-supervised deep learning pipelines for whole slide classification in computational pathology. Med. Image Anal. 2022, 79, 102474. [Google Scholar] [CrossRef]
- Mac Namee, B.; Cunningham, P.; Byrne, S.; Corrigan, O.I. The problem of bias in training data in regression problems in medical decision support. Artif. Intell. Med. 2002, 24, 51–70. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef]
- Rubens, U.; Mormont, R.; Paavolainen, L.; Bäcker, V.; Pavie, B.; Scholz, L.A.; Michiels, G.; Maška, M.; Ünay, D.; Ball, G.; et al. BIAFLOWS: A collaborative framework to reproducibly deploy and benchmark bioimage analysis workflows. Patterns 2020, 1, 100040. [Google Scholar] [CrossRef] [PubMed]
- Paul-Gilloteaux, P.; Tosi, S.; Hériché, J.K.; Gaignard, A.; Ménager, H.; Marée, R.; Baecker, V.; Klemm, A.; Kalaš, M.; Zhang, C.; et al. Bioimage analysis workflows: Community resources to navigate through a complex ecosystem. F1000Research 2021, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Mormont, R.; Geurts, P.; Marée, R. Comparison of deep transfer learning strategies for digital pathology. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–22 June 2018; pp. 2262–2271. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Hartley, M.; Kleywegt, G.J.; Patwardhan, A.; Sarkans, U.; Swedlow, J.R.; Brazma, A. The BioImage archive–building a home for life-sciences microscopy data. J. Mol. Biol. 2022, 434, 167505. [Google Scholar] [CrossRef] [PubMed]
- Ras, G.; Xie, N.; van Gerven, M.; Doran, D. Explainable Deep Learning: A Field Guide for the Uninitiated. J. Artif. Int. Res. 2022, 73, 329–396. [Google Scholar] [CrossRef]
- Sun, M.; Yang, X.; Xie, Y. Deep learning in aquaculture: A review. J. Comput. 2020, 31, 294–319. [Google Scholar]
- Rastegari, H.; Nadi, F.; Lam, S.S.; Abdullah, M.I.; Kasan, N.A.; Rahmat, R.F.; Mahari, W.A.W. Internet of Things in aquaculture: A review of the challenges and potential solutions based on current and future trends. Smart Agric. Technol. 2023, 4, 100187. [Google Scholar] [CrossRef]
- Kelasidi, E.; Svendsen, E. Robotics for Sea-Based Fish Farming. In Encyclopedia of Smart Agriculture Technologies; Springer: Berlin/Heidelberg, Germany, 2023; pp. 1–20. [Google Scholar]
- Wang, C.; Li, Z.; Wang, T.; Xu, X.; Zhang, X.; Li, D. Intelligent fish farm—The future of aquaculture. Aquac. Int. 2021, 29, 2681–2711. [Google Scholar] [CrossRef]
- Sawaki, R.; Sato, D.; Nakayama, H.; Nakagawa, Y.; Shimada, Y. ZF-AutoML: An Easy Machine-Learning-Based Method to Detect Anomalies in Fluorescent-Labelled Zebrafish. Inventions 2019, 4, 72. [Google Scholar] [CrossRef]
- Carreira, M.J.; Vila-Blanco, N.; Cabezas-Sainz, P.; Sánchez, L. Zftool: A software for automatic quantification of cancer cell mass evolution in zebrafish. Appl. Sci. 2021, 11, 7721. [Google Scholar] [CrossRef]
- Reischl, M.; Bartschat, A.; Liebel, U.; Gehrig, J.; Müller, F.; Mikut, R. ZebrafishMiner: An open source software for interactive evaluation of domain-specific fluorescence in zebrafish. Curr. Dir. Biomed. Eng. 2017, 3, 199–202. [Google Scholar] [CrossRef]
- Navarro, A.; Lee-Montero, I.; Santana, D.; Henríquez, P.; Ferrer, M.A.; Morales, A.; Soula, M.; Badilla, R.; Negrín-Báez, D.; Zamorano, M.J.; et al. IMAFISH_ML: A fully-automated image analysis software for assessing fish morphometric traits on gilthead seabream (Sparus aurata L.), meagre (Argyrosomus regius) and red porgy (Pagrus pagrus). Comput. Electron. Agric. 2016, 121, 66–73. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, N.; Marée, R.; Geurts, P.; Muller, M. Recent Advances in Bioimage Analysis Methods for Detecting Skeletal Deformities in Biomedical and Aquaculture Fish Species. Biomolecules 2023, 13, 1797. https://doi.org/10.3390/biom13121797
Kumar N, Marée R, Geurts P, Muller M. Recent Advances in Bioimage Analysis Methods for Detecting Skeletal Deformities in Biomedical and Aquaculture Fish Species. Biomolecules. 2023; 13(12):1797. https://doi.org/10.3390/biom13121797
Chicago/Turabian StyleKumar, Navdeep, Raphaël Marée, Pierre Geurts, and Marc Muller. 2023. "Recent Advances in Bioimage Analysis Methods for Detecting Skeletal Deformities in Biomedical and Aquaculture Fish Species" Biomolecules 13, no. 12: 1797. https://doi.org/10.3390/biom13121797
APA StyleKumar, N., Marée, R., Geurts, P., & Muller, M. (2023). Recent Advances in Bioimage Analysis Methods for Detecting Skeletal Deformities in Biomedical and Aquaculture Fish Species. Biomolecules, 13(12), 1797. https://doi.org/10.3390/biom13121797







