Molecular Breeding for Incorporation of Submergence Tolerance and Durable Bacterial Blight Resistance into the Popular Rice Variety ‘Ranidhan’
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Hybridization
2.2. DNA Isolation and Polymerase Chain Reaction
2.3. Marker Analysis
2.4. Evaluation for Submergence Tolerance
2.5. Bioassay for Resistance against BB Pathogen
2.6. Evaluation of Pyramided Lines for Yield, Agro-Morphology, and Quality Traits
3. Results
3.1. Molecular Marker Analysis of the Parental Lines for Submergence Tolerance and Bacterial Blight Resistance
3.2. Foreground Selection in Backcross-Derived Progenies
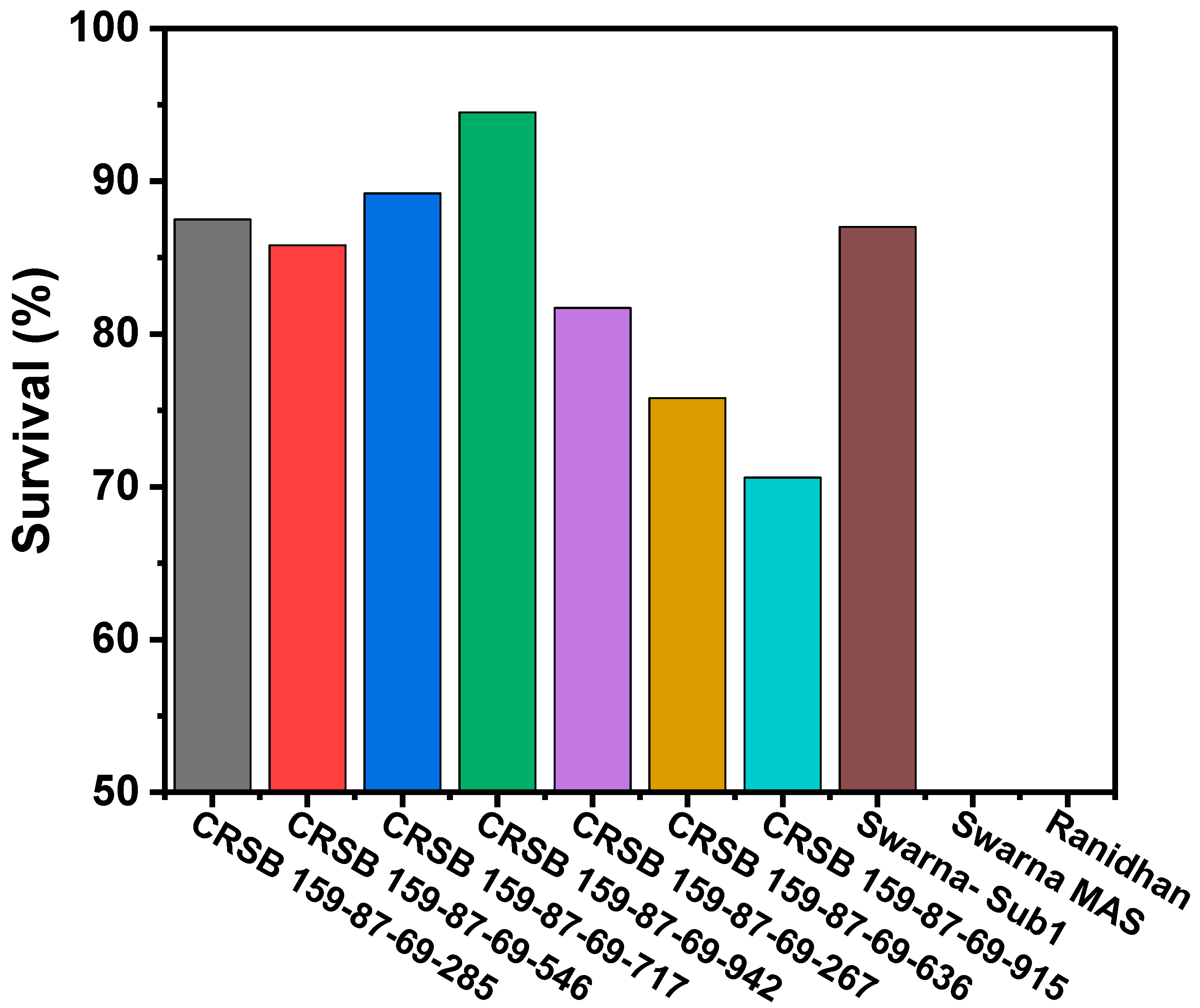
3.3. Evaluation of the Pyramided and Parental Lines for Submergence Tolerance
3.4. Bioassay of the Pyramided and Parental Lines for BB Disease Resistance
3.5. Agro-Morphology, Grain Yield, and Quality Traits of the Developed Pyramided and Parental Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Baksh, S.Y.; Mukherjee, A.K.; Mohanty, S.P. Development of flash-flood tolerant and durable bacterial blight resistant versions of mega rice variety ‘Swarna’ through marker-assisted backcross breeding. Sci. Rep. 2019, 9, 12810. [Google Scholar] [CrossRef]
- Mohapatra, S.; Panda, A.K.; Bastia, A.K.; Mukherjee, A.K.; Sanghamitra, P.; Meher, J.; Mohanty, S.P.; Pradhan, S.K. Development of Submergence-Tolerant, Bacterial Blight-Resistant, and High-Yielding Near Isogenic Lines of Popular Variety, ‘Swarna’ through Marker-Assisted Breeding Approach. Front. Plant Sci. 2021, 12, 672618. [Google Scholar] [CrossRef]
- Pandit, E.; Pawar, S.; Barik, S.R.; Mohanty, S.P.; Meher, J.; Pradhan, S.K. Marker-Assisted Backcross Breeding for Improvement of Submergence Tolerance and Grain Yield in the Popular Rice Variety ‘Maudamani’. Agronomy 2021, 11, 1263. [Google Scholar] [CrossRef]
- Ismail, A.M.; Singh, U.S.; Singh, S.; Dar, M.H.; Mackill, D.J. Contribution of submergence-tolerant (Sub1) rice varieties to food security in food prone rainfed lowland areas in Asia. Field Crops Res. 2013, 152, 83–93. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Chakraborti, M.; Chakraborty, K.; Behera, L.; Meher, J.; Subudhi, H.N. Genetic Improvement of Rainfed Shallow lowland Rice for Higher Yield and Climate Resilience. In Rice Research for Productivity, Profitability and Climate Resilience; ICAR-NRRI: Cuttack, India, 2018; pp. 107–121. [Google Scholar]
- Pradhan, S.K.; Pandit, E.; Barik, S.R.; Mohanty, S.P.; Nayak, D.K.; Sah, R.P.; Behera, L.; Sanghamitra, P.; Bose, L.K.; Das, S.R. Climate-Smart Rice Breeding: Progress and Challenges for the Rain-fed ecologies in India. In Advances in Rice Breeding: Stress Tolerance, Climate Resilience, Quality and High Yield; ICAR-NRRI: Cuttack, India, 2021; pp. 144–162. [Google Scholar]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S.; Anandan, A. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deep water rice variety, Jalmagna. Rice 2015, 8, 19. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Barik, S.; Nayak, D.; Pradhan, A.; Pandit, E.; Nayak, P.; Das, S.; Pathak, H. Genetics, Molecular Mechanisms and Deployment of Bacterial Blight Resistance Genes in Rice. Crit. Rev. Plant Sci. 2020, 39, 360–385. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Barik, S.R.; Mohanty, S.P.; Anandan, A.; Reddy, J.N. Characterization of morpho-quality traits and validation of bacterial blight resistance in pyramided rice genotypes under various hot spots of India. Aust. J. Crop Sci. 2015, 9, 127–134. [Google Scholar]
- Mishra, A.; Barik, S.R.; Pandit, E.; Yadav, S.S.; Das, S.R.; Pradhan, S.K. Genetics, Mechanisms and Deployment of Brown Planthopper Resistance Genes in Rice. Crit. Rev. Plant Sci. 2022, 41, 91–127. [Google Scholar] [CrossRef]
- Singh, S.; Pradhan, S.K.; Singh, A.K.; Singh, O.N. Marker validation in recombinant in bred lines and random varieties of rice for drought tolerance. Aust. J. Crop Sci. 2012, 6, 606–612. [Google Scholar]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Naveenkumar, R.; Barik, S.R.; Mohanty, S.P.; Nayak, D.K.; Ghritlahre, S.K.; Sanjiba, R.D.; Reddy, J.N.; et al. Linkage disequilibrium mapping for grain Fe and Zn enhancing QTLs useful for nutrient dense rice breeding. BMC Plant Biol. 2020, 20, 57. [Google Scholar] [CrossRef]
- Barik, S.R.; Pandit, E.; Pradhan, S.K.; Singh, S.; Swain, P.; Mohapatra, T. QTL mapping for relative water content trait at reproductive stage drought stress in rice. Indian J Genet. 2018, 78, 401–408. [Google Scholar]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Pandit, E.; Sahoo, A.; Panda, R.K.; Mohanty, D.P.; Pani, D.R.; Anandan, A.; Pradhan, S.K. Survey of rice cultivars and landraces of upland ecology for phosphorous uptake 1(pup1) QTL using linked and gene specific molecular markers. Oryza 2016, 53, 1–9. [Google Scholar]
- Pandit, E.; Panda, R.K.; Pani, D.R.; Chandra, R.; Singh, S.; Pradhan, S.K. Molecular marker and phenotypic analyses for low phosphorus stress tolerance in cultivars and landraces of upland rice under irrigated and drought situations. Indian J. Genet. Plant Breed. 2018, 78, 59–68. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Bharati, B.; Chatopadhyay, K.; Singh, S.; Dash, P.; Reddy, J.N. Association mapping reveals multiple QTLs for grain protein content in rice useful for biofortification. Mol. Genet. Genom. 2019, 294, 963–983. [Google Scholar] [CrossRef]
- Barik, S.R.; Pandit, E.; Sanghamitra, P.; Mohanty, S.P.; Behera, A.; Mishra, J.; Nayak, D.K.; Bastia, R.; Moharana, A.; Sahoo, A. Unraveling the genomic regions controlling the seed vigour index, root growth parameters and germination per cent in rice. PLoS ONE 2022, 17, e0267303. [Google Scholar] [CrossRef]
- Pradhan, K.C.; Pandit, E.; Mohanty, S.P.; Moharana, A.; Sanghamitra, P.; Meher, J.; Jena, B.K.; Dash, P.K.; Behera, L.; Mohapatra, P.M.; et al. Development of Broad Spectrum and Durable Bacterial Blight Resistant Variety through Pyramiding of Four Resistance Genes in Rice. Agronomy 2022, 12, 1903. [Google Scholar] [CrossRef]
- Sanghamitra, P.; Barik, S.R.; Bastia, R.; Mohanty, S.P.; Pandit, E.; Behera, A.; Mishra, J.; Kumar, G.; Pradhan, S.K. Detection of Genomic Regions Controlling the Antioxidant Enzymes, Phenolic Content, and Antioxidant Activities in Rice Grain through Association Mapping. Plants 2022, 11, 1463. [Google Scholar] [CrossRef]
- Wu, K.S.; Tanksley, S.D. Abundance, polymorphism and genetic mapping of micro satellites in rice. Mol. Gen. Genet. 1993, 241, 225–235. [Google Scholar] [CrossRef]
- Chen, X.; Temnykh, S.; Xu, Y.; Cho, Y.G.; McCouch, S.R. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Teor. Appl. Genet. 1997, 95, 553–567. [Google Scholar] [CrossRef]
- Chu, Z.; Fu, B.; Yang, H.; Xu, C.; Li, Z.; Sanchez, A.; Park, Y.J.; Bennetzen, J.L.; Zhang, Q.; Wang, S. Targeting xa13, a recessive gene for bacterial blight resistance in rice. Teor. Appl. Genet. 2006, 112, 455–461. [Google Scholar] [CrossRef]
- Hajira, S. Development durable bacterial blight resistant lines of Samba Mahsuri possessing Xa33, Xa21, Xa13 & Xa5. Prog. Res. 2014, 9, 1224–1227. [Google Scholar]
- Huang, N.; Angeles, E.; Domingo, J.; Magpantay, G.; Singh, S.; Zhang, G.; Kumaravadivel, N.; Bennett, J.; Khush, G. Pyramiding of bacterial blight resistance genes in rice: Marker assisted selection using RFLP and PCR. Teor. Appl. Genet. 1997, 95, 313–320. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Barik, S.R.; Sahoo, J.; Pandit, E.; Nayak, D.K.; Pani, D.R.; Anandan, A. Comparison of Sub1 markers and their combinations for submergence tolerance and analysis of adaptation strategies of rice in rainfed lowland ecology. Comptes. Rendus. Biol. 2015, 338, 650–659. [Google Scholar] [CrossRef]
- IRRI. Standard evaluation system for rice. In International Network for Genetic Evaluation of Rice, 5th ed.; International Rice Research Institute: Los Banos, CA, USA, 2014. [Google Scholar]
- Nayak, D.K.; Pandit, E.; Mohanty, S.; Barik, D.P.; Pradhan, S.K. Marker-assisted selection in back cross progenies for transfer of bacterial leaf blight resistance genes into a popular lowland rice cultivar. Oryza 2015, 52, 163–172. [Google Scholar]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Behera, L.; Anandan, A.; Mukherjee, A.K.; Lenka, S.; Barik, D.P. Incorporation of bacterial blight resistance genes into lowland rice cultivar through marker-assisted backcross breeding. Phytopathology 2016, 106, 710–718. [Google Scholar] [CrossRef]
- Pradhan, K.C.; Barik, S.R.; Mohapatra, S.; Nayak, D.K.; Pandit, E.; Jena, B.K.; Sangeeta, S.; Pradhan, A.; Samal, A.; Meher, J.; et al. Incorporation of Two Bacterial Blight Resistance Genes into the Popular Rice Variety, Ranidhan through Marker-Assisted Breeding. Agriculture 2022, 12, 1287. [Google Scholar] [CrossRef]
- Tan, Y.F.; Li, J.X.; Yu, S.B.; Xing, Y.Z.; Xu, C.G.; Zhang, Q.F. The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor. Appl. Genet. 1999, 99, 642–648. [Google Scholar] [CrossRef]
- Juliano, B.O. Rice Quality Screening with the Rapid Visco Analyser; Walker, C.E., Hazelton, J.L., Eds.; Newport Scientific: Sydney, Australia, 1996; pp. 19–24. [Google Scholar]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- International Rice Research Institute (IRRI). CropStat for Windows 7.2; IRRI: Manila, Philippines, 2007. [Google Scholar]
- Barik, S.R.; Pandit, E.; Pradhan, S.K.; Mohanty, S.P.; Mohapatra, T. Genetic mapping of morpho-physiological traits involved during reproductive stage drought tolerance in rice. PLoS ONE 2019, 14, e0214979. [Google Scholar] [CrossRef]
- Pandit, E.; Panda, R.K.; Sahoo, A.; Pani, D.R.; Pradhan, S.K. Genetic Relationship and Structure Analysis of Root Growth Angle for Improvement of Drought Avoidance in Early and Mid-Early Maturing Rice Genotypes. Rice Sci. 2020, 27, 124–132. [Google Scholar] [CrossRef]
- Pawar, S.; Pandit, E.; Mohanty, I.; Saha, D.; Pradhan, S.K. Population genetic structure and association mapping for iron toxicity tolerance in rice. PLoS ONE 2021, 16, e0246232. [Google Scholar] [CrossRef]
- Nayak, D.K.; Sahoo, S.; Barik, S.R.; Sanghamitra, P.; Sangeeta, S.; Pandit, E.; Reshmi Raj, K.R.; Basak, N.; Pradhan, S.K. Association mapping for protein, total soluble sugars, starch, amylose and chlorophyll content in rice. BMC Plant Biol. 2022, 22, 620. [Google Scholar] [CrossRef]
- Dokku, P.; Das, K.; Rao, G. Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker-assisted selection. Euphytica 2013, 192, 87–96. [Google Scholar] [CrossRef]
- Arunakumari, K.; Durgarani, C.V.; Satturu, V.; Sarikonda, K.R.; Chittoor, P.D.R.; Vutukuri, B.; Laha, G.S.; Nelli, A.P.K.; Gattu, S.; Jamal, M.; et al. Marker-Assisted Pyramiding of Genes Conferring Resistance Against Bacterial Blight and Blast Diseases into Indian Rice Variety MTU1010. Rice Sci. 2016, 23, 306–316. [Google Scholar] [CrossRef]
- Neeraja, C.N.; Maghirang-Rodriguez, R.; Pamplona, A.; Heuer, S.; Collard, B. A marker-assisted backcross approach for developing submergence tolerant rice cultivars. Theor. Appl. Genet. 2007, 115, 767–776. [Google Scholar] [CrossRef]
- Ifekharuddaula, K.M. Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into BR11, the rainfed lowland rice mega variety of Bangladesh. Euphytica 2012, 178, 83–97. [Google Scholar] [CrossRef]
- Echeverri-Rico, J.; Petro, E.; Fory, P.A.; Mosquera, G.M.; Lang, J.M.; Leach, J.E.; Lobaton, J.D.; Garcés, G.; Perafán, R.; Amezquita, N.; et al. Understanding the complexity of disease-climate interactions for rice bacterial panicle blight under tropical conditions. PLoS ONE 2021, 16, e0252061. [Google Scholar] [CrossRef]
- Niones, J.T.; Sharp, R.T.; Donayre, D.K.M. Dynamics of bacterial blight disease in resistant and susceptible rice varieties. Eur. J. Plant Pathol. 2022, 163, 1–17. [Google Scholar] [CrossRef]
- Azharudheen, T.P.; Sah, R.P.; Moharana, D.; Behera, S.; Pradhan, S.K. Genetic Improvement of Rice for Lowlands; ICAR-National Rice Research Institute: Cuttack, India, 2021; pp. 63–85. [Google Scholar]
- Sundaram, R.; Vishnupriya, M.; Biradar, S.; Laha, G.S.; Reddy, G.; Rani, N.; Sarma, N.; Sonti, R. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 2007, 160, 411–422. [Google Scholar] [CrossRef]
- Suh, J.P.; Jeung, J.U.; Noh, T.H.; Cho, Y.C.; Park, S.H.; Park, H.S.; Shin, M.S.; Kim, C.K.; Jena, K.K. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 2013, 6, 5. [Google Scholar] [CrossRef]
- Das, G.; Rao, G. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015, 6, 698. [Google Scholar] [CrossRef]
- Rajpurohit, D.; Kumar, R.; Kumar, M.; Paul, P.; Awasthi, A.; Basha, P.O.; Puri, A.; Jhang, T.; Singh, K.; Dhaliwal, H.S. Pyramiding of two bacterial blight resistance and a semi dwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica 2011, 178, 111–126. [Google Scholar] [CrossRef]
- Bhasin, H.; Bhatia, D.; Raghuvanshi, S.; Lore, J.S.; Sahi, G.K.; Kaur, B.; Vikal, Y.; Singh, K. New PCR-based sequence-tagged site marker for bacterial blight resistance gene Xa38 of rice. Mol. Breed. 2012, 30, 607–611. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Das, S.R.; Patra, B.C. Advances in Rice Breeding: Stress Tolerance, Climate Resilience, Quality & High Yield; ICAR-National Rice Research Institute: Cuttack, India, 2021; pp. 1–426. [Google Scholar]
- Mohapatra, S.; Bastia, A.K.; Panda, A.K.; Pradhan, S.K. Marker-assisted selection for transfer of submergence tolerance, bacterial blight resistance and yield enhancement in the rice backcross derivatives. Aust. J. Crop Sci. 2020, 14, 1288–1294. [Google Scholar] [CrossRef]

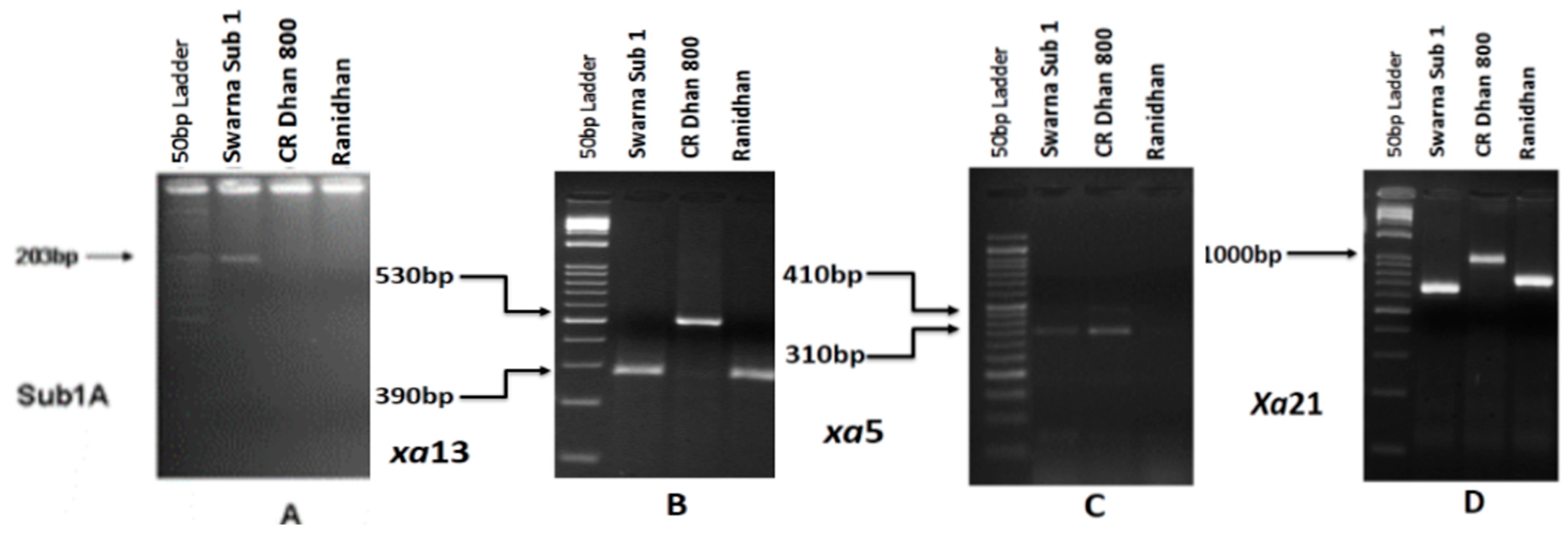

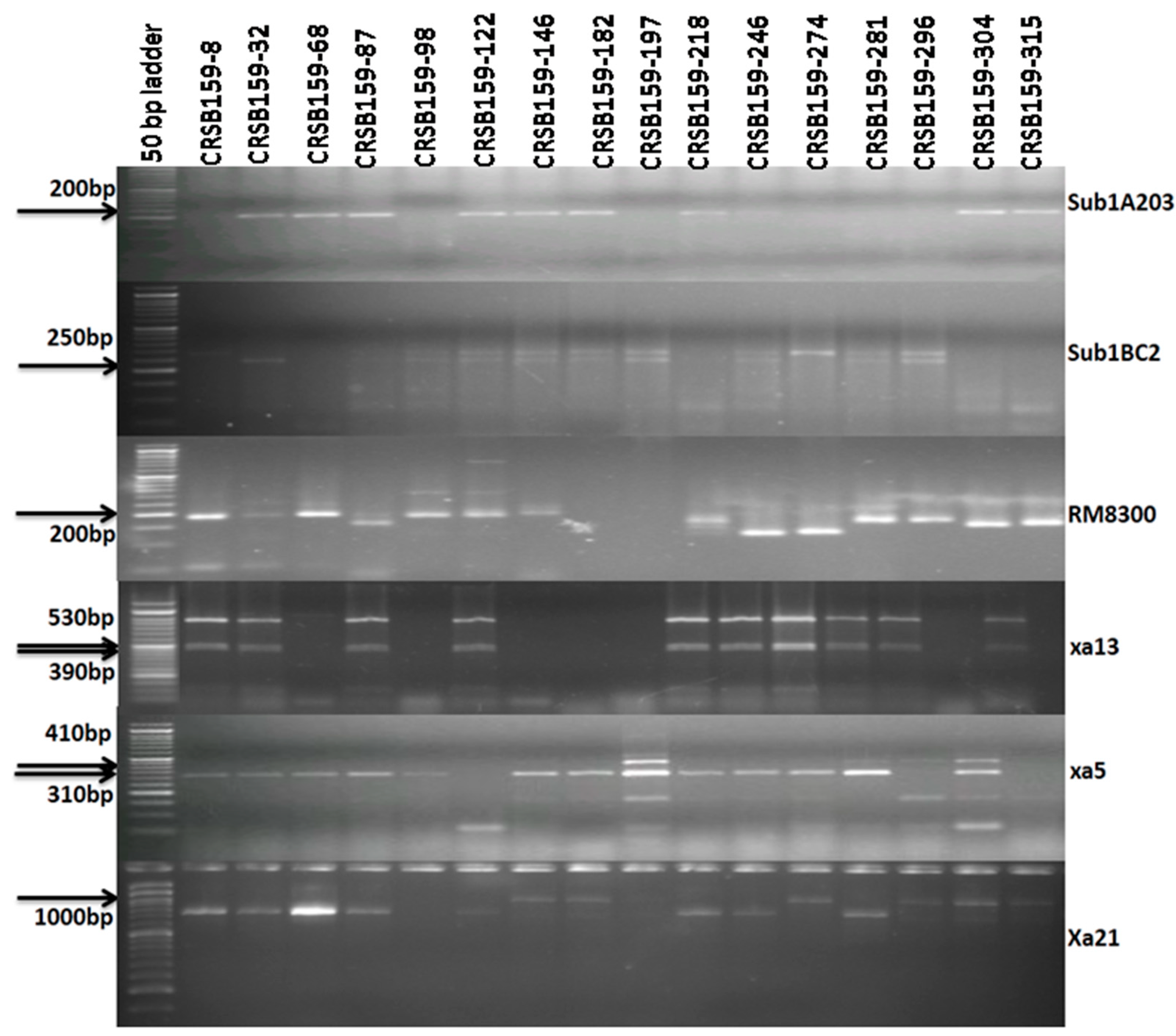
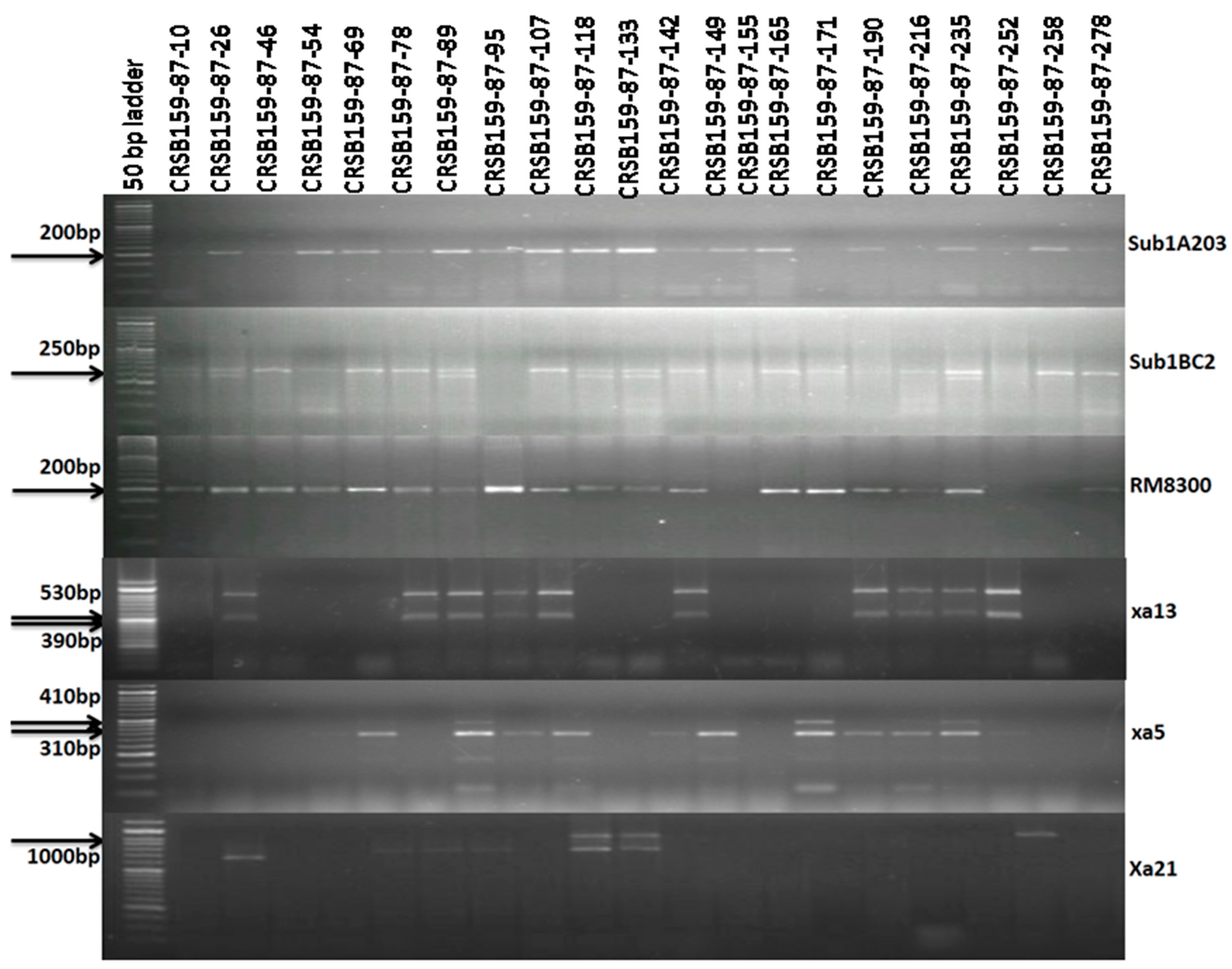



| Resistance Gene | Chromosome Number | Marker | Primer Sequence Used for Gene Detection | Expected Size (bp) | Marker Type | ||
|---|---|---|---|---|---|---|---|
| Forward (5′–3′) | Reverse (5′–3′) | ||||||
| xa5 | 5 | RM 122 | GAGTCGATGTAATGTCATCAGTGC | GAAGGAGGTATCGCTTTGTTGGAC | 260 bp | SSR | [21,22] |
| xa5S (multiplex) xa5SR/R (multiplex) | GTCTGGAATTTGCTCGCGTTCG AGCTCGCCATTCAAGTTCTTGAG | TGGTAAAGTAGATACCTTATCAAACTGGA TGACTTGGTTCTCCAAGGCTT | 160 bp | STS | [7] | ||
| xa13 | 8 | Xa13 prom | TCCCAGAAAGCTACTACAGC | GCAGACTCCAGTTTGACTTC | 500 bp | STS | [23,24] |
| Xa21 | 11 | pTA248 | AGACGCGGAAGGGTGGTTCCCGGA | AGACGCGGTAATCGAAGATGAAA | 1000 bp | STS | [25] |
| Sub1 | 9 | Sub1A203 | CTTCTTGCTCAACGACAACG | AGGCTCCAGATGTCCATGTC | 200 bp | STS | [26] |
| Sub1BC2 | AAAACAATGGTTCCATACGAGAC | GCCTATCAATGCGTGCTCTT | 240 bp | SRS | [26] | ||
| RM 8300 | GCTAGTGCAGGGTTGACACA | CTCTGGCCGTTTCATGGTAT | 205 bp | SSR | [26] | ||
| Sl. No. | Name of the Genotypes | % Survival after De-Submergence | Remarks |
|---|---|---|---|
| 1 | CRSB 159-87-69-285 | 87.5 | Tolerant |
| 2 | CRSB 159-87-69-546 | 85.8 | Tolerant |
| 3 | CRSB 159-87-69-717 | 89.2 | Tolerant |
| 4 | CRSB 159-87-69-942 | 94.5 | Tolerant |
| 5 | CRSB 159-87-69-267 | 81.7 | Tolerant |
| 6 | CRSB 159-87-69-636 | 75.8 | Tolerant |
| 7 | CRSB 159-87-69-915 | 70.6 | Tolerant |
| 8 | CR Dhan 800 | 0 | Susceptible |
| 9 | Swarna-Sub1 | 87.0 | Tolerant |
| 10 | Ranidhan | 0 | Susceptible |
| Sl. No. | Pyramided Lines | Gene Combination | Mean Lesion Length (MLL) in cm (Mean ± Standard Error) | Disease Reaction | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xoo Strains Inoculated | ||||||||||||
| Xa17 | Xa7 | xa2 | xb7 | xc4 | xd1 | xa1 | xa5 | MLL | ||||
| 1 | CRSB 159-87-69-285 | xa5+xa13+Xa21 | 2.03 ± 0.88 | 2.09 ± 0.86 | 2.16 ± 0.79 | 2.18 ± 0.77 | 2.31 ± 0.59 | 2.41 ± 0.56 | 2.36 ± 0.59 | 2.33 ± 0.62 | 2.23 ± 0.71 | R |
| 2 | CRSB 159-87-69-546 | xa5+xa13+Xa21 | 2.20 ± 0.75 | 2.25 ± 0.70 | 2.23 ± 0.63 | 2.13 ± 0.77 | 2.26 ± 0.59 | 2.36 ± 0.56 | 2.31 ± 0.59 | 2.28 ± 0.62 | 2.25 ± 0.65 | R |
| 3 | CRSB 159-87-69-717 | xa5+xa13+Xa21 | 2.05 ± 0.90 | 2.08 ± 0.84 | 2.13 ± 0.74 | 2.16 ± 0.69 | 2.21 ± 0.59 | 2.26 ± 0.56 | 2.23 ± 0.59 | 2.18 ± 0.62 | 2.16 ± 0.69 | R |
| 4 | CRSB 159-87-69-942 | xa5+xa13+Xa21 | 2.10 ± 0.85 | 2.05 ± 0.80 | 2.08 ± 0.82 | 2.11 ± 0.74 | 2.14 ± 0.59 | 2.21 ± 0.56 | 2.18 ± 0.59 | 2.23 ± 0.62 | 2.14 ± 0.70 | R |
| 5 | CRSB 159-87-69-267 | xa5 | 6.08 ± 0.87 | 6.17 ± 0.82 | 6.13 ± 0.72 | 6.26 ± 0.64 | 6.23 ± 0.52 | 6.28 ± 0.53 | 6.18 ± 0.52 | 5.93 ± 0.57 | 6.07 ± 0.65 | MS |
| 6 | CRSB 159-87-69-636 | Xa13 | 6.13 ± 0.92 | 6.21 ± 0.89 | 6.45 ± 0.75 | 6.45 ± 0.70 | 6.43 ± 0.63 | 6.33 ± 0.62 | 6.46 ± 0.69 | 6.38 ± 0.67 | 6.34 ± 0.73 | MS |
| 7 | CRSB 159-87-69-915 | Xa21 | 4.83 ± 1.08 | 4.78 ± 1.02 | 4.64 ± 1.11 | 4.84 ± 1.01 | 5.03 ± 0.73 | 5.13 ± 0.68 | 4.93 ± 0.73 | 5.10 ± 0.70 | 4.91 ± 0.88 | MR |
| 8 | CR Dhan 800 | xa5+xa13+Xa21 | 1.88 ± 0.42 | 1.83 ± 0.40 | 1.85 ± 0.30 | 1.97 ± 0.32 | 2.13 ± 0.28 | 2.03 ± 0.27 | 1.93 ± 0.28 | 1.84 ± 0.31 | 1.93 ± 0.32 | R |
| 9 | Swarna-Sub1 | - | 10.13 ± 1.17 | 10.13 ± 1.12 | 10.11 ± 1.04 | 10.12 ± 0.93 | 10.33 ± 0.73 | 10.23 ± 0.72 | 10.13 ± 0.72 | 10.70 ± 0.70 | 10.24 ± 0.89 | S |
| 10 | Ranidhan | - | 9.50 ± 1.65 | 9.83 ± 1.62 | 9.80 ± 1.45 | 9.78 ± 1.47 | 10.08 ± 1.18 | 9.88 ± 1.18 | 9.86 ± 1.16 | 9.73 ± 1.17 | 9.81 ± 1.39 | S |
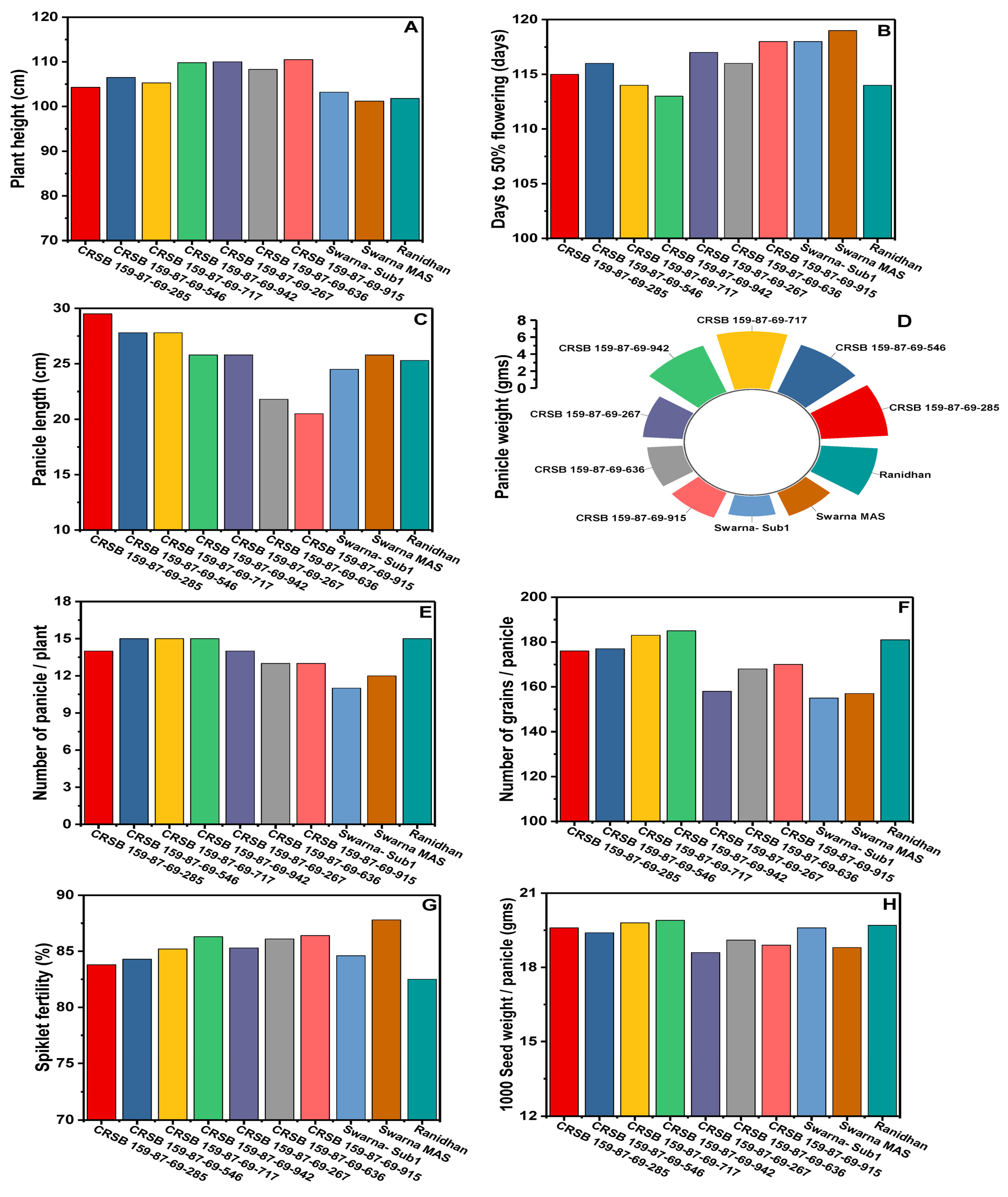
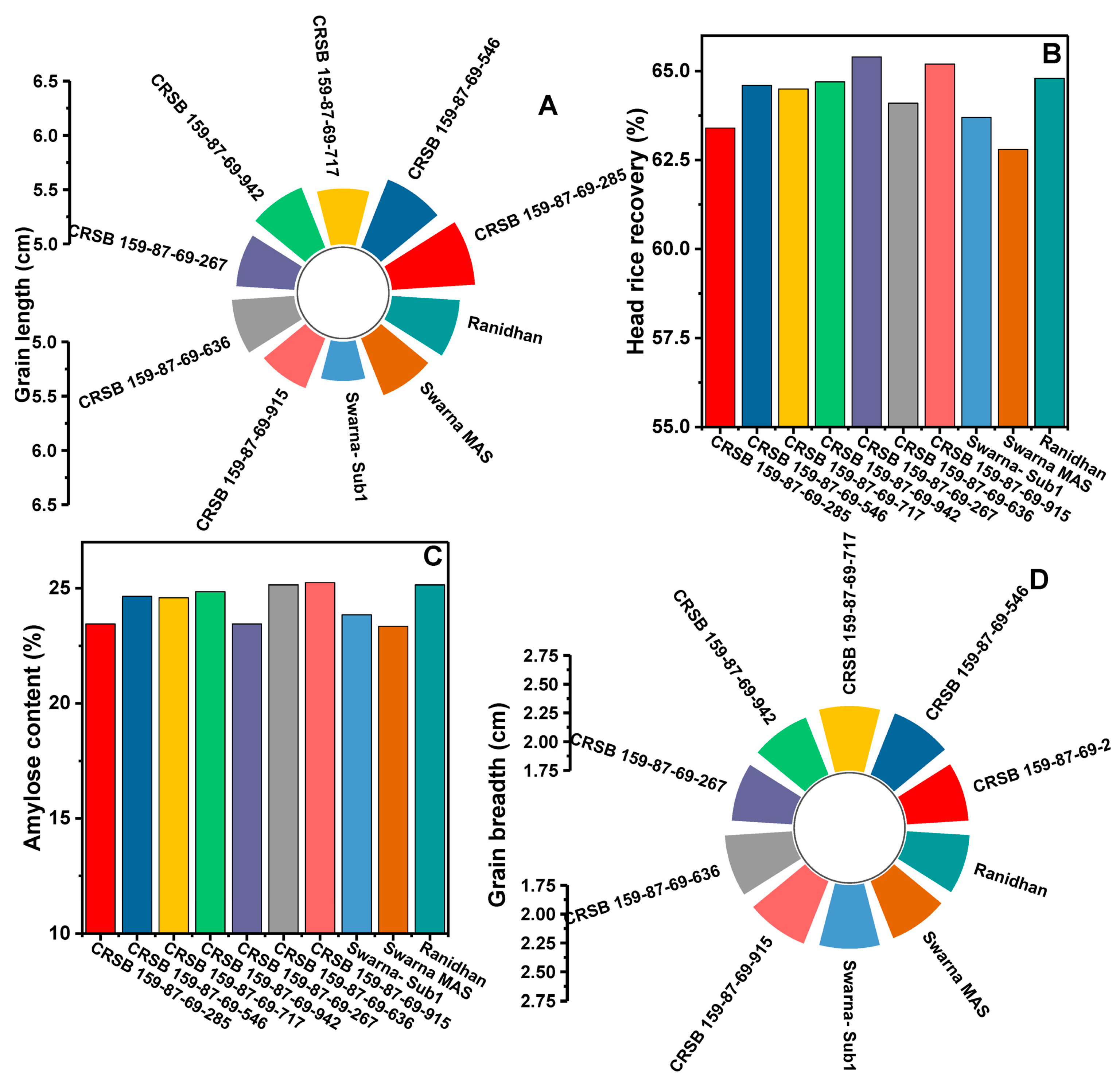
| Sl. No. | Pyramided Lines | PH (cm) | DFF (day) | PL (cm) | PW (g) | NPP | NGP | SW (g) | SF (%) | GL (cm) | GB (cm) | HRR (%) | AC (%) | PY (q/ha) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRSB 159-87-69-285 | 104.3 | 115 | 29.5 | 6.15 | 14 | 176 | 19.6 | 83.8 | 5.76 | 2.28 | 63.4 | 23.45 | 67.65 |
| 2 | CRSB 159-87-69-546 | 106.5 | 116 | 27.8 | 5.94 | 15 | 177 | 19.4 | 84.3 | 5.67 | 2.32 | 64.6 | 24.65 | 68.45 |
| 3 | CRSB 159-87-69-717 | 105.3 | 114 | 27.8 | 6.68 | 15 | 183 | 19.8 | 85.2 | 5.51 | 2.31 | 64.5 | 24.59 | 69.90 |
| 4 | CRSB 159-87-69-942 | 109.8 | 113 | 25.8 | 5.85 | 15 | 185 | 19.9 | 86.3 | 5.59 | 2.28 | 64.7 | 24.85 | 71.70 |
| 5 | CRSB 159-87-69-267 | 110.0 | 117 | 25.8 | 3.65 | 14 | 158 | 18.6 | 85.3 | 5.53 | 2.27 | 65.4 | 23.45 | 63.50 |
| 6 | CRSB 159-87-69-636 | 108.3 | 116 | 21.8 | 3.25 | 13 | 168 | 19.1 | 86.1 | 5.57 | 2.33 | 64.1 | 25.15 | 60.45 |
| 7 | CRSB 159-87-69-915 | 110.5 | 118 | 20.5 | 3.15 | 13 | 170 | 18.9 | 86.4 | 5.49 | 2.33 | 65.2 | 25.25 | 63.65 |
| 8 | Swarna- Sub1 | 103.2 | 118 | 24.5 | 2.34 | 11 | 155 | 19.6 | 84.6 | 5.36 | 2.30 | 63.7 | 23.85 | 60.03 |
| 9 | Swarna MAS | 101.2 | 119 | 25.8 | 2.92 | 12 | 157 | 18.8 | 87.8 | 5.56 | 2.28 | 62.8 | 23.35 | 65.60 |
| 10 | Ranidhan | 101.8 | 114 | 25.3 | 5.20 | 15 | 181 | 19.7 | 82.5 | 5.62 | 2.29 | 64.8 | 25.15 | 58.90 |
| CD5% df | 9.57 | 3.23 | 2.47 | 0.75 | 2.18 | 44.57 | 3.07 | 7.84 | 0.25 | 0.07 | 7.614 | 2.816 | 12.13 | |
| CV (%) | 5.45 | 1.69 | 5.87 | 10.03 | 8.49 | 10.58 | 6.68 | 9.52 | 2.66 | 1.81 | 9.428 | 6.923 | 11.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohapatra, S.; Barik, S.R.; Dash, P.K.; Lenka, D.; Pradhan, K.C.; Raj K. R, R.; Mohanty, S.P.; Mohanty, M.R.; Sahoo, A.; Jena, B.K.; et al. Molecular Breeding for Incorporation of Submergence Tolerance and Durable Bacterial Blight Resistance into the Popular Rice Variety ‘Ranidhan’. Biomolecules 2023, 13, 198. https://doi.org/10.3390/biom13020198
Mohapatra S, Barik SR, Dash PK, Lenka D, Pradhan KC, Raj K. R R, Mohanty SP, Mohanty MR, Sahoo A, Jena BK, et al. Molecular Breeding for Incorporation of Submergence Tolerance and Durable Bacterial Blight Resistance into the Popular Rice Variety ‘Ranidhan’. Biomolecules. 2023; 13(2):198. https://doi.org/10.3390/biom13020198
Chicago/Turabian StyleMohapatra, Shibani, Saumya Ranjan Barik, Prasanta K. Dash, Devidutta Lenka, Kartika Chandra Pradhan, Reshmi Raj K. R, Shakti Prakash Mohanty, Mihir Ranjan Mohanty, Ambika Sahoo, Binod Kumar Jena, and et al. 2023. "Molecular Breeding for Incorporation of Submergence Tolerance and Durable Bacterial Blight Resistance into the Popular Rice Variety ‘Ranidhan’" Biomolecules 13, no. 2: 198. https://doi.org/10.3390/biom13020198
APA StyleMohapatra, S., Barik, S. R., Dash, P. K., Lenka, D., Pradhan, K. C., Raj K. R, R., Mohanty, S. P., Mohanty, M. R., Sahoo, A., Jena, B. K., Panda, A. K., Panigrahi, D., Dash, S. K., Meher, J., Sahoo, C. R., Mukherjee, A. K., Das, L., Behera, L., & Pradhan, S. K. (2023). Molecular Breeding for Incorporation of Submergence Tolerance and Durable Bacterial Blight Resistance into the Popular Rice Variety ‘Ranidhan’. Biomolecules, 13(2), 198. https://doi.org/10.3390/biom13020198









