Systemic Metabolomic Profiles in Adult Patients with Bacterial Sepsis: Characterization of Patient Heterogeneity at the Time of Diagnosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
- Our present study included 60 patients with bacterial sepsis; 30 patients had infections with Gram-positive and 30 patients with Gram-negative bacteria. We wanted to include 30 patients in each of these two groups to allow reliable bioinformatical comparisons between various patient subsets. Thus, our present study included only 60 out of the 80 patients in the original study that fulfilled the Sepsis-2 SOFA score-based criteria [21]. The 20 excluded patients were:
- Five patients with mixed infections, i.e., evidence for two infecting bacteria.
- Eight patients with exceptional bacterial etiology, i.e., one patient each with Enterobacter cloacae, Acinubaculum schalii, Bacteroides fragilis, Fusobacterium necroforum, Kingella kingae, Neiseria meningitidis, Klebsiella pneumoniae and Clostridium infection.
- Six (randomly selected from eight) patients for which the bacterial diagnosis was based on detection of bacterial antigens alone but with no bacterial growth for any patient samples.
- One randomly selected patient with Gram-negative infection (Escherichia coli blood culture) that was removed so that we had two equal groups with 30 patients with Gram-negative and Gram-positive infection, separately.
2.2. Metabolomic Analyses
2.3. Statistical and Bioinformatical Analyses
3. Results
3.1. Identified Lipid Metabolites in Serum Samples Derived from Sepsis Patients
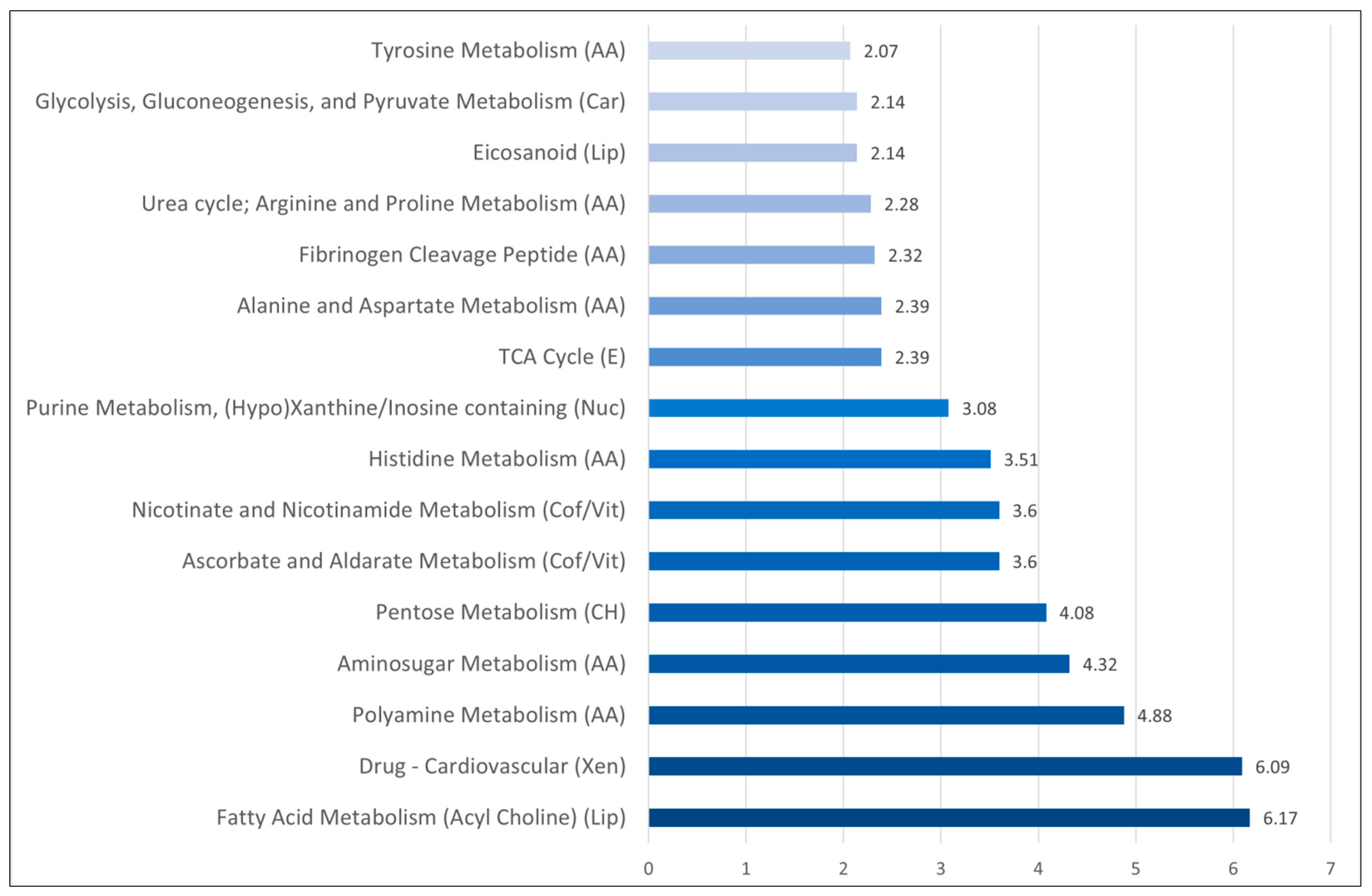
3.2. Comparison of Metabolomic Profiles for Sepsis-3 Patients versus Patients only Fulfilling the Criteria for Sepsis 2: Differences of Amino Acid Metabolite Levels Is a Main Characteristic
- Several terms reflecting altered amino acid metabolism also received a high score in this analysis, including a specific amino acid metabolism, as well as the urea cycle and polyamine metabolism.
- The differences in fibrinogen cleavage peptides possibly reflect acute inflammation and the acute phase reaction.
- There were differences in fatty acid metabolism (acyl cholines, eicosanoid).
- There was a difference in several xenobiotics and especially cardiovascular drugs; this reflects the cardiovascular comorbidity of a Sepsis-3 patient subset.
- There were also differences in pentose/aminosugar metabolism and glycolysis/gluconeogenesis/pyruvate metabolism suggesting that the carbohydrate metabolism is altered together with the amino acid metabolism.
- Vitamin and cofactor metabolism showed differences in nicotinate and nicotinamide metabolism as well as ascorbate and aldarate metabolism.
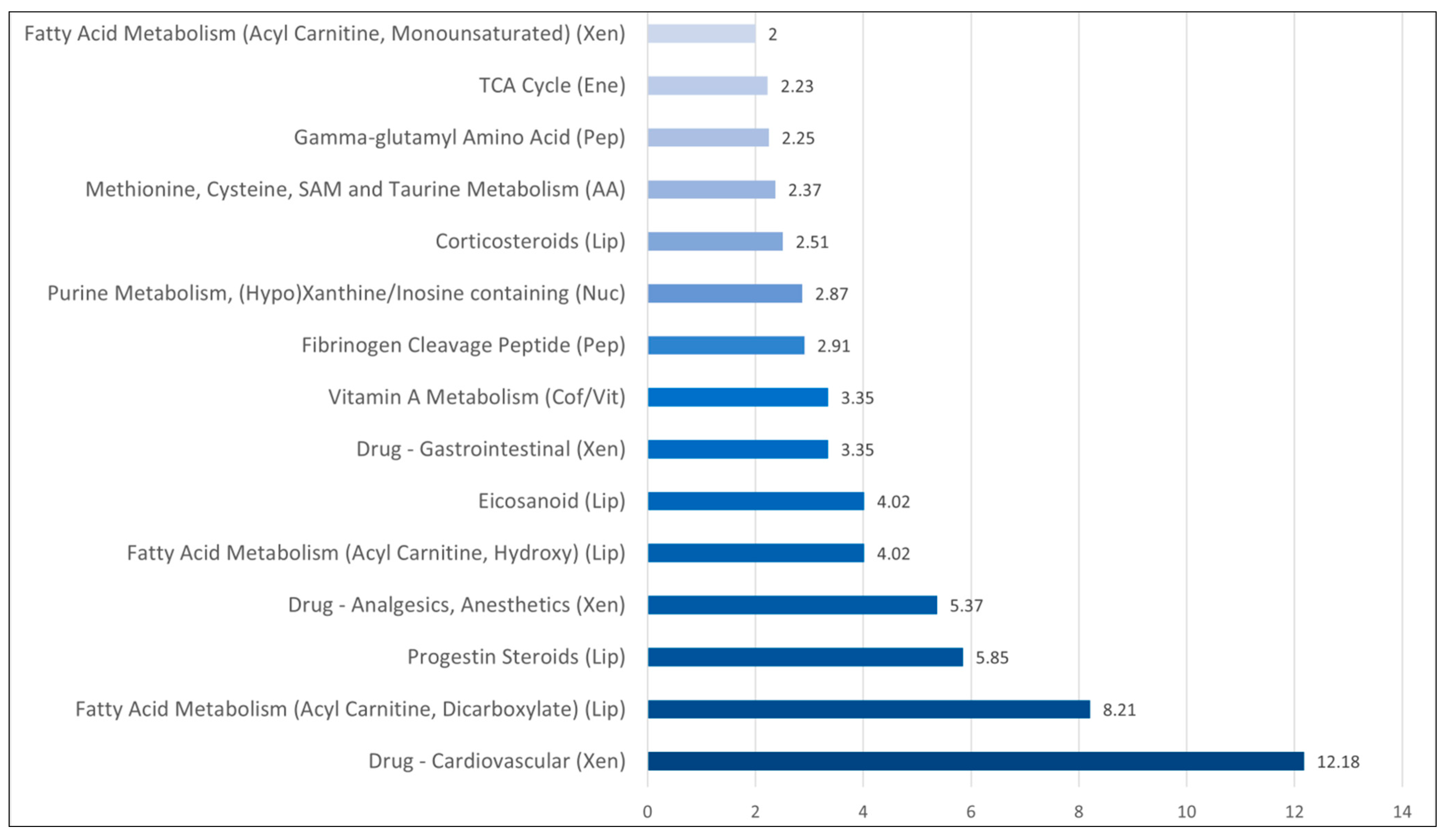
3.3. Comparison of Metabolomic Profiles for Sepsis Patients with and without Bacteremia: Differences in Systemic Levels of Lipid and Amino Acid Metabolites Are Main Characteristics
3.4. Comparison of Metabolomic Profiles for Patients with Gram-Negative and Gram-Positive Infections: Only Weak Associations Are Detected
3.5. Metabolic Heterogeneity of Sepsis Patients: A Clustering Analysis Based on Amino Acid Metabolites Showing a Strong Correlation with the Total SOFA Score
3.6. A Minor Subset of Lipid Metabolites Show a Strong Correlation with Total SOFA Score
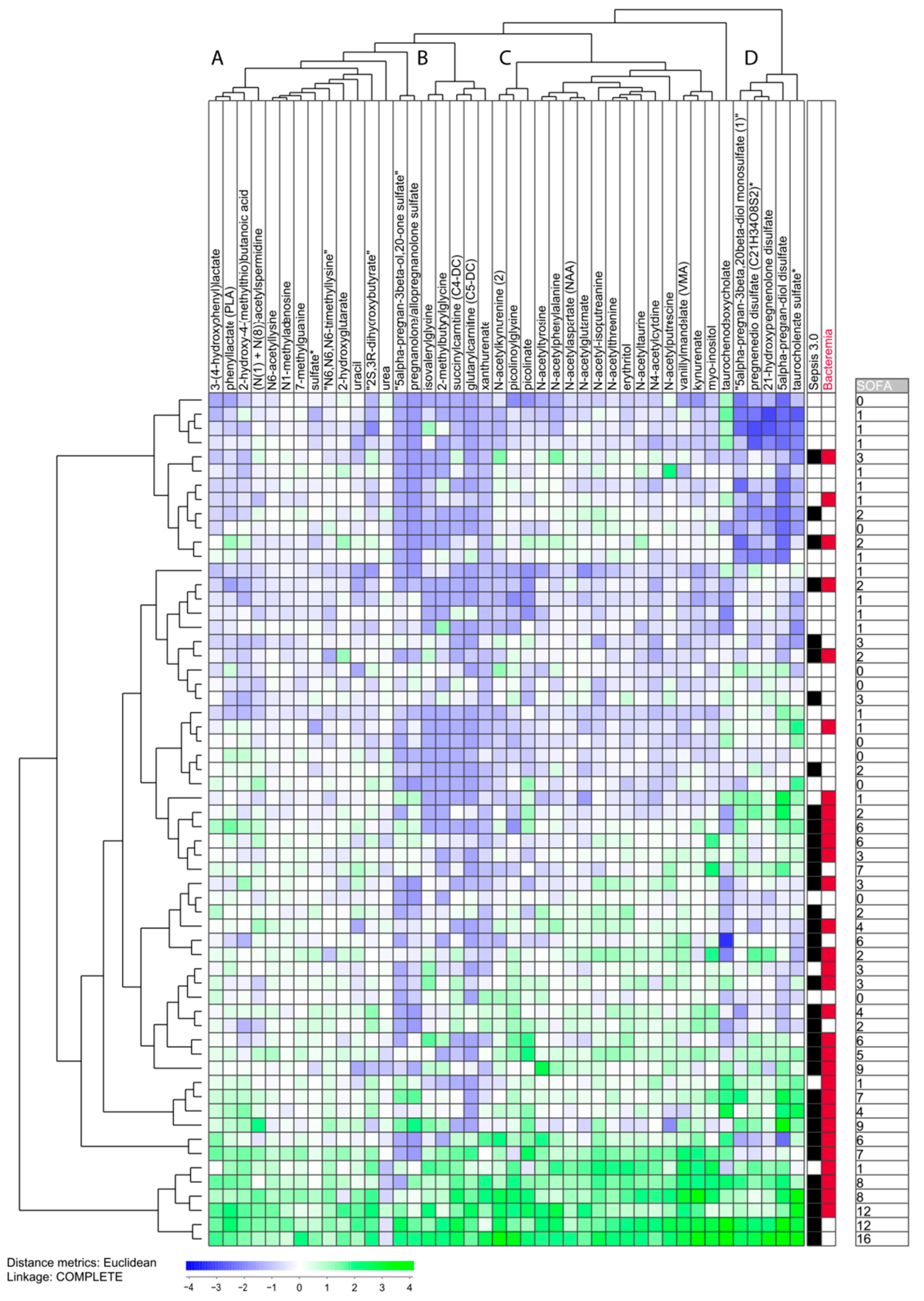
3.7. The Heterogeneity of Sepsis Patients Characterized by Unsupervised Hierarchical Clustering Analysis Based on Heterogeneous Metabolites Showing Strong Association with Total SOFA Score
- Only a minority of our sepsis patients showed increased levels of most of the 42 metabolites; these patients clustered close to each other and represent the eight patients in the lowest part of the cluster analysis.
- All the other 52 patients showed relatively low levels for several of the 42 metabolites, and especially low levels of metabolites belonging to metabolite cluster B (middle left; see Table S7), which seems to be a common characteristic both for the other Sepsis-3 patients and patients only fulfilling the Sepsis-2 criteria. Cluster B is a small and heterogeneous cluster including seven metabolites (four of them being amino acid metabolites).
- The patients in the upper part of the diagram also showed generally low levels for metabolite clusters A and C. Cluster A (left) is a relatively large and heterogeneous metabolite cluster whereas cluster C (middle right) includes 15 metabolites and 12 of them are amino acid metabolites.
- Cluster D (right) includes only lipid metabolites and decreased levels were seen, especially for the upper 12 patients.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Cecconi, M.; Evans, L.; Levy, M.; Rhodes, A. Sepsis and septic shock. Lancet 2018, 392, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Gotts, J.E.; Matthay, M.A. Sepsis: Pathophysiology and clinical management. BMJ 2016, 353, i1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, N.M.; Boothby, M.R.; Chi, H. Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 2020, 20, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Maciolek, J.A.; Pasternak, J.A.; Wilson, H.L. Metabolism of activated T lymphocytes. Curr. Opin. Immunol. 2014, 27, 60–74. [Google Scholar] [CrossRef]
- 6Geltink, R.I.K.; Kyle, R.L.; Pearce, E.L. Unraveling the Complex Interplay Between T Cell Metabolism and Function. Annu. Rev. Immunol. 2018, 36, 461–488. [Google Scholar] [CrossRef]
- Yan, J.; Horng, T. Lipid Metabolism in Regulation of Macrophage Functions. Trends Cell Biol. 2020, 30, 979–989. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, K.; Yang, L.; Liu, R.; Chu, Y.; Qin, X.; Yang, P.; Yu, H. Lipid metabolism in inflammation-related diseases. Analyst 2018, 143, 4526–4536. [Google Scholar] [CrossRef]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [Green Version]
- Gaber, T.; Strehl, C.; Buttgereit, F. Metabolic regulation of inflammation. Nat. Rev. Rheumatol. 2017, 13, 267–279. [Google Scholar] [CrossRef]
- 11Arts, R.J.W.; Gresnigt, M.S.; Joosten, L.A.B.; Netea, M.G. Cellular metabolism of myeloid cells in sepsis. J. Leukoc. Biol. 2017, 101, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, G.; Riquelme, S.; Prince, A.; Avraham, R. Immunometabolic crosstalk during bacterial infection. Nat. Microbiol. 2022, 7, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Toller-Kawahisa, J.E.; O’Neill, L.A.J. How neutrophil metabolism affects bacterial killing. Open Biol. 2022, 12, 220248. [Google Scholar] [CrossRef]
- Bettencourt, I.A.; Powell, J.D. Targeting Metabolism as a Novel Therapeutic Approach to Autoimmunity, Inflammation, and Transplantation. J. Immunol. 2017, 198, 999–1005. [Google Scholar] [CrossRef] [Green Version]
- Galli, G.; Saleh, M. Immunometabolism of Macrophages in Bacterial Infections. Front. Cell. Infect. Microbiol. 2021, 10, 607650. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Aarstad, H.H.; Tvedt, T.H.A. Combined C-Reactive Protein and Novel Inflammatory Parameters as a Predictor in Cancer-What Can We Learn from the Hematological Experience? Cancers 2020, 12, 1966. [Google Scholar] [CrossRef] [PubMed]
- Hamar, P. A New Role of Acute Phase Proteins: Local Production Is an Ancient, General Stress-Response System of Mammalian Cells. Int. J. Mol. Sci. 2022, 23, 2972. [Google Scholar] [CrossRef]
- Beloborodova, N.V.; Olenin, A.Y.; Pautova, A.K. Metabolomic findings in sepsis as a damage of host-microbial metabolism integration. J. Crit. Care 2018, 43, 246–255. [Google Scholar] [CrossRef]
- Lee, J.; Banerjee, D. Metabolomics and the Microbiome as Biomarkers in Sepsis. Crit. Care Clin. 2020, 36, 105–113. [Google Scholar] [CrossRef]
- Lin, S.-H.; Fan, J.; Zhu, J.; Zhao, Y.-S.; Wang, C.-J.; Zhang, M.; Xu, F. Exploring plasma metabolomic changes in sepsis: A clinical matching study based on gas chromatography–mass spectrometry. Ann. Transl. Med. 2020, 8, 1568. [Google Scholar] [CrossRef] [PubMed]
- Mosevoll, K.A.; Skrede, S.; Markussen, D.L.; Fanebust, H.R.; Flaatten, H.K.; Aßmus, J.; Reikvam, H.; Bruserud, Ø. Inflam-matory Mediator Profiles Differ in Sepsis Patients with and Without Bacteremia. Front. Immunol. 2018, 9, 691. [Google Scholar] [CrossRef]
- Levy, M.M.; Fink, M.P.; Marshall, J.C.; Abraham, E.; Angus, D.; Cook, D.; Cohen, J.; Opal, S.M.; Vincent, J.-L.; Ramsay, G.; et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit. Care Med. 2003, 31, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Lambden, S.; Laterre, P.F.; Levy, M.M.; Francois, B. The SOFA score-development, utility and challenges of accurate as-sessment in clinical trials. Crit. Care 2019, 23, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerra, F.E.; Borgogna, T.R.; Patel, D.M.; Sward, E.W.; Voyich, J.M. Epic Immune Battles of History: Neutrophils vs. Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2017, 7, 286. [Google Scholar] [CrossRef] [Green Version]
- Koppe, U.; Suttorp, N.; Opitz, B. Recognition of Streptococcus pneumoniae by the innate immune system. Cell. Microbiol. 2012, 14, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Lee, J.-O. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 2013, 45, e66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarczak, D.; Kluge, S.; Nierhaus, A. Sepsis-Pathophysiology and Therapeutic Concepts. Front. Med. 2021, 8, 628302. [Google Scholar] [CrossRef] [PubMed]
- Leligdowicz, A.; Matthay, M.A. Heterogeneity in sepsis: New biological evidence with clinical applications. Crit Care 2019, 23, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janicova, A.; Relja, B. Neutrophil Phenotypes and Functions in Trauma and Trauma-Related Sepsis. Shock 2021, 56, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Lee, J.; Jung, Y.S.; Kwon, W.Y.; Oh, D.K.; Park, M.H.; Lim, C.-M.; Lee, S.-M. Preexisting Clinical Frailty Is Associated with Worse Clinical Outcomes in Patients with Sepsis. Crit. Care Med. 2020, 50, 780–790. [Google Scholar] [CrossRef]
- Marzetti, E.; Picca, A.; Marini, F.; Biancolillo, A.; Coelho-Junior, H.J., Jr.; Gervasoni, J.; Bossola, M.; Cesari, M.; Onder, G.; Landi, F.; et al. Inflammatory signatures in older persons with physical frailty and sarcopenia: The frailty “cytokinome” at its core. Exp. Gerontol. 2019, 122, 129–138. [Google Scholar] [CrossRef]
- Fernández-Garrido, J.; Navarro-Martínez, R.; Buigues-González, C.; Martínez-Martínez, M.; Ruiz-Ros, V.; Cauli, O. The value of neutrophil and lymphocyte count in frail older women. Exp. Gerontol. 2014, 54, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Samson, L.D.; Boots, A.M.H.; Verschuren, W.M.; Picavet, H.S.J.; Engelfriet, P.; Buisman, A.-M. Frailty is associated with elevated CRP trajectories and higher numbers of neutrophils and monocytes. Exp. Gerontol. 2019, 125, 110674. [Google Scholar] [CrossRef] [PubMed]
- Leng, S.X.; Xue, Q.-L.; Tian, J.; Huang, Y.; Yeh, S.-H.; Fried, L.P. Associations of neutrophil and monocyte counts with frailty in community-dwelling disabled older women: Results from the Women’s Health and Aging Studies I. Exp. Gerontol. 2009, 44, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Vo, A.K.; Rekvam, H. Hematopoiesis, Inflammation and Aging—The Biological Background and Clinical Impact of Anemia and Increased C-Reactive Protein Levels on Elderly Individuals. J. Clin. Med. 2022, 11, 706. [Google Scholar] [CrossRef] [PubMed]
- Hennigs, J.K.; Matuszcak, C.; Trepel, M.; Körbelin, J. Vascular Endothelial Cells: Heterogeneity and Targeting Approaches. Cells 2021, 10, 2712. [Google Scholar] [CrossRef]
- Sirnio, P.; Vayrynen, J.P.; Klintrup, K.; Mäkelä, J.; Karhu, T.; Herzig, K.H.; Minkkinen, I.; Mäkinen, M.J.; Karttunen, T.J.; Tuomisto, A. Alterations in serum amino-acid profile in the progression of colorectal cancer: Associations with systemic in-flammation, tumour stage and patient survival. Br. J. Cancer 2019, 120, 238–246. [Google Scholar] [CrossRef] [Green Version]
- Mustafa, A.; Gupta, S.; Hudes, G.R.; Egleston, B.L.; Uzzo, R.G.; Kruger, W.D. Serum Amino Acid Levels as a Biomarker for Renal Cell Carcinoma. J. Urol. 2011, 186, 1206–1212. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Ma, X.; Li, H.; Guo, G.; Li, P.; Li, H.; Gu, L.; Li, X.; Chen, L.; Zhang, X. The predictive and prognostic values of serum amino acid levels for clear cell renal cell carcinoma. Urol. Oncol. Semin. Orig. Investig. 2017, 35, 392–400. [Google Scholar] [CrossRef]
- Shingyoji, M.; Iizasa, T.; Higashiyama, M.; Imamura, F.; Saruki, N.; Imaizumi, A.; Yamamoto, H.; Daimon, T.; Tochikubo, O.; Mitsushima, T.; et al. The significance and robustness of a plasma free amino acid (PFAA) profile-based multiplex function for detecting lung cancer. BMC Cancer 2013, 13, 77. [Google Scholar] [CrossRef]
- Cadoni, G.; Giraldi, L.; Chiarla, C.; Gervasoni, J.; Persichilli, S.; Primiano, A.; Settimi, S.; Galli, J.; Paludetti, G.; Arzani, D.; et al. Prognostic Role of Serum Amino Acids in Head and Neck Cancer. Dis. Markers 2020, 2020, 2291759. [Google Scholar] [CrossRef] [PubMed]
- Vsiansky, V.; Svobodova, M.; Gumulec, J.; Cernei, N.; Sterbova, D.; Zitka, O.; Kostrica, R.; Smilek, P.; Plzak, J.; Betka, J.; et al. Prognostic Significance of Serum Free Amino Acids in Head and Neck Cancers. Cells 2019, 8, 428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, H.; Vutipongsatorn, K.; Jiménez, B.; Antcliffe, D.B. Patient Stratification in Sepsis: Using Metabolomics to Detect Clinical Phenotypes, Sub-Phenotypes and Therapeutic Response. Metabolites 2022, 12, 376. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Reikvam, H.; Brenner, A.K. Toll-like Receptor 4, Osteoblasts and Leukemogenesis; the Lesson from Acute Myeloid Leukemia. Molecules 2022, 27, 735. [Google Scholar] [CrossRef] [PubMed]
- Rundgren, I.M.; Ryningen, A.; Anderson Tvedt, T.H.; Bruserud, Ø.; Ersvær, E. Immunomodulatory Drugs Alter the Me-tabolism and the Extracellular Release of Soluble Mediators by Normal Monocytes. Molecules 2020, 25, 367. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Liu, X.; Li, X. Peptidoglycan-based immunomodulation. Appl. Microbiol. Biotechnol. 2022, 106, 981–993. [Google Scholar] [CrossRef]
- de Jong, N.W.M.; van Kessel, K.P.M.; van Strijp, J.A.G. Immune Evasion by Staphylococcus aureus. Microbiol. Spectr. 2019, 7, 2. [Google Scholar] [CrossRef]
- Postma, B.; Kleibeuker, W.; Poppelier, M.J.J.G.; Boonstra, M.; Van Kessel, K.P.M.; Van Strijp, J.A.G.; de Haas, C.J.C. Residues 10–18 within the C5a Receptor N Terminus Compose a Binding Domain for Chemotaxis Inhibitory Protein of Staphylococcus aureus. J. Biol. Chem. 2005, 280, 2020–2027. [Google Scholar] [CrossRef] [Green Version]
- Laarman, A.J.; Mijnheer, G.; Mootz, J.M.; Van Rooijen, W.J.M.; Ruyken, M.; Malone, C.L.; Heezius, E.C.; Ward, R.; Milligan, G.; Van Strijp, J.A.G.; et al. Staphylococcus aureusStaphopain A inhibits CXCR2-dependent neutrophil activation and chemotaxis. EMBO J. 2012, 31, 3607–3619. [Google Scholar] [CrossRef] [Green Version]
- Wilson, G.J.; Seo, K.S.; Cartwright, R.A.; Connelley, T.; Chuang-Smith, O.N.; Merriman, J.A.; Guinane, C.M.; Park, J.Y.; Bohach, G.A.; Schlievert, P.M.; et al. A novel core genome-encoded superantigen contributes to lethality of communi-ty-associated MRSA necrotizing pneumonia. PLoS Pathog. 2011, 7, e1002271. [Google Scholar] [CrossRef]
- Bardoel, B.W.; Vos, R.; Bouman, T.; Aerts, P.C.; Bestebroer, J.; Huizinga, E.G.; Brondijk, T.H.C.; van Strijp, J.A.G.; de Haas, C.J.C. Evasion of Toll-like receptor 2 activation by staphylococcal superantigen-like protein 3. J. Mol. Med. 2012, 90, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Itoh, S.; Kamoshida, G.; Takii, T.; Fujii, S.; Tsuji, T.; Onozaki, K. Staphylococcal Superantigen-Like Protein 3 Binds to the Toll-Like Receptor 2 Extracellular Domain and Inhibits Cytokine Production Induced by Staphylococcus aureus, Cell Wall Component, or Lipopeptides in Murine Macrophages. Infect. Immun. 2012, 80, 2816–2825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koymans, K.J.; Bisschop, A.; Vughs, M.M.; van Kessel, K.P.; de Haas, C.J.; van Strijp, J.A. Staphylococcal Superantigen-Like Protein 1 and 5 (SSL1 & SSL5) Limit Neutrophil Chemotaxis and Migration through MMP-Inhibition. Int. J. Mol. Sci. 2016, 17, 1072. [Google Scholar] [PubMed] [Green Version]
- Kumar, S.; Sunagar, R.; Gosselin, E. Bacterial Protein Toll-Like-Receptor Agonists: A Novel Perspective on Vaccine Adju-vants. Front. Immunol. 2019, 10, 1144. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-Q.; Cao, G.; Chen, H.; Argyopoulos, C.P.; Yu, H.; Su, W.; Chen, L.; Samuels, D.C.; Zhuang, S.; Bayliss, G.P.; et al. Identification of serum metabolites associating with chronic kidney disease progression and anti-fibrotic effect of 5-methoxytryptophan. Nat. Commun. 2019, 10, 1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajaj, J.S.; Garcia-Tsao, G.; Reddy, K.R.; O’Leary, J.G.; Vargas, H.E.; Lai, J.C.; Kamath, P.S.; Tandon, P.; Subramanian, R.M.; Thuluvath, P.; et al. Admission Urinary and Serum Metabolites Predict Renal Outcomes in Hospitalized Patients with Cirrhosis. Hepatology 2021, 74, 2699–2713. [Google Scholar] [CrossRef]
- Saito, A.; Niwa, T.; Maeda, K.; Kobayashi, K.; Yamamoto, Y.; Ohta, K. Tryptophan and indolic tryptophan metabolites in chronic renal failure. Am. J. Clin. Nutr. 1980, 33, 1402–1406. [Google Scholar] [CrossRef]
- Sun, J.; Cao, Z.; Schnackenberg, L.; Pence, L.; Yu, L.R.; Choudhury, D.; Palevsky, P.M.; Portilla, D.; Beger, R.D. Serum me-tabolite profiles predict outcomes in critically ill patients receiving renal replacement therapy. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2021, 1187, 123024. [Google Scholar] [CrossRef]
- Perna, A.F.; Pane, F.; Sepe, N.; Fontanarosa, C.; Pinto, G.; Zacchia, M.; Trepiccione, F.; Anishchenko, E.; Ingrosso, D.; Pucci, P.; et al. Lanthionine and Other Relevant Sulfur Amino Acid Metabolites: Detection of Prospective Uremic Toxins in Serum by Multiple Reaction Monitoring Tandem Mass Spec-trometry. Methods Mol. Biol. 2019, 2007, 9–17. [Google Scholar] [PubMed]
- Pool, R.; Gomez, H.; Kellum, J.A. Mechanisms of Organ Dysfunction in Sepsis. Crit. Care Clin. 2018, 34, 63–80. [Google Scholar] [CrossRef]
- Barker, G.; Leeuwenburgh, C.; Brusko, T.; Moldawer, L.; Reddy, S.T.; Guirgis, F.W. Lipid and Lipoprotein Dysregulation in Sepsis: Clinical and Mechanistic Insights into Chronic Critical Illness. J. Clin. Med. 2021, 10, 1693. [Google Scholar] [CrossRef] [PubMed]
- Svahn, S.L.; Gutiérrez, S.; Ulleryd, M.A.; Nookaew, I.; Osla, V.; Beckman, F.; Nilsson, S.; Karlsson, A.; Jansson, J.-O.; Johansson, M.E. Dietary Polyunsaturated Fatty Acids Promote Neutrophil Accumulation in the Spleen by Altering Chemotaxis and Delaying Cell Death. Infect. Immun. 2019, 87, e00270-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svahn, S.L.; Ulleryd, M.A.; Grahnemo, L.; Ståhlman, M.; Borén, J.; Nilsson, S.; Jansson, J.-O.; Johansson, M.E. Dietary Omega-3 Fatty Acids Increase Survival and Decrease Bacterial Load in Mice Subjected to Staphylococcus aureus-Induced Sepsis. Infect. Immun. 2016, 84, 1205–1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svahn, S.L.; Grahnemo, L.; Pálsdóttir, V.; Nookaew, I.; Wendt, K.; Gabrielsson, B.; Schéle, E.; Benrick, A.; Andersson, N.; Nilsson, S.; et al. Dietary polyunsaturated fatty acids increase survival and decrease bacterial load during septic Staphylo-coccus aureus infection and improve neutrophil function in mice. Infect. Immun. 2015, 83, 514–521. [Google Scholar] [CrossRef] [Green Version]
- Mosevoll, K.A.; Hansen, B.A.; Gundersen, I.M.; Reikvam, H.; Bruserud, Ø.; Bruserud, Ø.; Wendelbo, Ø. Patients with Bac-terial Sepsis Are Heterogeneous with Regard to Their Systemic Lipidomic Profiles. Metabolites 2023, 13, 52. [Google Scholar] [CrossRef]
- Téblick, A.; Gunst, J.; Berghe, G.V.D. Critical Illness–induced Corticosteroid Insufficiency: What It Is Not and What It Could Be. J. Clin. Endocrinol. Metab. 2022, 107, 2057–2064. [Google Scholar] [CrossRef]
- Van den Berghe, G.; Téblick, A.; Langouche, L.; Gunst, J. The hypothalamus-pituitary-adrenal axis in sepsis- and hyperin-flammation-induced critical illness: Gaps in current knowledge and future translational research directions. EBioMedicine 2022, 84, 104284. [Google Scholar] [CrossRef]
- Boonen, E.; Vervenne, H.; Meersseman, P.; Andrew, R.; Mortier, L.; Declercq, P.E.; Vanwijngaerden, Y.-M.; Spriet, I.; Wouters, P.J.; Perre, S.V.; et al. Reduced Cortisol Metabolism during Critical Illness. N. Engl. J. Med. 2013, 368, 1477–1488. [Google Scholar] [CrossRef]

| Subpathway | Identity |
|---|---|
| AMINO ACIDS AND PEPTIDES (40/228 metabolites altered, 31 correlated with SOFA score) | |
| Alanine and Aspartate Metabolism (2/9) | N-acetylalanine, N-carbamoylalanine |
| Glutamate Metabolism (1/12) | carboxyethyl-GABA |
| Histidine Metabolism (6/19) | 1-methylhistidine, N-acetyl-1-methylhistidine *, imidazole lactate, 1-methyl-4-imidazoleacetate, 1-methyl-5-imidazoleacetate, 1-methyl-5-imidazolelactate |
| Lysine Metabolism (1/19) | 5-(galactosylhydroxy)-lysine |
| Tyrosine Metabolism (4/) | N-acetyltyrosine, 3-(4-hydroxyphenyl)lactate, phenol sulfate, N-formylphenylalanine, |
| Tryptophan Metabolism (2/25) | Kynurenine, indolelactate |
| Leucine, Isoleucine and Valine Metabolism (6/27) | beta-hydroxyisovaleroylcarnitine, 3-methylglutaconate, Isoleucine, tiglylcarnitine (C5:1-DC), 3-hydroxy-2-ethylpropionate, N-carbamoylvaline |
| Methionine, Cysteine, SAM and Taurine Metabolism (3/26) | cysteine, N-acetylmethionine sulfoxide, 2,3-dihydroxy-5-methylthio-4-pentenoate (DMTPA) * |
| Urea cycle; Arginine and Proline Metabolism (5/24) | Arginine, urea, homocitrulline, N2,N5-diacetylornithine, argininate |
| Polyamine Metabolism (4/9) | N(1) + N(8))-acetylspermidine, Acisoga, N1,N12-diacetylspermine, 4-acetamidobutanoate |
| Guanidino and Acetamido Metabolism (1/4) | guanidinosuccinate |
| Peptides (5/50) | gamma-glutamylserine, isoleucylglycine, fibrinopeptide A (3–15) *, fibrinopeptide B (1–11) * fibrinopeptide B (1–12) * |
| CARBOHYDRATE AND ENERGY (all increased, all associated with the SOFA score) | |
| Carbohydrate (6/76) | Lactate, ribonate, arabinose, arabitol/xylitol, galactonate, erythronate *, N-acetylglucosamine/N-acetylgalactosamine |
| Energy metabolism (2/10) | alpha-ketoglutarate, fumarate |
| LIPIDS (7 increased including the acyl cholines, 8 associated with thevSOFA score including the acylcholines) | |
| Fatty Acid metabolites (10/145) | N-acetyl-2-aminooctanoate *, palmitoylcholine, dihomo-linolenoyl-choline, linoleoylcholine * stearoylcholine *, arachidonoylcholine, 12-HETE, N-stearoylserine *, 3-hydroxy-3-methylglutarate, glycocholate |
| NUCLEOTIDE (all 7 increased, six associated with the SOFA score) | |
| Purine Metabolism, (3/20) | Urate, allantoin, N6-succinyladenosine |
| Pyrimidine Metabolism (4/23) | Orotidine, uracil, 3-(3-amino-3-carboxypropyl)uridine *, 5,6-dihydrothymine |
| COFACTORS AND VITAMINS (6/38; all 6 increased, and 5 were associated with the SOFA score) | |
| Nicotinate and Nicotinamide Metabolism | N1-methyl-2-pyridone-5-carboxamide, N1-methyl-4-pyridone-3-carboxamide |
| Pantothenate and CoA Metabolism | pantoate |
| Ascorbate and Aldarate Metabolism | ascorbic acid 3-sulfate *, 2-O-methylascorbic acid |
| Vitamin B6 Metabolism | Pyridoxate |
| ADDITIONAL (25 identified metabolites, two of them increased, 10 associated with the SOFA score) | |
| Xenobiotics (29/275) | 6 food components, 11 drug metabolites, 4 chemicals, 3 bilirubin degradation products, 1 glycine conjugate, 4 unidentified |
| Subpathway | Identity |
|---|---|
| AMINO ACIDS AND PEPTIDES (8 out of 11 metabolites correlated with SOFA | |
| Lysine Metabolism | N,N-dimethyl-5-aminovalerate |
| Tryptophan Metabolism | Picolinate, 6-bromotryptophan |
| Methionine, Cysteine, SAM and Taurine Metabolism | Cystathionine, alpha-ketobutyrate |
| Urea cycle; Arginine and Proline Metabolism | N,N,N-trimethyl-alanylproline betaine (TMAP) |
| Gamma-glutamyl Amino Acid | gamma-glutamylphenylalanine, gamma-glutamyl-2-aminobutyrate |
| Fibrinogen Cleavage Peptide | fibrinopeptide B (1-11) *, fibrinopeptide B (1-12) * |
| Modified Peptides | N,N-dimethyl-pro-pro |
| ENERGY (the metabolite was associated with SOFA score) | |
| TCA Cycle | 2-methylcitrate/homocitrate |
| LIPIDS (11 out of 16 correlated with SOFA score) | |
| Fatty Acid Synthesis | Malonylcarnitine |
| Short Chain Fatty Acid | butyrate/isobutyrate (4:0) |
| Fatty Acid, Dicarboxylate | decadienedioic acid (C10:2-DC) * |
| Fatty Acid Metabolism (Acyl Carnitine, Monounsaturated) | ximenoylcarnitine (C26:1) * |
| Fatty Acid Metabolism (Acyl Carnitine, Dicarboxylate) | octadecanedioylcarnitine (C18-DC) *, octadecenedioylcarnitine (C18:1-DC) * |
| Fatty Acid Metabolism (Acyl Carnitine, Hydroxy) | 3-hydroxyoleoylcarnitine |
| Fatty Acid, Monohydroxy | 2-hydroxydecanoate, 3-hydroxyhexanoate |
| Eicosanoid | leukotriene B4 |
| Progestin Steroids | 5alpha-pregnan-3beta, 20alpha-diol disulfate, pregnanediol-3-glucuronide |
| Corticosteroids | cortisol 21-sulfate |
| Primary Bile Acid Metabolism | Taurocholate |
| Secondary Bile Acid Metabolism | taurochenodeoxycholic acid 3-sulfate |
| NUCLEOTIDES (none of the three metabolites associated with SOFA score) | |
| Purine Metabolism, (Hypo)Xanthine/Inosine containing | Xanthine |
| Pyrimidine Metabolism, Uracil containing | 5-methyluridine (ribothymidine) |
| Pyrimidine Metabolism, Thymine containing | 3-aminoisobutyrate |
| COFACTORS AND VITAMINS (one metabolite associated with SOFA score) | |
| Vitamin A Metabolism | Retinol (vitamin A) |
| XENOBIOTICS (only 3 out of 19 metabolites associated with SOFA score) | |
| Food Component/Plant | methyl glucopyranoside (alpha + beta), vanillic acid glycine, 4-vinylguaiacol glucuronide |
| Drug—Analgesics, Anesthetics | 3-(N-acetyl-cystein-S-yl) acetaminophen, 4-acetamidophenylglucuronide, 2-hydroxyacetaminophen sulfate *, 2-methoxyacetaminophen sulfate *, 2-methoxyacetaminophen glucuronide *, 3-(cystein-S-yl)acetaminophen *, 3-(methylthio)acetaminophen sulfate * |
| Drug—Cardiovascular | Metoprolol, metoprolol acid metabolite *, alpha-hydroxymetoprolol, warfarin, 6-hydroxywarfarin, 7-hydroxywarfarin, 10-hydroxywarfarin |
| Drug—Gastrointestinal | Pantoprazole |
| Chemical | 2-acrylamidoglycolic acid |
| Parameter | Upper Subcluster (n = 26) | Lower Subcluster Plus Lower Main Cluster (n = 34) | p-Value |
|---|---|---|---|
| Number of Sepsis-3 patients | 8 | 27 | 0.002 |
| Total SOFA score | 1 (1–12) | 3 (0–16) | 0.0074 |
| Number of patients with cardiovascular comorbidity | 0 | 9 | 0.0038 |
| Age (rears) | 54 (20–83) | 72 (23–96) | 0.0076 |
| Peripheral blood neutrophil count (×109/L) | 12.9 (6.4–1.1) | 10.6 (2.4–26.6) | 0.55 |
| Peripheral blood platelet count (×109/L) | 237 (58–568) | 176 (24—407) | 0.080 |
| Serum creatinine (mmol/L) | 66 (27–417) | 121 (51–706) | 0.0010 |
| Parameter | Sepsis-3 Patients in the Upper Subcluster (n = 8) | Sepsis-3 Patients in the Lower Subcluster Plus Lower Main Cluster (n = 27) | p-Value |
|---|---|---|---|
| Total SOFA score | 3 (2–7) | 4 (1–16) | 0.32 |
| Number of patients with cardiovascular comorbidity | 0 | 9 | 0.037 |
| Empty row | |||
| Age (rears) | 62 (34–66) | 75 (23–96) | 0.048 |
| Peripheral blood neutrophil count (×109/L) | 10.1 (6.4–41.1) | 10.3 (3.4–26.6) | 0.55 |
| Peripheral blood platelet count (×109/L) | 231 (58.538) | 162 (24–407) | 0.46 |
| Serum creatinine (mmol/L) | 77 (54–107) | 130 (51–475) | 0.0139 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mosevoll, K.A.; Hansen, B.A.; Gundersen, I.M.; Reikvam, H.; Bruserud, Ø.; Bruserud, Ø.; Wendelbo, Ø. Systemic Metabolomic Profiles in Adult Patients with Bacterial Sepsis: Characterization of Patient Heterogeneity at the Time of Diagnosis. Biomolecules 2023, 13, 223. https://doi.org/10.3390/biom13020223
Mosevoll KA, Hansen BA, Gundersen IM, Reikvam H, Bruserud Ø, Bruserud Ø, Wendelbo Ø. Systemic Metabolomic Profiles in Adult Patients with Bacterial Sepsis: Characterization of Patient Heterogeneity at the Time of Diagnosis. Biomolecules. 2023; 13(2):223. https://doi.org/10.3390/biom13020223
Chicago/Turabian StyleMosevoll, Knut Anders, Bent Are Hansen, Ingunn Margareetta Gundersen, Håkon Reikvam, Øyvind Bruserud, Øystein Bruserud, and Øystein Wendelbo. 2023. "Systemic Metabolomic Profiles in Adult Patients with Bacterial Sepsis: Characterization of Patient Heterogeneity at the Time of Diagnosis" Biomolecules 13, no. 2: 223. https://doi.org/10.3390/biom13020223







