Pan-Cancer Analysis of the TRP Family, Especially TRPV4 and TRPC4, and Its Expression Correlated with Prognosis, Tumor Microenvironment, and Treatment Sensitivity
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Differential Expression of TRP Family Genes in Human Pan-Cancer Tissues
2.2. Survival Analyses Based on the Expression Level of TRP Family Genes in Human Cancer
2.3. Correlation Analysis of the Expression of TRP Family Genes with TME and Stemness Score in Pan-Cancer Tissues
2.4. Correlation Analysis of TRP Family Genes with Drug Sensitivity and Immune Subtypes
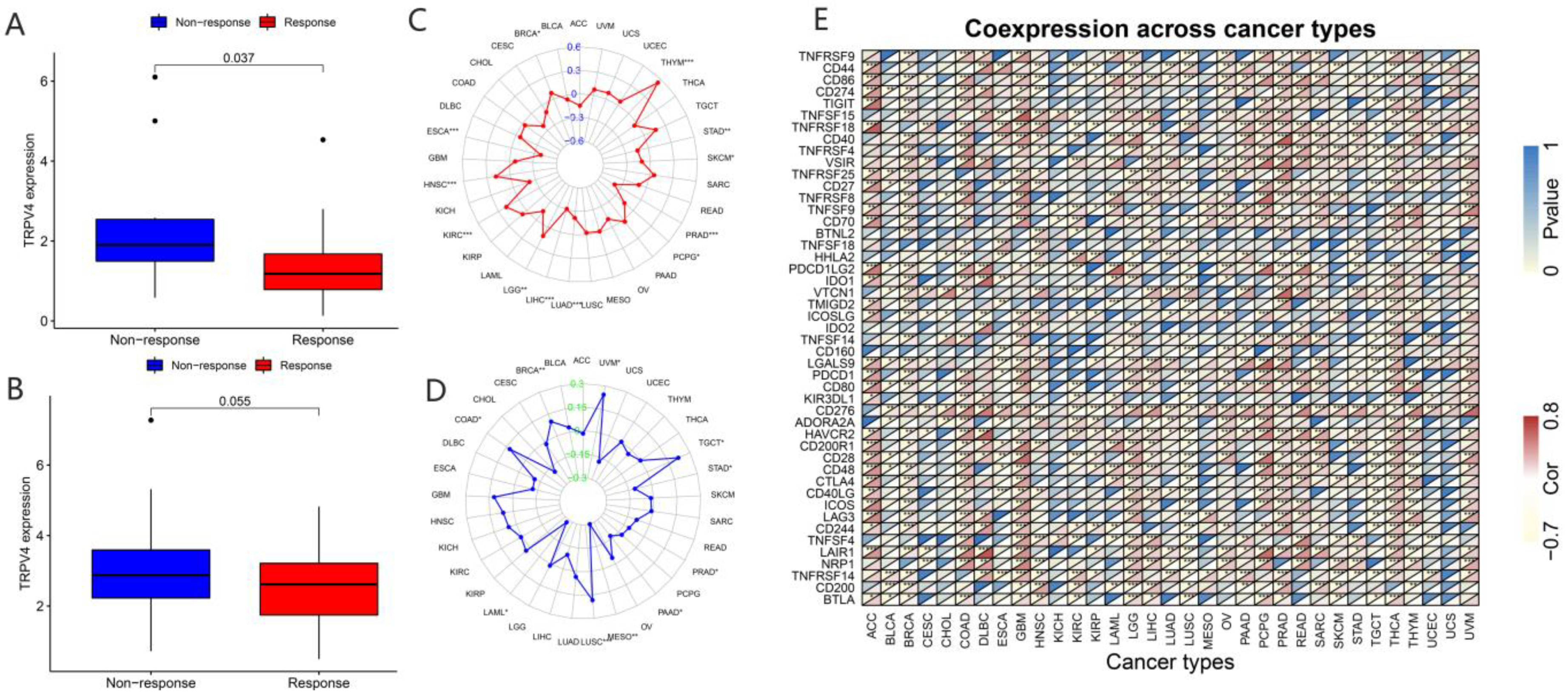
2.5. Correlation Analysis of TRP Family Genes and Immunotherapy Outcome
2.6. Correlation Analysis of TRPC4 and TRPV4 Expression with TMB and MSI
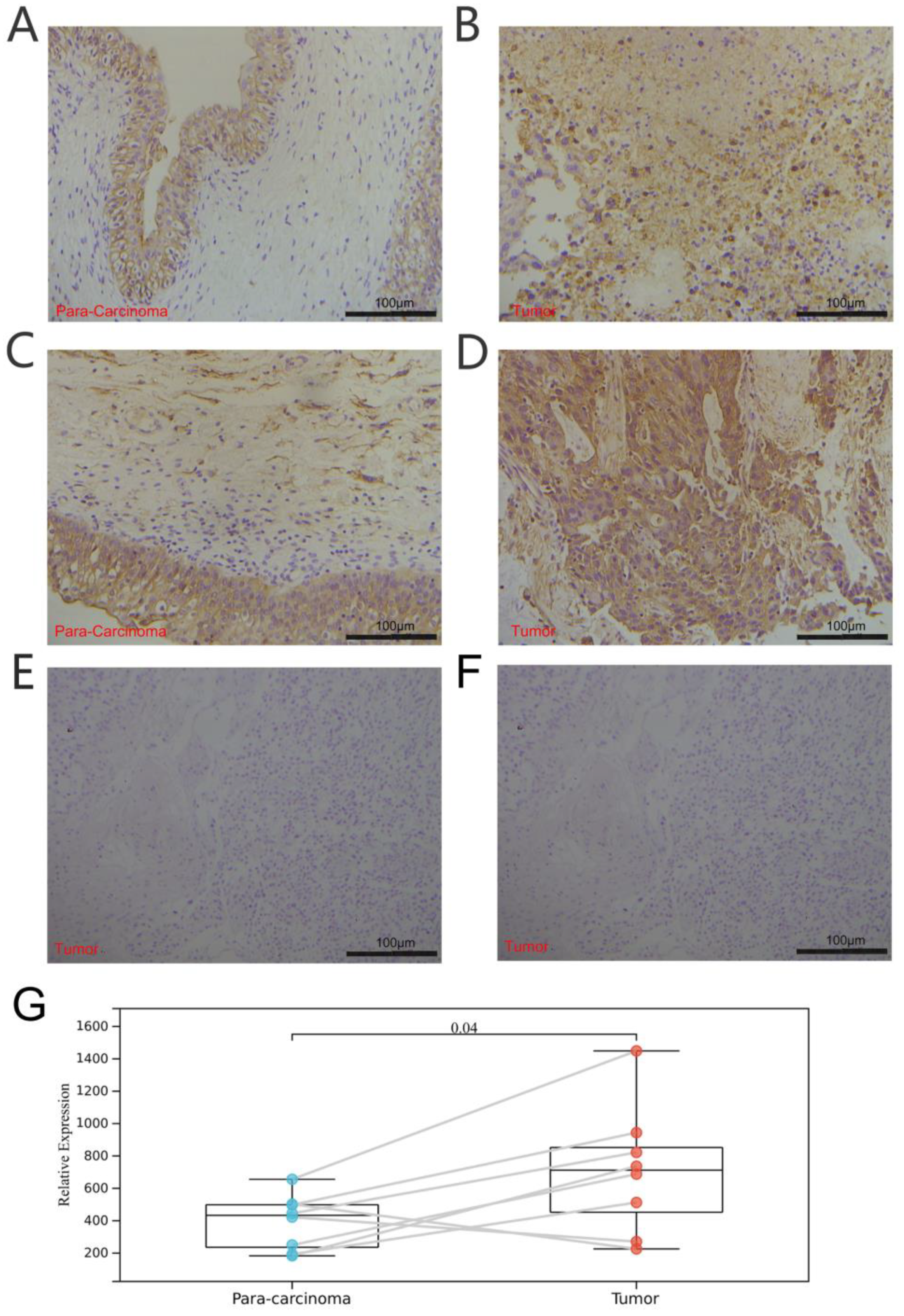
2.7. Tissue Specimens and Immunohistochemistry
2.8. Statistical Analyses
3. Results
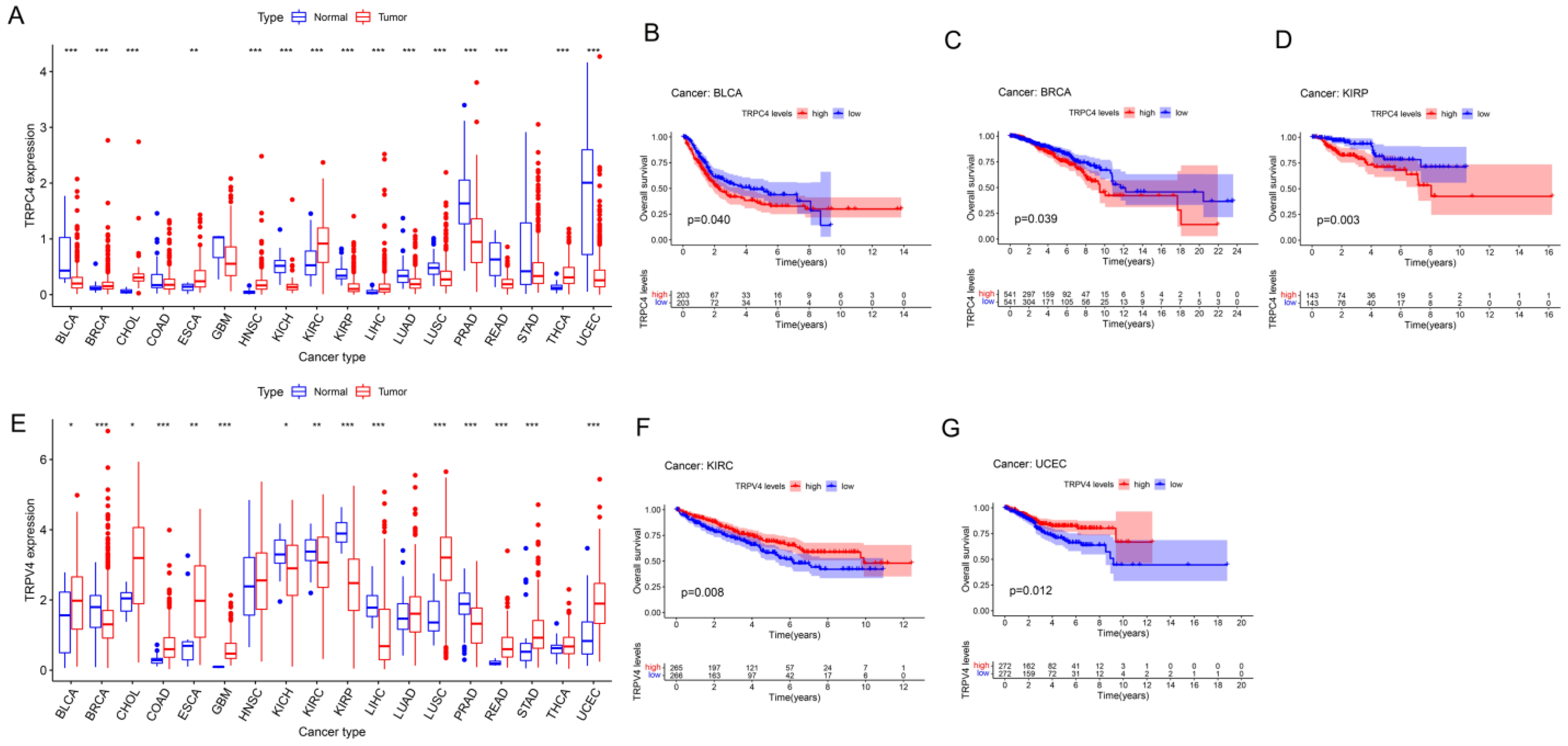
3.1. Expression and Correlation of TRP Family Genes in Pan-Cancer Tissues
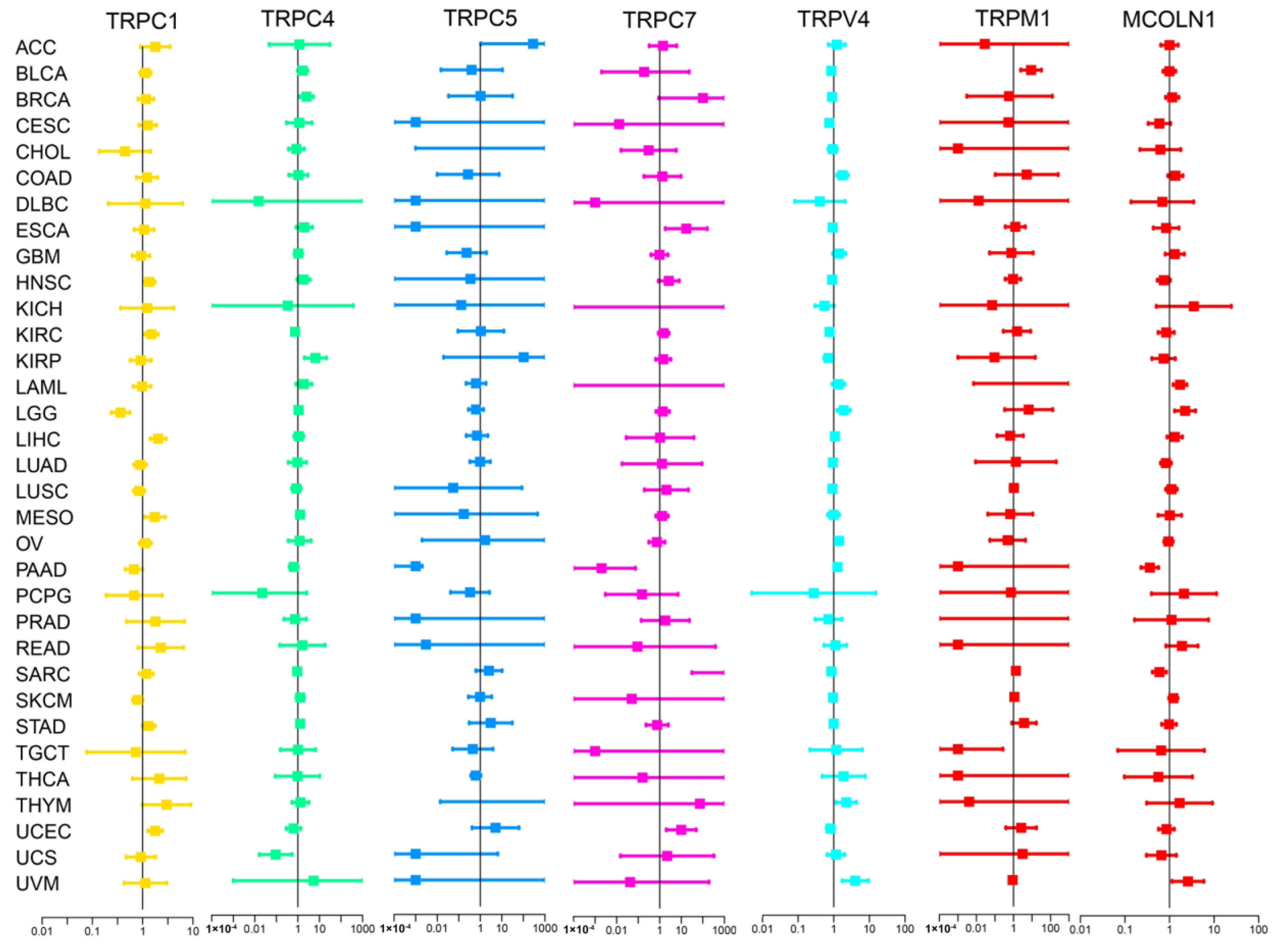
3.2. Different Expression and Correlation of TRP Family Genes and Prognosis in Pan-Cancer Tissues
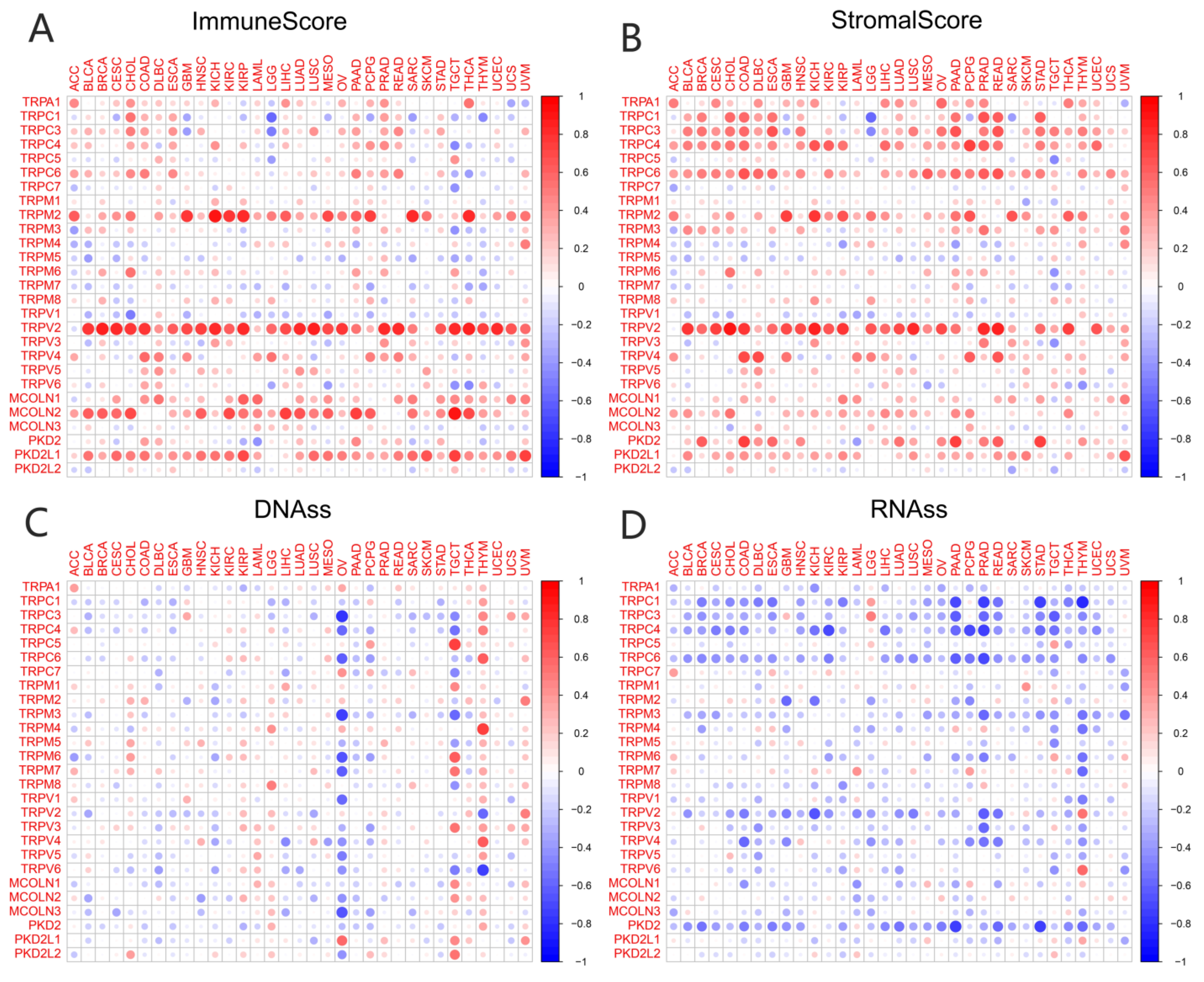
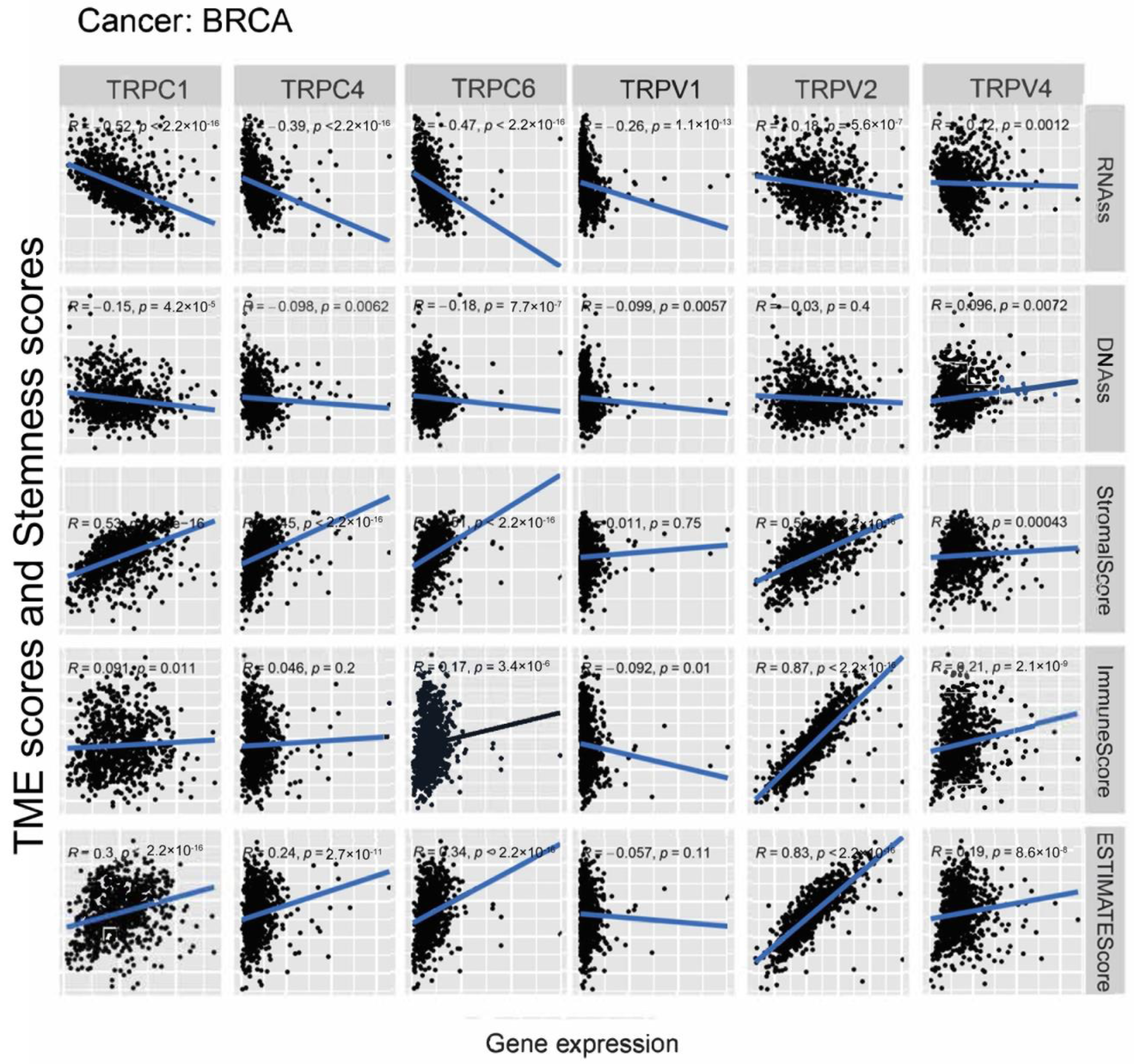
3.3. Association of TRP Family Genes with TME and Stemness Score in Pan-Cancer Tissues
3.4. Association of TRP Family Genes with Immune Subtypes in Pan-Cancer Tissues
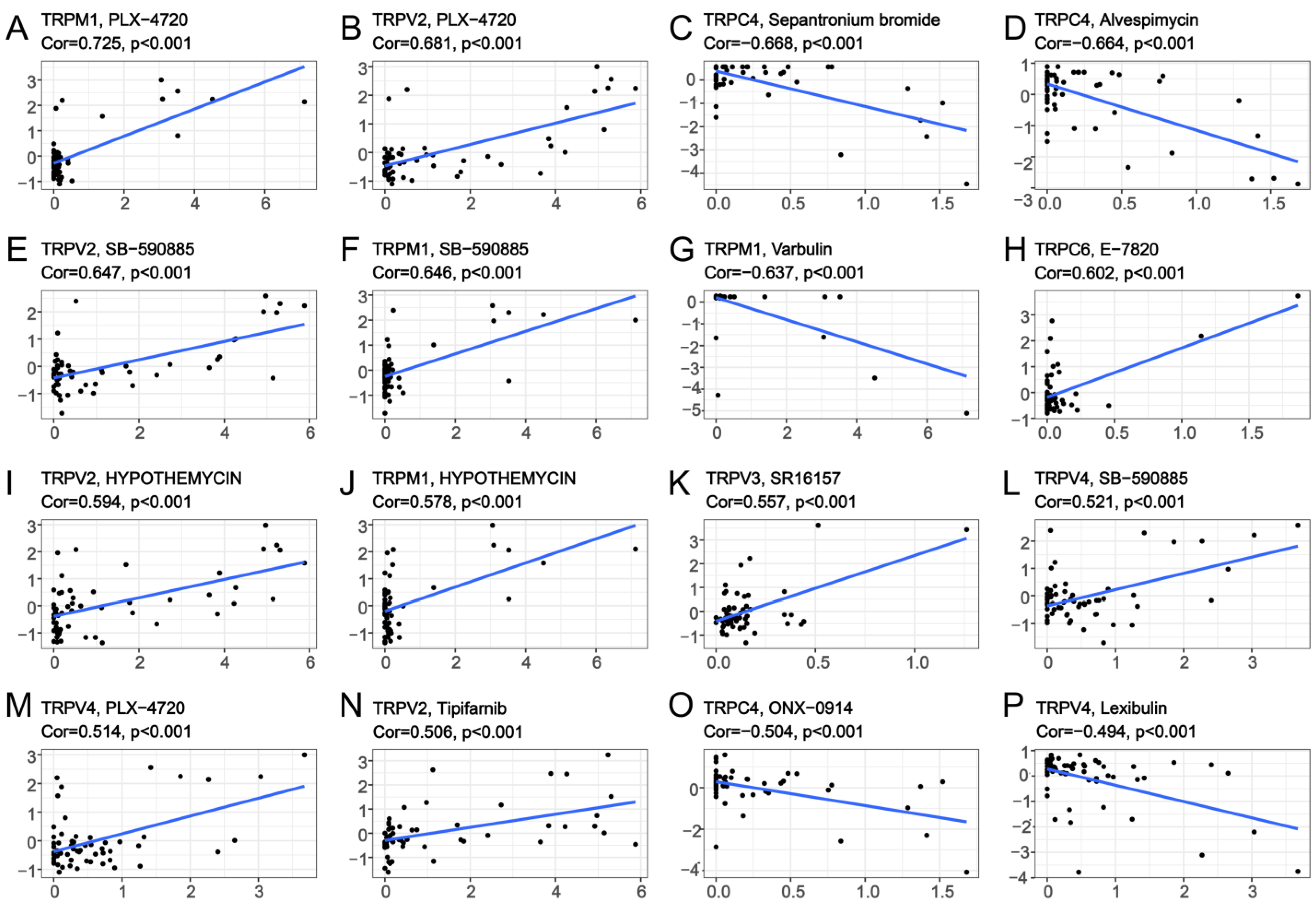
3.5. Association of TRP Family Genes with Pan-Cancer Treatments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krause, J.E.; Chenard, B.L.; Cortright, D.N. Transient receptor potential ion channels as targets for the discovery of pain therapeutics. Curr. Opin. Investig. Drugs 2005, 6, 48–57. [Google Scholar] [PubMed]
- Gupta, G.P.; Massague, J. Cancer metastasis: Building a framework. Cell 2006, 127, 679–695. [Google Scholar] [CrossRef] [PubMed]
- Nilius, B.; Owsianik, G.; Voets, T.; Peters, J.A. Transient receptor potential cation channels in disease. Physiol. Rev. 2007, 87, 165–217. [Google Scholar] [CrossRef] [PubMed]
- Kiselyov, K.; Soyombo, A.; Muallem, S. TRPpathies. J. Physiol. 2007, 578, 641–653. [Google Scholar] [CrossRef]
- Yang, D.; Kim, J. Emerging role of transient receptor potential (TRP) channels in cancer progression. BMB Rep. 2020, 53, 125–132. [Google Scholar] [CrossRef]
- Prevarskaya, N.; Zhang, L.; Barritt, G. TRP channels in cancer. Biochim. Biophys. Acta 2007, 1772, 937–946. [Google Scholar] [CrossRef]
- Gkika, D.; Prevarskaya, N. Molecular mechanisms of TRP regulation in tumor growth and metastasis. Biochim. Biophys. Acta 2009, 1793, 953–958. [Google Scholar] [CrossRef]
- Santoni, G.; Farfariello, V. TRP channels and cancer: New targets for diagnosis and chemotherapy. Endocr. Metab. Immune Disord. Drug Targets 2011, 11, 54–67. [Google Scholar] [CrossRef]
- Huang, J.K.; Chou, C.T.; Chang, H.T.; Shu, S.S.; Kuo, C.C.; Tsai, J.Y.; Liao, W.C.; Wang, J.L.; Lin, K.L.; Lu, Y.C.; et al. Effect of thapsigargin on Ca(2)+ fluxes and viability in human prostate cancer cells. J. Recept. Signal Transduct. 2011, 31, 247–255. [Google Scholar] [CrossRef]
- Vriens, J.; Janssens, A.; Prenen, J.; Nilius, B.; Wondergem, R. TRPV channels and modulation by hepatocyte growth factor/scatter factor in human hepatoblastoma (HepG2) cells. Cell Calcium 2004, 36, 19–28. [Google Scholar] [CrossRef]
- Potter, D.A.; Tirnauer, J.S.; Janssen, R.; Croall, D.E.; Hughes, C.N.; Fiacco, K.A.; Mier, J.W.; Maki, M.; Herman, I.M. Calpain regulates actin remodeling during cell spreading. J. Cell Biol. 1998, 141, 647–662. [Google Scholar] [CrossRef]
- Yamada, T.; Ugawa, S.; Ueda, T.; Ishida, Y.; Kajita, K.; Shimada, S. Differential localizations of the transient receptor potential channels TRPV4 and TRPV1 in the mouse urinary bladder. J. Histochem. Cytochem. 2009, 57, 277–287. [Google Scholar] [CrossRef]
- Everaerts, W.; Vriens, J.; Owsianik, G.; Appendino, G.; Voets, T.; De Ridder, D.; Nilius, B. Functional characterization of transient receptor potential channels in mouse urothelial cells. Am. J. Physiol. Renal Physiol. 2010, 298, F692–F701. [Google Scholar] [CrossRef]
- Mochizuki, T.; Sokabe, T.; Araki, I.; Fujishita, K.; Shibasaki, K.; Uchida, K.; Naruse, K.; Koizumi, S.; Takeda, M.; Tominaga, M. The TRPV4 cation channel mediates stretch-evoked Ca2+ influx and ATP release in primary urothelial cell cultures. J. Biol. Chem. 2009, 284, 21257–21264. [Google Scholar] [CrossRef]
- Miao, Y.; Wang, J.; Li, Q.; Quan, W.; Wang, Y.; Li, C.; Wu, J.; Mi, D. Prognostic value and immunological role of PDCD1 gene in pan-cancer. Int. Immunopharmacol. 2020, 89, 107080. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e411. [Google Scholar] [CrossRef]
- Wen, J.; Zu, S.; Chen, Z.; Daugherty, S.L.; de Groat, W.C.; Liu, Y.; Yuan, M.; Cheng, G.; Zhang, X. Reduced bladder responses to capsaicin and GSK-1016790A in retired-breeder female rats with diminished volume sensitivity. Am. J. Physiol. Renal Physiol. 2018, 315, F1217–F1227. [Google Scholar] [CrossRef]
- Chen, C.; He, Z.C.; Shi, Y.; Zhou, W.; Zhang, X.; Xiao, H.L.; Wu, H.B.; Yao, X.H.; Luo, W.C.; Cui, Y.H.; et al. Microvascular fractal dimension predicts prognosis and response to chemotherapy in glioblastoma: An automatic image analysis study. Lab. Investig. 2018, 98, 924–934. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef]
- Picard, E.; Verschoor, C.P.; Ma, G.W.; Pawelec, G. Relationships Between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer. Front. Immunol. 2020, 11, 369. [Google Scholar] [CrossRef]
- Rizzo, A.; Ricci, A.D.; Brandi, G. PD-L1, TMB, MSI, and Other Predictors of Response to Immune Checkpoint Inhibitors in Biliary Tract Cancer. Cancers 2021, 13, 558. [Google Scholar] [CrossRef] [PubMed]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Liu, Y.; Zeng, S.; Han, Y.; Shen, H. Intratumor heterogeneity: The hidden barrier to immunotherapy against MSI tumors from the perspective of IFN-gamma signaling and tumor-infiltrating lymphocytes. J. Hematol. Oncol. 2021, 14, 160. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Bai, Y.; Hebda, L.; Zhong, X.; Liu, J.; Kao, J.; Duan, C. Calcium deficiency-induced and TRP channel-regulated IGF1R-PI3K-Akt signaling regulates abnormal epithelial cell proliferation. Cell Death Differ. 2014, 21, 568–581. [Google Scholar] [CrossRef]
- Almasi, S.; Kennedy, B.E.; El-Aghil, M.; Sterea, A.M.; Gujar, S.; Partida-Sanchez, S.; El Hiani, Y. TRPM2 channel-mediated regulation of autophagy maintains mitochondrial function and promotes gastric cancer cell survival via the JNK-signaling pathway. J. Biol. Chem. 2018, 293, 3637–3650. [Google Scholar] [CrossRef]
- Zhong, T.; Zhang, W.; Guo, H.; Pan, X.; Chen, X.; He, Q.; Yang, B.; Ding, L. The regulatory and modulatory roles of TRP family channels in malignant tumors and relevant therapeutic strategies. Acta Pharm. Sin. B 2022, 12, 1761–1780. [Google Scholar] [CrossRef]
- Saldias, M.P.; Maureira, D.; Orellana-Serradell, O.; Silva, I.; Lavanderos, B.; Cruz, P.; Torres, C.; Caceres, M.; Cerda, O. TRP Channels Interactome as a Novel Therapeutic Target in Breast Cancer. Front. Oncol. 2021, 11, 621614. [Google Scholar] [CrossRef]
- Cui, C.; Merritt, R.; Fu, L.; Pan, Z. Targeting calcium signaling in cancer therapy. Acta Pharm. Sin. B 2017, 7, 3–17. [Google Scholar] [CrossRef]
- Qi, J.; Xing, Y.; Liu, Y.; Wang, M.M.; Wei, X.; Sui, Z.; Ding, L.; Zhang, Y.; Lu, C.; Fei, Y.H.; et al. MCOLN1/TRPML1 finely controls oncogenic autophagy in cancer by mediating zinc influx. Autophagy 2021, 17, 4401–4422. [Google Scholar] [CrossRef]
- Xing, Y.; Wei, X.; Liu, Y.; Wang, M.M.; Sui, Z.; Wang, X.; Zhu, W.; Wu, M.; Lu, C.; Fei, Y.H.; et al. Autophagy inhibition mediated by MCOLN1/TRPML1 suppresses cancer metastasis via regulating a ROS-driven TP53/p53 pathway. Autophagy 2022, 18, 1932–1954. [Google Scholar] [CrossRef]
- Boudaka, A.; Saito, C.T.; Tominaga, M. Deletion of TRPV4 enhances in vitro wound healing of murine esophageal keratinocytes. Sci. Rep. 2020, 10, 11349. [Google Scholar] [CrossRef]
- Cohen, D.M. TRPV4 and the mammalian kidney. Pflugers Arch. 2005, 451, 168–175. [Google Scholar] [CrossRef]
- Ramirez, A.; Vazquez-Sanchez, A.Y.; Carrion-Robalino, N.; Camacho, J. Ion Channels and Oxidative Stress as a Potential Link for the Diagnosis or Treatment of Liver Diseases. Oxidative Med. Cell. Longev. 2016, 2016, 3928714. [Google Scholar] [CrossRef]
- Simonsen, U.; Wandall-Frostholm, C.; Olivan-Viguera, A.; Kohler, R. Emerging roles of calcium-activated K channels and TRPV4 channels in lung oedema and pulmonary circulatory collapse. Acta Physiol. 2017, 219, 176–187. [Google Scholar] [CrossRef]
- Wu, Y.; Qi, J.; Wu, C.; Rong, W. Emerging roles of the TRPV4 channel in bladder physiology and dysfunction. J. Physiol. 2021, 599, 39–47. [Google Scholar] [CrossRef]
- Li, M.; Zheng, J.; Wu, T.; He, Y.; Guo, J.; Xu, J.; Gao, C.; Qu, S.; Zhang, Q.; Zhao, J.; et al. Activation of TRPV4 Induces Exocytosis and Ferroptosis in Human Melanoma Cells. Int. J. Mol. Sci. 2022, 23, 4146. [Google Scholar] [CrossRef]
- Lee, W.H.; Choong, L.Y.; Mon, N.N.; Lu, S.; Lin, Q.; Pang, B.; Yan, B.; Krishna, V.S.; Singh, H.; Tan, T.Z.; et al. TRPV4 Regulates Breast Cancer Cell Extravasation, Stiffness and Actin Cortex. Sci. Rep. 2016, 6, 27903. [Google Scholar] [CrossRef]
- Janssen, D.A.; Hoenderop, J.G.; Jansen, K.C.; Kemp, A.W.; Heesakkers, J.P.; Schalken, J.A. The mechanoreceptor TRPV4 is localized in adherence junctions of the human bladder urothelium: A morphological study. J. Urol. 2011, 186, 1121–1127. [Google Scholar] [CrossRef]
- Wong, K.K.; Hussain, F.A. TRPM4 is overexpressed in breast cancer associated with estrogen response and epithelial-mesenchymal transition gene sets. PLoS ONE 2020, 15, e0233884. [Google Scholar] [CrossRef]
- Stoklosa, P.; Borgstrom, A.; Hauert, B.; Baur, R.; Peinelt, C. Investigation of Novel Small Molecular TRPM4 Inhibitors in Colorectal Cancer Cells. Cancers 2021, 13, 5400. [Google Scholar] [CrossRef]
- Sagredo, A.I.; Sagredo, E.A.; Cappelli, C.; Baez, P.; Andaur, R.E.; Blanco, C.; Tapia, J.C.; Echeverria, C.; Cerda, O.; Stutzin, A.; et al. TRPM4 regulates Akt/GSK3-beta activity and enhances beta-catenin signaling and cell proliferation in prostate cancer cells. Mol. Oncol. 2018, 12, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liao, P. TRPM4 channel and cancer. Cancer Lett. 2019, 454, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Borgstrom, A.; Peinelt, C.; Stoklosa, P. TRPM4 in Cancer-A New Potential Drug Target. Biomolecules 2021, 11, 229. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.A. TRPM2 in Cancer. Cell Calcium 2019, 80, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Bao, X.; Wang, H.; Zhu, S.; Liu, Z.; Chen, Q.; Ai, K.; Shi, B. TRPM2 promotes pancreatic cancer by PKC/MAPK pathway. Cell Death Dis. 2021, 12, 585. [Google Scholar] [CrossRef]
- Ji, D.; Luo, Z.W.; Ovcjak, A.; Alanazi, R.; Bao, M.H.; Feng, Z.P.; Sun, H.S. Role of TRPM2 in brain tumours and potential as a drug target. Acta Pharmacol. Sin. 2022, 43, 759–770. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Wu, T.; Dai, Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017, 387, 61–68. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Reinhold, W.C.; Sunshine, M.; Varma, S.; Doroshow, J.H.; Pommier, Y. Using CellMiner 1.6 for Systems Pharmacology and Genomic Analysis of the NCI-60. Clin. Cancer Res. 2015, 21, 3841–3852. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Zhao, Y.; Tian, Y.; Cao, R.; Shang, D. Pan-Cancer Analysis of the TRP Family, Especially TRPV4 and TRPC4, and Its Expression Correlated with Prognosis, Tumor Microenvironment, and Treatment Sensitivity. Biomolecules 2023, 13, 282. https://doi.org/10.3390/biom13020282
Chen Z, Zhao Y, Tian Y, Cao R, Shang D. Pan-Cancer Analysis of the TRP Family, Especially TRPV4 and TRPC4, and Its Expression Correlated with Prognosis, Tumor Microenvironment, and Treatment Sensitivity. Biomolecules. 2023; 13(2):282. https://doi.org/10.3390/biom13020282
Chicago/Turabian StyleChen, Zhenghao, Youquan Zhao, Ye Tian, Rui Cao, and Donghao Shang. 2023. "Pan-Cancer Analysis of the TRP Family, Especially TRPV4 and TRPC4, and Its Expression Correlated with Prognosis, Tumor Microenvironment, and Treatment Sensitivity" Biomolecules 13, no. 2: 282. https://doi.org/10.3390/biom13020282
APA StyleChen, Z., Zhao, Y., Tian, Y., Cao, R., & Shang, D. (2023). Pan-Cancer Analysis of the TRP Family, Especially TRPV4 and TRPC4, and Its Expression Correlated with Prognosis, Tumor Microenvironment, and Treatment Sensitivity. Biomolecules, 13(2), 282. https://doi.org/10.3390/biom13020282







