Metabolic Patterns of High-Invasive and Low-Invasive Oral Squamous Cell Carcinoma Cells Using Quantitative Metabolomics and 13C-Glucose Tracing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Sample Collection
2.2. Cell Culture
2.3. Transwell Invasion Assay
2.4. Metabolite Extraction
2.5. Methyl Chloroformate (MCF) Derivatization and Gas Chromatography–Mass Spectrometry (GC–MS) Analysis
2.6. GC–MS Data Processing and Targeted Quantitative Analysis
2.7. [U-13C6] Glucose Labeling Experiment
2.8. Protein Quantification
2.9. Glucose Assay
2.10. Statistical Analysis
3. Results
3.1. Tumor Tissue/Normal Tissue Ratios of Metabolites versus Clinical Stage
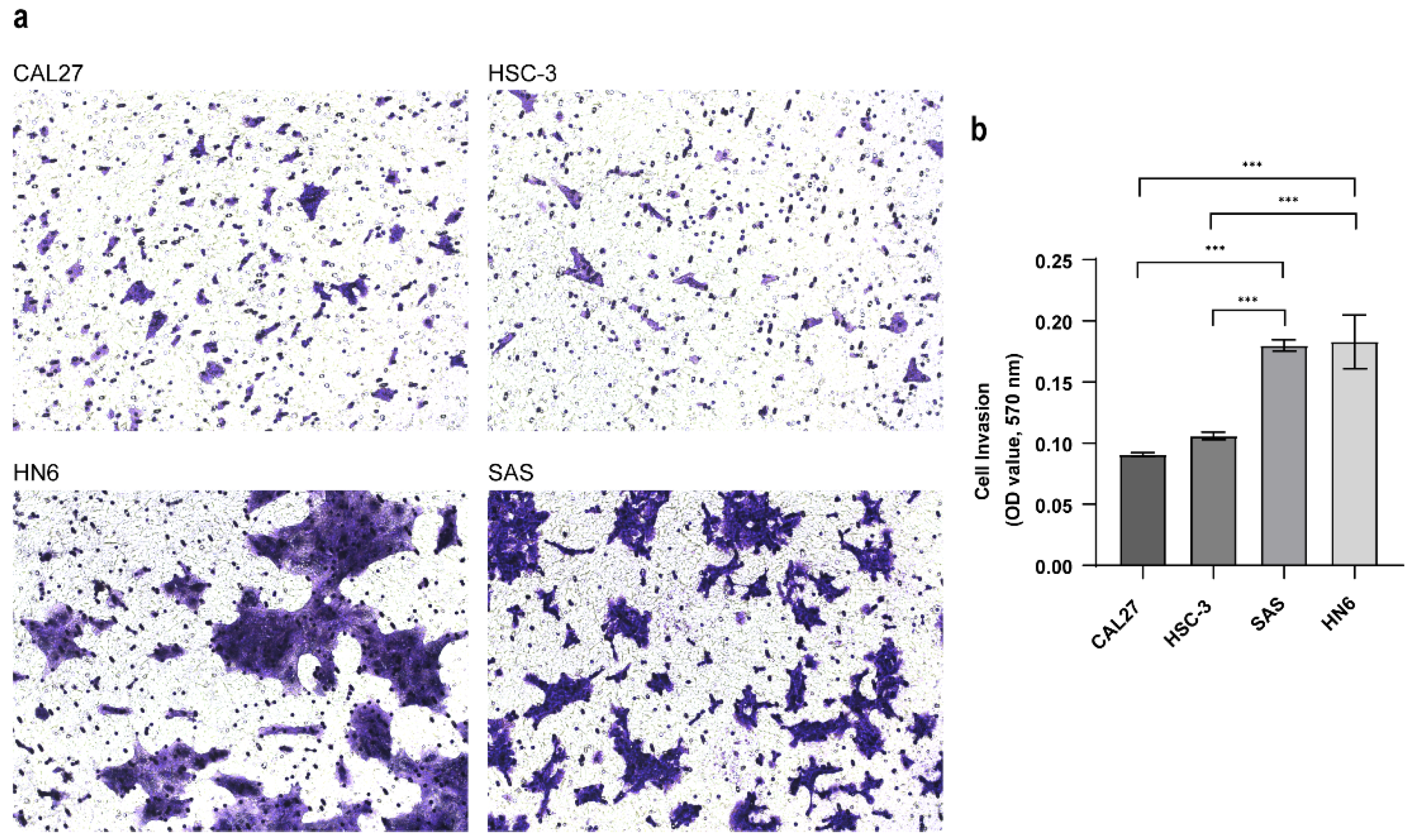
3.2. Establishing the Invasive Capacity of OSCC Cell Lines
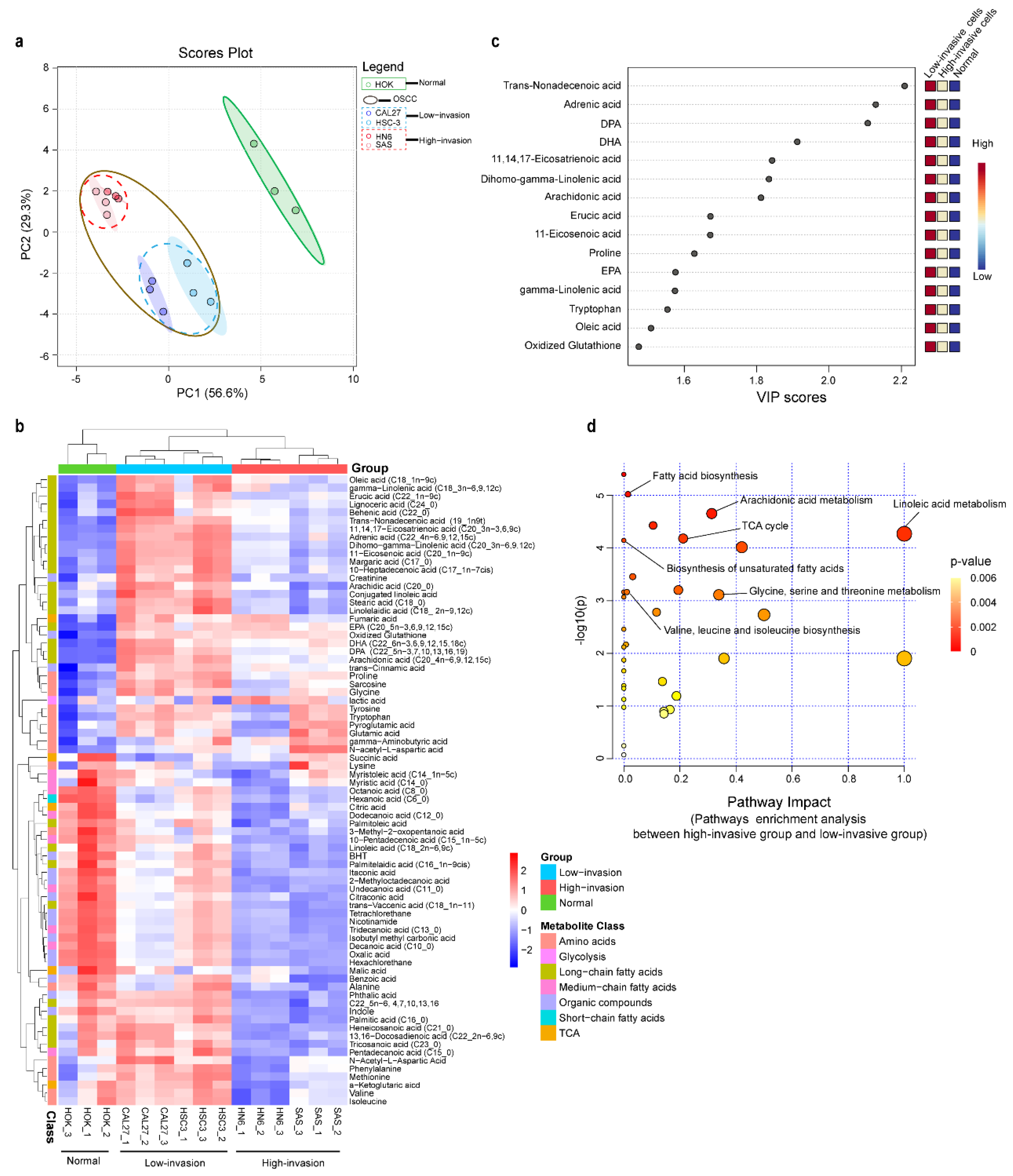
3.3. Metabolic Profiling Alterations of Normal Cells and Low-Invasive and High-Invasive Cancer Cells Based on Untargeted Metabolomics
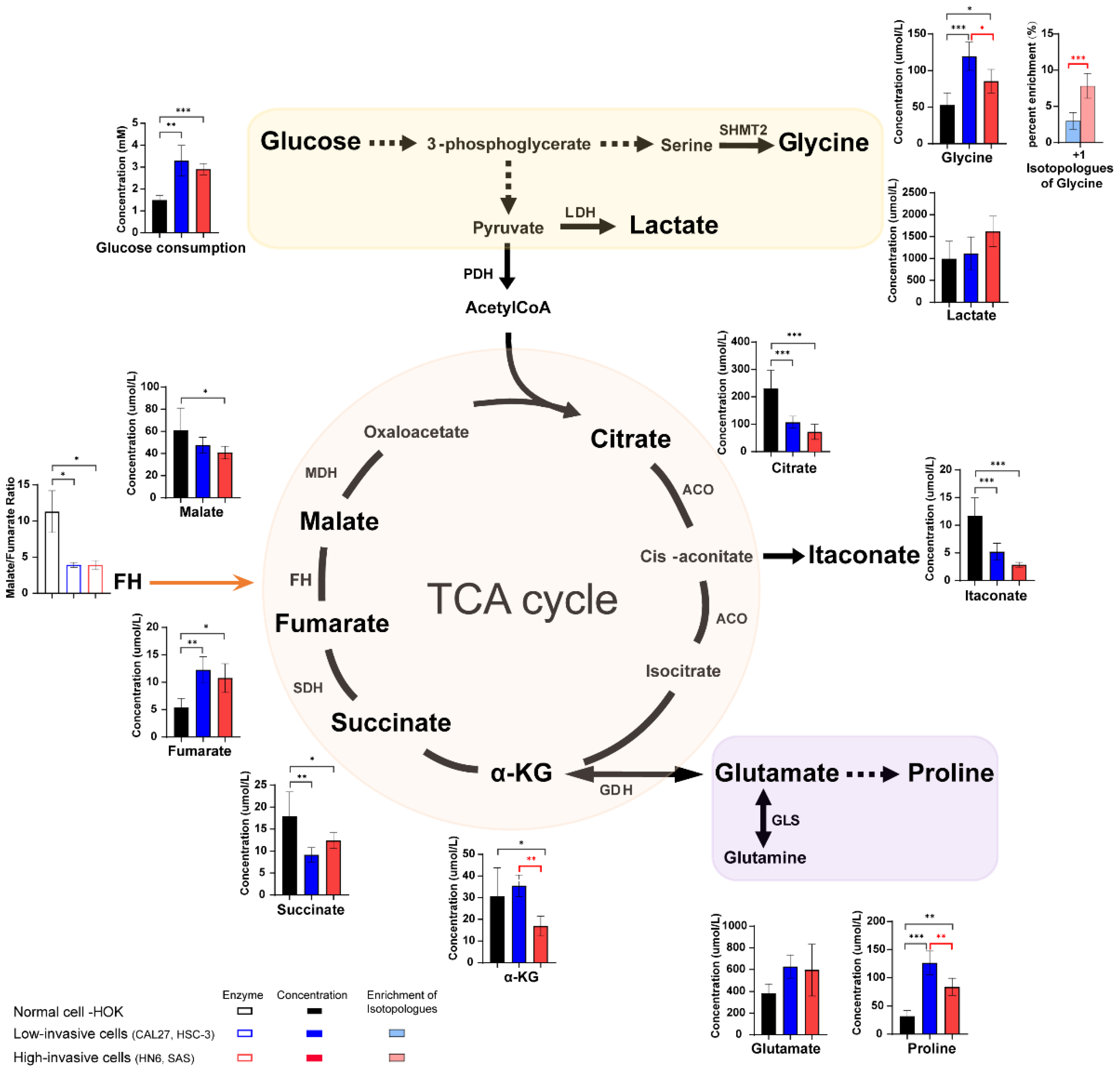
3.4. The Different Concentration of Glucose, TCA Cycle Intermediates and Amino Acids between High-Invasive and Low-Invasive Cancer Cells
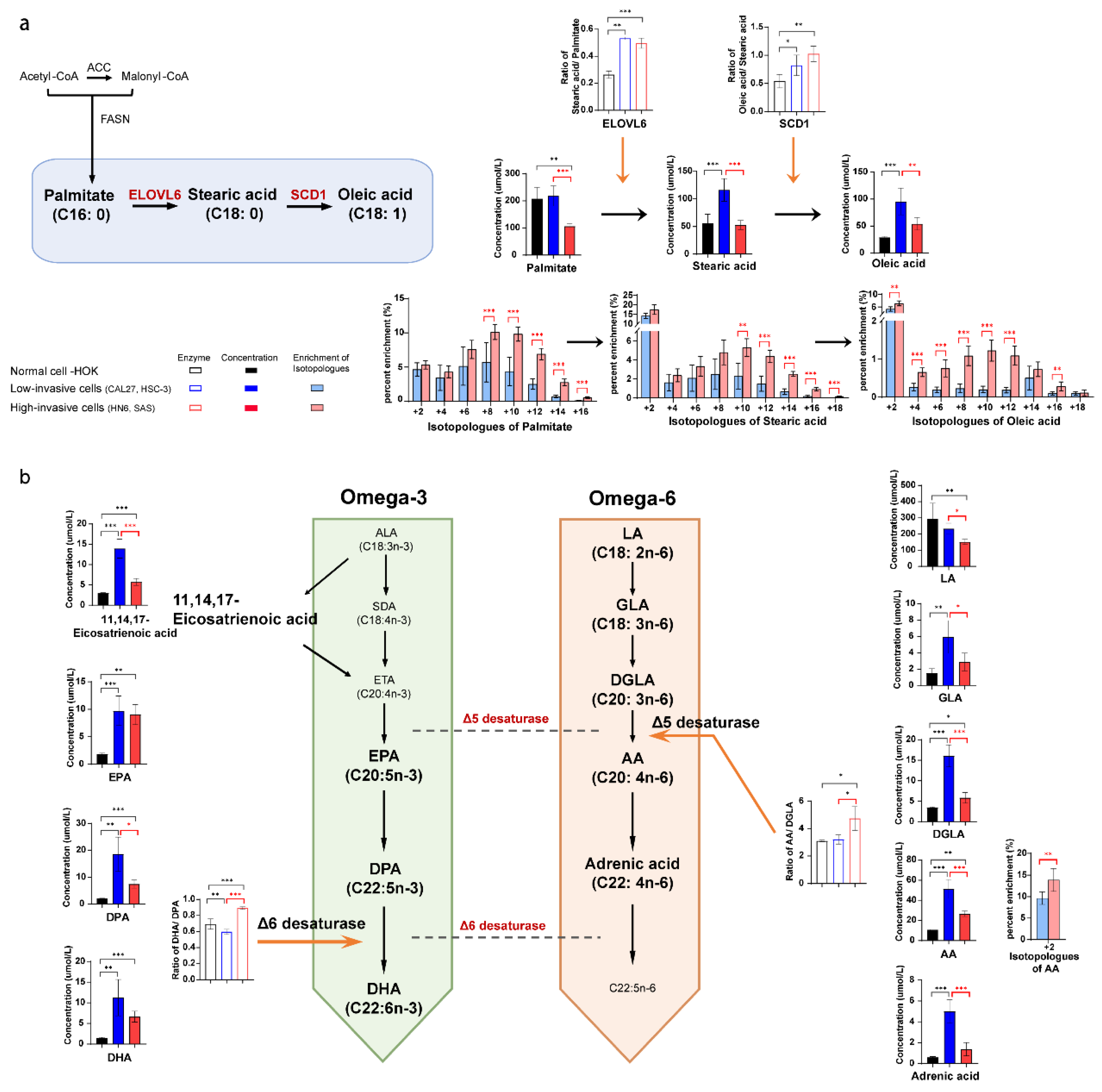
3.5. Long-Chain Fatty Acid Metabolism between High-Invasive and Low-Invasive Cancer Cells
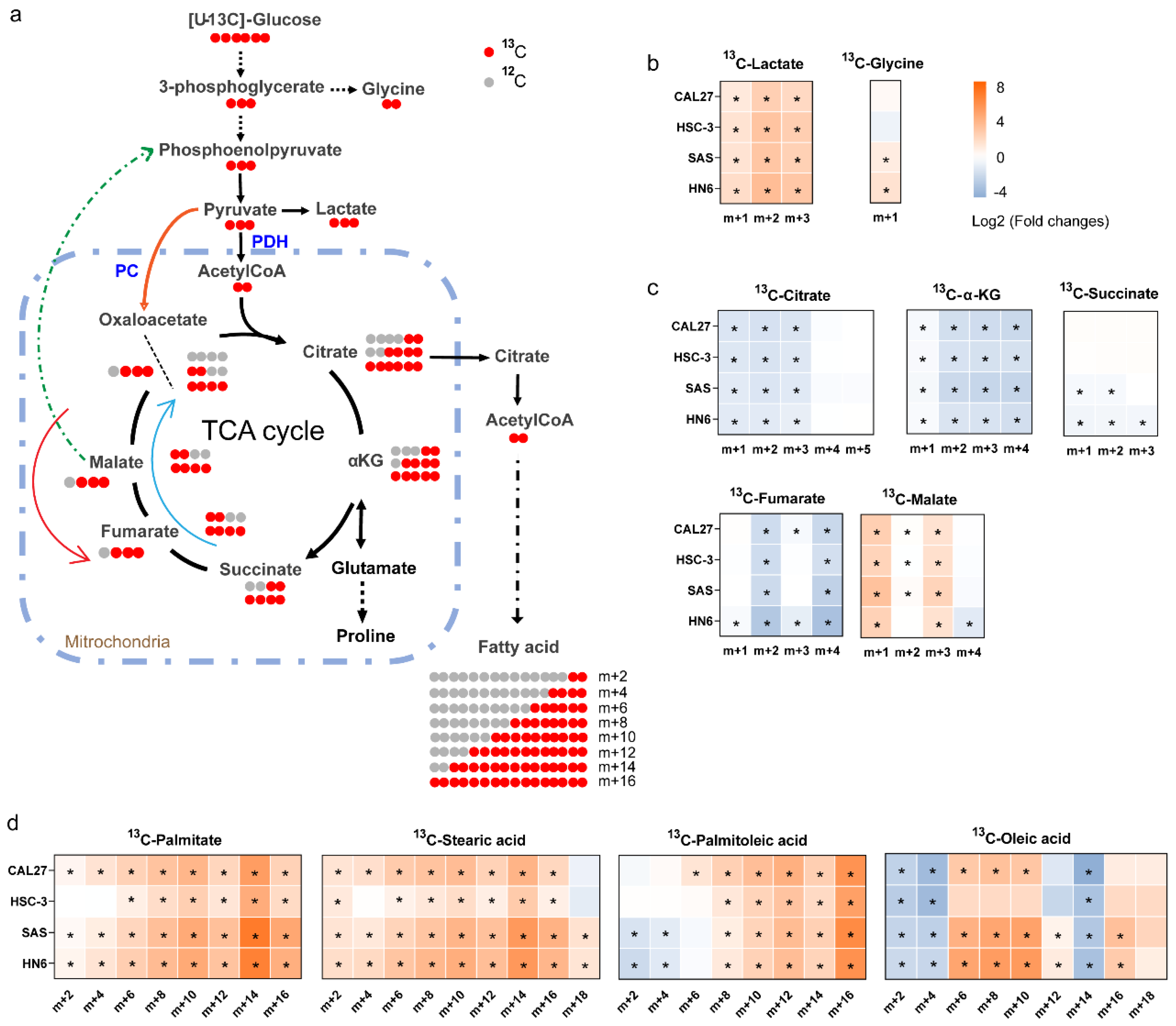
3.6. 13C-Enrichment of Metabolites Derived from Glucose in Normal Cells and High-Invasive and Low-Invasive Cells
3.7. Concentration of Extracellular Metabolites between High-Invasive and Low-Invasive Cancer Cells
4. Discussion
4.1. Metabolic Reprogramming of Low-Invasive and High-Invasive Cancer Cells Compared with Normal Cells
4.2. Metabolic Alterations between Low-Invasive Cancer Cells and High-Invasive Cancer Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, C.-C.; Lee, W.-T.; Hsiao, J.-R.; Ou, C.-Y.; Huang, C.-C.; Tsai, S.-T.; Chen, K.-C.; Huang, J.-S.; Wong, T.-Y.; Lai, Y.-H.; et al. Oral hygiene and the overall survival of head and neck cancer patients. Cancer Med. 2019, 8, 1854–1864. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, D.K.; Montero, P.H.; Migliacci, J.C.; Shah, J.P.; Wong, R.J.; Ganly, I.; Patel, S.G. Survival outcomes after treatment of cancer of the oral cavity (1985–2015). Oral. Oncol. 2019, 90, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Duprez, F.; Berwouts, D.; de Neve, W.; Bonte, K.; Boterberg, T.; Deron, P.; Huvenne, W.; Rottey, S.; Mareel, M. Distant metastases in head and neck cancer. Head. Neck 2017, 39, 1733–1743. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef]
- Zhu, L.; Zhu, X.; Wu, Y. Effects of Glucose Metabolism, Lipid Metabolism, and Glutamine Metabolism on Tumor Microenvironment and Clinical Implications. Biomolecules 2022, 12, 580. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.A.; El-Derany, M.O.; Anwar, A.M.; Saied, E.M.; Magdeldin, S. Metabolomics and Lipidomics Screening Reveal Reprogrammed Signaling Pathways toward Cancer Development in Non-Alcoholic Steatohepatitis. Int. J. Mol. Sci. 2022, 24, 210. [Google Scholar] [CrossRef]
- Sarhadi, V.K.; Armengol, G. Molecular Biomarkers in Cancer. Biomolecules 2022, 12, 1021. [Google Scholar] [CrossRef]
- Al-Ishaq, R.K.; Overy, A.J.; Büsselberg, D. Phytochemicals and Gastrointestinal Cancer: Cellular Mechanisms and Effects to Change Cancer Progression. Biomolecules 2020, 10, 105. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Wang, S.; Li, Z.; Hu, X.; Yang, X.; Song, Y.; Jing, Y.; Hu, Q.; Ni, Y. Identification of Metabolism-Associated Biomarkers for Early and Precise Diagnosis of Oral Squamous Cell Carcinoma. Biomolecules 2022, 12, 400. [Google Scholar] [CrossRef]
- Fu, Y.; Ding, L.; Yang, X.; Ding, Z.; Huang, X.; Zhang, L.; Chen, S.; Hu, Q.; Ni, Y. Asparagine Synthetase-Mediated l-Asparagine Metabolism Disorder Promotes the Perineural Invasion of Oral Squamous Cell Carcinoma. Front. Oncol. 2021, 11, 637226. [Google Scholar] [CrossRef]
- Pascual, G.; Domínguez, D.; Elosúa-Bayes, M.; Beckedorff, F.; Laudanna, C.; Bigas, C.; Douillet, D.; Greco, C.; Symeonidi, A.; Hernández, I.; et al. Dietary palmitic acid promotes a prometastatic memory via Schwann cells. Nature 2021, 599, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna-Silva, A.C.B.; Santos, G.C.; Campos, S.P.C.; Oliveira Gomes, A.M.; Pérez-Valencia, J.A.; Rumjanek, F.D. Metabolic Profile of Oral Squamous Carcinoma Cell Lines Relies on a Higher Demand of Lipid Metabolism in Metastatic Cells. Front. Oncol. 2018, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Kalyankrishna, S.; Grandis, J.R. Epidermal growth factor receptor biology in head and neck cancer. J. Clin. Oncol. 2006, 24, 2666–2672. [Google Scholar] [CrossRef] [PubMed]
- Brusevold, I.J.; Aasrum, M.; Bryne, M.; Christoffersen, T. Migration induced by epidermal and hepatocyte growth factors in oral squamous carcinoma cells in vitro: Role of MEK/ERK, p38 and PI-3 kinase/Akt. J. Oral. Pathol. Med. 2012, 41, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Caswell, D.R.; Swanton, C. The role of tumour heterogeneity and clonal cooperativity in metastasis, immune evasion and clinical outcome. BMC Med. 2017, 15, 133. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Gao, W.; Xu, T.; Liu, C.; Wu, D.; Tang, W. A UPLC Q-Exactive Orbitrap Mass Spectrometry-Based Metabolomic Study of Serum and Tumor Tissue in Patients with Papillary Thyroid Cancer. Toxics 2022, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Ho, C.-L.; Chen, P.-K.; Chen, Y.-L.; Chang, C.-F. Anterior gradient 2: A novel sensitive tumor marker for metastatic oral cancer. Cancer Lett. 2013, 339, 270–278. [Google Scholar] [CrossRef]
- Weiße, J.; Rosemann, J.; Müller, L.; Kappler, M.; Eckert, A.W.; Glaß, M.; Misiak, D.; Hüttelmaier, S.; Ballhausen, W.G.; Hatzfeld, M.; et al. Identification of lymphocyte cell-specific protein-tyrosine kinase (LCK) as a driver for invasion and migration of oral cancer by tumor heterogeneity exploitation. Mol. Cancer 2021, 20, 88. [Google Scholar] [CrossRef]
- Chiu, A.M.; Mitra, M.; Boymoushakian, L.; Coller, H.A. Integrative analysis of the inter-tumoral heterogeneity of triple-negative breast cancer. Sci. Rep. 2018, 8, 11807. [Google Scholar] [CrossRef]
- Sexton, R.E.; Hallak, M.N.A.; Uddin, M.H.; Diab, M.; Azmi, A.S. Gastric Cancer Heterogeneity and Clinical Outcomes. Technol. Cancer Res. Treat. 2020, 19, 1533033820935477. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed]
- Bader, J.E.; Voss, K.; Rathmell, J.C. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol. Cell 2020, 78, 1019–1033. [Google Scholar] [CrossRef] [PubMed]
- Bujak, R.; Struck-Lewicka, W.; Markuszewski, M.J.; Kaliszan, R. Metabolomics for laboratory diagnostics. J. Pharm. Biomed. Anal. 2015, 113, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Kamarajan, P.; Somashekar, B.S.; MacKinnon, N.; Chinnaiyan, A.M.; Kapila, Y.L.; Rajendiran, T.M.; Ramamoorthy, A. Delineating metabolic signatures of head and neck squamous cell carcinoma: Phospholipase A2, a potential therapeutic target. Int. J. Biochem. Cell Biol. 2012, 44, 1852–1861. [Google Scholar] [CrossRef]
- Rangel-Huerta, O.D.; Pastor-Villaescusa, B.; Gil, A. Are we close to defining a metabolomic signature of human obesity? A systematic review of metabolomics studies. Metabolomics 2019, 15, 93. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Fan, T.W.-M.; Higashi, R.M. Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Methods Cell Biol. 2008, 84, 541–588. [Google Scholar] [CrossRef] [PubMed]
- Gkiouli, M.; Biechl, P.; Eisenreich, W.; Otto, A.M. Diverse Roads Taken by 13C-Glucose-Derived Metabolites in Breast Cancer Cells Exposed to Limiting Glucose and Glutamine Conditions. Cells 2019, 8, 1113. [Google Scholar] [CrossRef]
- Zhang, G.-F.; Jensen, M.V.; Gray, S.M.; El, K.; Wang, Y.; Lu, D.; Becker, T.C.; Campbell, J.E.; Newgard, C.B. Reductive TCA cycle metabolism fuels glutamine- and glucose-stimulated insulin secretion. Cell Metab. 2021, 33, 804–817.e5. [Google Scholar] [CrossRef]
- Pacold, M.E.; Brimacombe, K.R.; Chan, S.H.; Rohde, J.M.; Lewis, C.A.; Swier, L.J.Y.M.; Possemato, R.; Chen, W.W.; Sullivan, L.B.; Fiske, B.P.; et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 2016, 12, 452–458. [Google Scholar] [CrossRef]
- Christen, S.; Lorendeau, D.; Schmieder, R.; Broekaert, D.; Metzger, K.; Veys, K.; Elia, I.; Buescher, J.M.; Orth, M.F.; Davidson, S.M.; et al. Breast Cancer-Derived Lung Metastases Show Increased Pyruvate Carboxylase-Dependent Anaplerosis. Cell Rep. 2016, 17, 837–848. [Google Scholar] [CrossRef]
- Momose, F.; Araida, T.; Negishi, A.; Ichijo, H.; Shioda, S.; Sasaki, S. Variant sublines with different metastatic potentials selected in nude mice from human oral squamous cell carcinomas. J. Oral. Pathol. Med. 1989, 18, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Gioanni, J.; Fischel, J.L.; Lambert, J.C.; Demard, F.; Mazeau, C.; Zanghellini, E.; Ettore, F.; Formento, P.; Chauvel, P.; Lalanne, C.M. Two new human tumor cell lines derived from squamous cell carcinomas of the tongue: Establishment, characterization and response to cytotoxic treatment. Eur. J. Cancer Clin. Oncol. 1988, 24, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Easty, D.M.; Easty, G.C.; Carter, R.L.; Monaghan, P.; Butler, L.J. Ten human carcinoma cell lines derived from squamous carcinomas of the head and neck. Br. J. Cancer 1981, 43, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Kanazawa, H.; Akiyama, Y.; Tazaki, S.; Takahara, M.; Muto, T.; Tanzawa, H.; Sato, K. Establishment and characterization of a cell line (SAS) from poorly defferentiated human squamous cell carcinoma of the tongue. J. Jpn. Stomatol. Soc. 1989, 38, 20–28. [Google Scholar]
- Smart, K.F.; Aggio, R.B.M.; van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef] [PubMed]
- Karpievitch, Y.V.; Taverner, T.; Adkins, J.N.; Callister, S.J.; Anderson, G.A.; Smith, R.D.; Dabney, A.R. Normalization of peak intensities in bottom-up MS-based proteomics using singular value decomposition. Bioinformatics 2009, 25, 2573–2580. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Washio, J.; Takahashi, T.; Echigo, S.; Takahashi, N. Glucose and glutamine metabolism in oral squamous cell carcinoma: Insight from a quantitative metabolomic approach. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 2014, 118, 218–225. [Google Scholar] [CrossRef]
- Hsieh, Y.-T.; Chen, Y.-F.; Lin, S.-C.; Chang, K.-W.; Li, W.-C. Targeting Cellular Metabolism Modulates Head and Neck Oncogenesis. Int. J. Mol. Sci. 2019, 20, 3960. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Funchain, P.; Retuerto, M.; Jurevic, R.J.; Fowler, N.; Burkey, B.; Eng, C.; Ghannoum, M.A. Metabolomic analysis identifies differentially produced oral metabolites, including the oncometabolite 2-hydroxyglutarate, in patients with head and neck squamous cell carcinoma. BBA Clin. 2017, 7, 8–15. [Google Scholar] [CrossRef]
- Noguchi, S.; Ishikawa, H.; Wakita, K.; Matsuda, F.; Shimizu, H. Direct and quantitative analysis of altered metabolic flux distributions and cellular ATP production pathway in fumarate hydratase-diminished cells. Sci. Rep. 2020, 10, 13065. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, E.; Sciacovelli, M.; Costa, A.S.H.; Tran, M.G.B.; Johnson, T.I.; Machado, D.; Frezza, C.; Saez-Rodriguez, J. Post-translational regulation of metabolism in fumarate hydratase deficient cancer cells. Metab. Eng. 2018, 45, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Sandulache, V.C.; Ow, T.J.; Pickering, C.R.; Frederick, M.J.; Zhou, G.; Fokt, I.; Davis-Malesevich, M.; Priebe, W.; Myers, J.N. Glucose, not glutamine, is the dominant energy source required for proliferation and survival of head and neck squamous carcinoma cells. Cancer 2011, 117, 2926–2938. [Google Scholar] [CrossRef] [PubMed]
- Geeraerts, S.L.; Heylen, E.; de Keersmaecker, K.; Kampen, K.R. The ins and outs of serine and glycine metabolism in cancer. Nat. Metab. 2021, 3, 131–141. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Z.; Xue, D.; Zhang, Q.; Liu, X.; Luh, F.; Hong, L.; Zhang, H.; Pan, F.; Liu, Y.; et al. Prognostic and therapeutic value of mitochondrial serine hydroxyl-methyltransferase 2 as a breast cancer biomarker. Oncol. Rep. 2016, 36, 2489–2500. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wanggou, S.; Li, X.; Liu, Q.; Xie, Y. Overexpression of mitochondrial serine hydroxyl-methyltransferase 2 is associated with poor prognosis and promotes cell proliferation and invasion in gliomas. Onco. Targets. Ther. 2017, 10, 3781–3788. [Google Scholar] [CrossRef]
- Kim, D.; Fiske, B.P.; Birsoy, K.; Freinkman, E.; Kami, K.; Possemato, R.L.; Chudnovsky, Y.; Pacold, M.E.; Chen, W.W.; Cantor, J.R.; et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature 2015, 520, 363–367. [Google Scholar] [CrossRef]
- Jäger, K.; Larribère, L.; Wu, H.; Weiss, C.; Gebhardt, C.; Utikal, J. Expression of Neural Crest Markers GLDC and ERRFI1 is Correlated with Melanoma Prognosis. Cancers 2019, 11, 76. [Google Scholar] [CrossRef]
- Berezowska, S.; Galván, J.A.; Langer, R.; Bubendorf, L.; Savic, S.; Gugger, M.; Schmid, R.A.; Marti, T.M. Glycine decarboxylase and HIF-1α expression are negative prognostic factors in primary resected early-stage non-small cell lung cancer. Virchows Arch. 2017, 470, 323–330. [Google Scholar] [CrossRef]
- Schmidt, J.; Kajtár, B.; Juhász, K.; Péter, M.; Járai, T.; Burián, A.; Kereskai, L.; Gerlinger, I.; Tornóczki, T.; Balogh, G.; et al. Lipid and protein tumor markers for head and neck squamous cell carcinoma identified by imaging mass spectrometry. Oncotarget 2020, 11, 2702–2717. [Google Scholar] [CrossRef]
- Benjamin, D.I.; Li, D.S.; Lowe, W.; Heuer, T.; Kemble, G.; Nomura, D.K. Diacylglycerol Metabolism and Signaling Is a Driving Force Underlying FASN Inhibitor Sensitivity in Cancer Cells. ACS Chem. Biol. 2015, 10, 1616–1623. [Google Scholar] [CrossRef] [PubMed]
- Louie, S.M.; Roberts, L.S.; Mulvihill, M.M.; Luo, K.; Nomura, D.K. Cancer cells incorporate and remodel exogenous palmitate into structural and oncogenic signaling lipids. Biochim. Biophys. Acta 2013, 1831, 1566–1572. [Google Scholar] [CrossRef] [PubMed]
- Hilvo, M.; Denkert, C.; Lehtinen, L.; Müller, B.; Brockmöller, S.; Seppänen-Laakso, T.; Budczies, J.; Bucher, E.; Yetukuri, L.; Castillo, S.; et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011, 71, 3236–3245. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Yépez, S.; Tirado-Rodriguez, A.B.; Hankinson, O. Role of diets rich in omega-3 and omega-6 in the development of cancer. Boletín Médico Del. Hosp. Infant. De. México 2016, 73, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Halczy-Kowalik, L.; Drozd, A.; Stachowska, E.; Drozd, R.; Żabski, T.; Domagała, W. Fatty acids distribution and content in oral squamous cell carcinoma tissue and its adjacent microenvironment. PLoS ONE 2019, 14, e0218246. [Google Scholar] [CrossRef] [PubMed]
- Brasky, T.M.; Till, C.; White, E.; Neuhouser, M.L.; Song, X.; Goodman, P.; Thompson, I.M.; King, I.B.; Albanes, D.; Kristal, A.R. Serum phospholipid fatty acids and prostate cancer risk: Results from the prostate cancer prevention trial. Am. J. Epidemiol. 2011, 173, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Markworth, J.F.; Kaur, G.; Miller, E.G.; Larsen, A.E.; Sinclair, A.J.; Maddipati, K.R.; Cameron-Smith, D. Divergent shifts in lipid mediator profile following supplementation with n-3 docosapentaenoic acid and eicosapentaenoic acid. FASEB J. 2016, 30, 3714–3725. [Google Scholar] [CrossRef]
- Manzo, T.; Prentice, B.M.; Anderson, K.G.; Raman, A.; Schalck, A.; Codreanu, G.S.; Nava Lauson, C.B.; Tiberti, S.; Raimondi, A.; Jones, M.A.; et al. Accumulation of long-chain fatty acids in the tumor microenvironment drives dysfunction in intrapancreatic CD8+ T cells. J. Exp. Med. 2020, 217, e20191920. [Google Scholar] [CrossRef]
- Kumagai, S.; Togashi, Y.; Sakai, C.; Kawazoe, A.; Kawazu, M.; Ueno, T.; Sato, E.; Kuwata, T.; Kinoshita, T.; Yamamoto, M.; et al. An Oncogenic Alteration Creates a Microenvironment that Promotes Tumor Progression by Conferring a Metabolic Advantage to Regulatory T Cells. Immunity 2020, 53, 187–203.e8. [Google Scholar] [CrossRef]
- Montecillo-Aguado, M.; Tirado-Rodriguez, B.; Tong, Z.; Vega, O.M.; Morales-Martínez, M.; Abkenari, S.; Pedraza-Chaverri, J.; Huerta-Yepez, S. Importance of the Role of ω-3 and ω-6 Polyunsaturated Fatty Acids in the Progression of Brain Cancer. Brain Sci. 2020, 10, 381. [Google Scholar] [CrossRef]
- Bojková, B.; Winklewski, P.J.; Wszedybyl-Winklewska, M. Dietary Fat and Cancer-Which Is Good, Which Is Bad, and the Body of Evidence. Int. J. Mol. Sci. 2020, 21, 4114. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.-H.; Liu, P.-S.; Tsai, M.-M.; Chen, J.-L.; Wang, S.-J.; Hsieh, H.-L. The COX-2-derived PGE2 autocrine contributes to bradykinin-induced matrix metalloproteinase-9 expression and astrocytic migration via STAT3 signaling. Cell Commun. Signal. 2020, 18, 185. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Tito, N.; Soto-Guzman, A.; Castro-Sanchez, L.; Martinez-Orozco, R.; Salazar, E.P. Oleic acid promotes migration on MDA-MB-231 breast cancer cells through an arachidonic acid-dependent pathway. Int. J. Biochem. Cell Biol. 2010, 42, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Tian, L.; Zhao, B.; Sun, Y.; Cao, J.; Chen, K.; Li, F.; Li, M.; Shang, D.; Liu, M. FADS1 promotes the progression of laryngeal squamous cell carcinoma through activating AKT/mTOR signaling. Cell Death Dis. 2020, 11, 272. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Gao, P.; Li, Y.; Tian, H.; Ma, D.; Sun, N.; Chen, C.; Zhang, Y.; Qi, X. Investigating the role of FADS family members in breast cancer based on bioinformatic analysis and experimental validation. Front. Immunol. 2023, 14, 1074242. [Google Scholar] [CrossRef]


| Parameters | |
|---|---|
| Gender | 12 males, 5 females |
| Age (y) | 63 ± 9.5 |
| TNM stage | No. of patients |
| I + II | 9 |
| III + IV | 8 |
| Tumor grade | |
| G1 | 9 |
| G2 | 6 |
| G3 | 2 |
| Primary sites | |
| Buccal mucosa | 3 |
| Tongue | 8 |
| Gum | 2 |
| Floor of mouth | 2 |
| Hard palate | 2 |
| Ratios of Tumor Tissue/ Normal Tissue | |||
|---|---|---|---|
| Amino Acids | Stage I and II (Non-Metastatic) | Stage III and IV (Metastatic) | p-Value |
| Proline | 4.51 | 1.90 | 0.036 |
| Glycine | 1.58 | 4.54 | 0.021 |
| Leucine | 1.74 | 3.24 | 0.200 |
| Methionine | 3.08 | 3.72 | 0.409 |
| Alanine | 1.13 | 1.22 | 0.412 |
| Isoleucine | 2.34 | 3.27 | 0.963 |
| Serine | 2.31 | 1.89 | 0.482 |
| Glutamic acid | 2.61 | 3.17 | 0.486 |
| Phenylalanine | 2.18 | 1.84 | 0.200 |
| Threonine | 2.21 | 2.38 | 0.788 |
| Tryptophan | 2.00 | 1.75 | 0.541 |
| Tyrosine | 1.96 | 1.83 | 0.888 |
| Histidine | 1.51 | 1.43 | 0.799 |
| Lysine | 1.65 | 1.56 | 0.822 |
| Ratios of Tumor Tissue/ Normal Tissue | |||
|---|---|---|---|
| TCA Cycle | Stage I and II (Non-Metastatic) | Stage III and IV (Metastatic) | p-Value |
| Malate | 1.17 | 1.69 | 0.075 |
| α-Ketoglutarate | 1.23 | 1.56 | 0.743 |
| Succinate | 3.04 | 1.92 | 0.139 |
| Cis-aconitate | 3.33 | 2.36 | 0.888 |
| Itaconate | 1.38 | 1.28 | 0.585 |
| Fumarate | 1.13 | 1.21 | 0.732 |
| Ratios of Tumor Tissue/ Adjacent Normal Tumor Tissue | |||
|---|---|---|---|
| Fatty Acids | Stage I and II (Non-Metastatic) | Stage III and IV (Metastatic) | p-Value |
| SFAs | |||
| Decanoic acid (C10:0) | 0.97 | 0.97 | 0.815 |
| Dodecanoic acid (C12:0) | 0.88 | 0.88 | >0.999 |
| Myristic acid (C14:0) | 1.08 | 2.11 | 0.093 |
| Palmitate (C16:0) | 0.81 | 1.00 | 0.339 |
| Stearate (C18:0) | 2.78 | 2.99 | 0.423 |
| Arachidic acid (C20:0) | 1.20 | 1.36 | 0.541 |
| Lignoceric acid (C24:0) | 1.87 | 2.27 | 0.481 |
| MUFAs | |||
| Myristoleic acid (C14:1) | 1.09 | 1.91 | 0.139 |
| Oleic acid (C18:1) | 0.87 | 1.38 | 0.093 |
| Gondoic acid (C20:1) | 0.99 | 1.84 | 0.073 |
| Erucic acid (C22:1) | 1.04 | 1.80 | 0.012 |
| Nervonic acid (C24:1) | 0.99 | 1.39 | 0.049 |
| Omega-3 PUFAs | |||
| 11,14,17-Eicosatrienoic acid (C20:3n-3) | 0.51 | 0.82 | 0.128 |
| EPA (C20:5n-3) | 1.33 | 2.27 | 0.101 |
| DPA (C22:5n-3) | 1.01 | 2.12 | 0.003 |
| DHA (C22:6n-3) | 1.13 | 2.32 | 0.004 |
| Omega-6 PUFAs | |||
| LA (C18:2n-6) | 0.81 | 2.11 | 0.009 |
| GLA (C18:3n-6) | 0.41 | 1.84 | 0.003 |
| 11,14-Eicosadienoic acid (C20:2n-6) | 1.70 | 1.84 | 0.746 |
| DGLA (C20:3n-6) | 0.97 | 1.85 | 0.034 |
| AA (C22:4n-6) | 1.01 | 2.37 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, W.; Zhang, T.; Zhang, H.; Han, T.; Ji, P.; Ou, Z. Metabolic Patterns of High-Invasive and Low-Invasive Oral Squamous Cell Carcinoma Cells Using Quantitative Metabolomics and 13C-Glucose Tracing. Biomolecules 2023, 13, 1806. https://doi.org/10.3390/biom13121806
Jiang W, Zhang T, Zhang H, Han T, Ji P, Ou Z. Metabolic Patterns of High-Invasive and Low-Invasive Oral Squamous Cell Carcinoma Cells Using Quantitative Metabolomics and 13C-Glucose Tracing. Biomolecules. 2023; 13(12):1806. https://doi.org/10.3390/biom13121806
Chicago/Turabian StyleJiang, Wenrong, Ting Zhang, Hua Zhang, Tingli Han, Ping Ji, and Zhanpeng Ou. 2023. "Metabolic Patterns of High-Invasive and Low-Invasive Oral Squamous Cell Carcinoma Cells Using Quantitative Metabolomics and 13C-Glucose Tracing" Biomolecules 13, no. 12: 1806. https://doi.org/10.3390/biom13121806






