Identification of an Autophagy-Related Signature for Prognosis and Immunotherapy Response Prediction in Ovarian Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Acquirement of Target Datasets
2.2. Gene Set Variation Analysis (GSVA)
2.3. Human Clinical Specimens
2.4. Immunohistochemical Staining and Analysis
2.5. Analysis of Functional Enrichment and Immune Cell Infiltration in TIME
2.6. Cluster Identification and Annotation of scRNA-Seq Data
2.7. Cell-Cell Interaction Analysis
2.8. Construction and Validation of Autophagy-Risk Signature
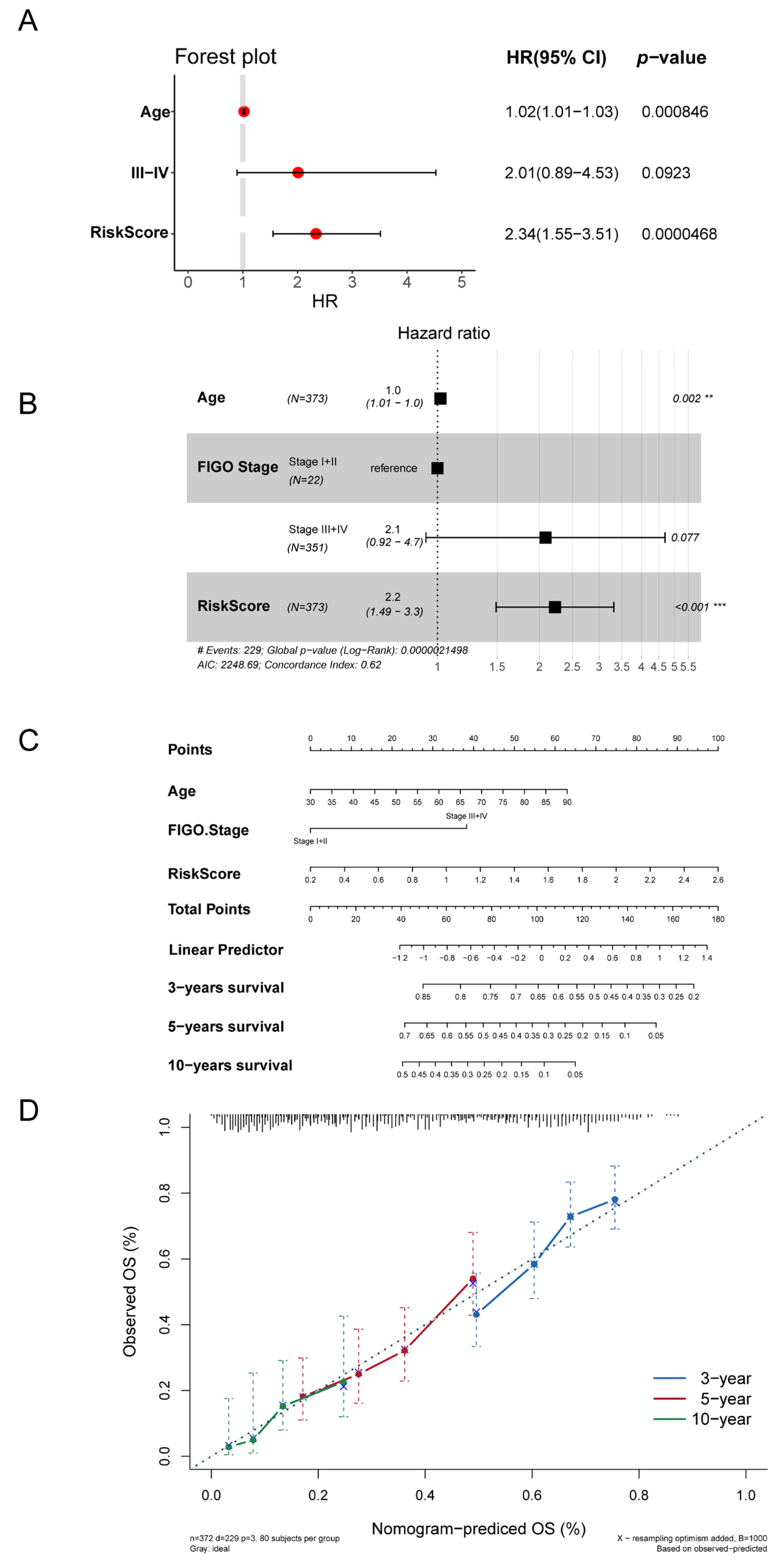
2.9. Prognostic Value and Clinical Applicability Assessment of Autophagy-Risk Signature
2.10. Statistical Analysis
3. Results
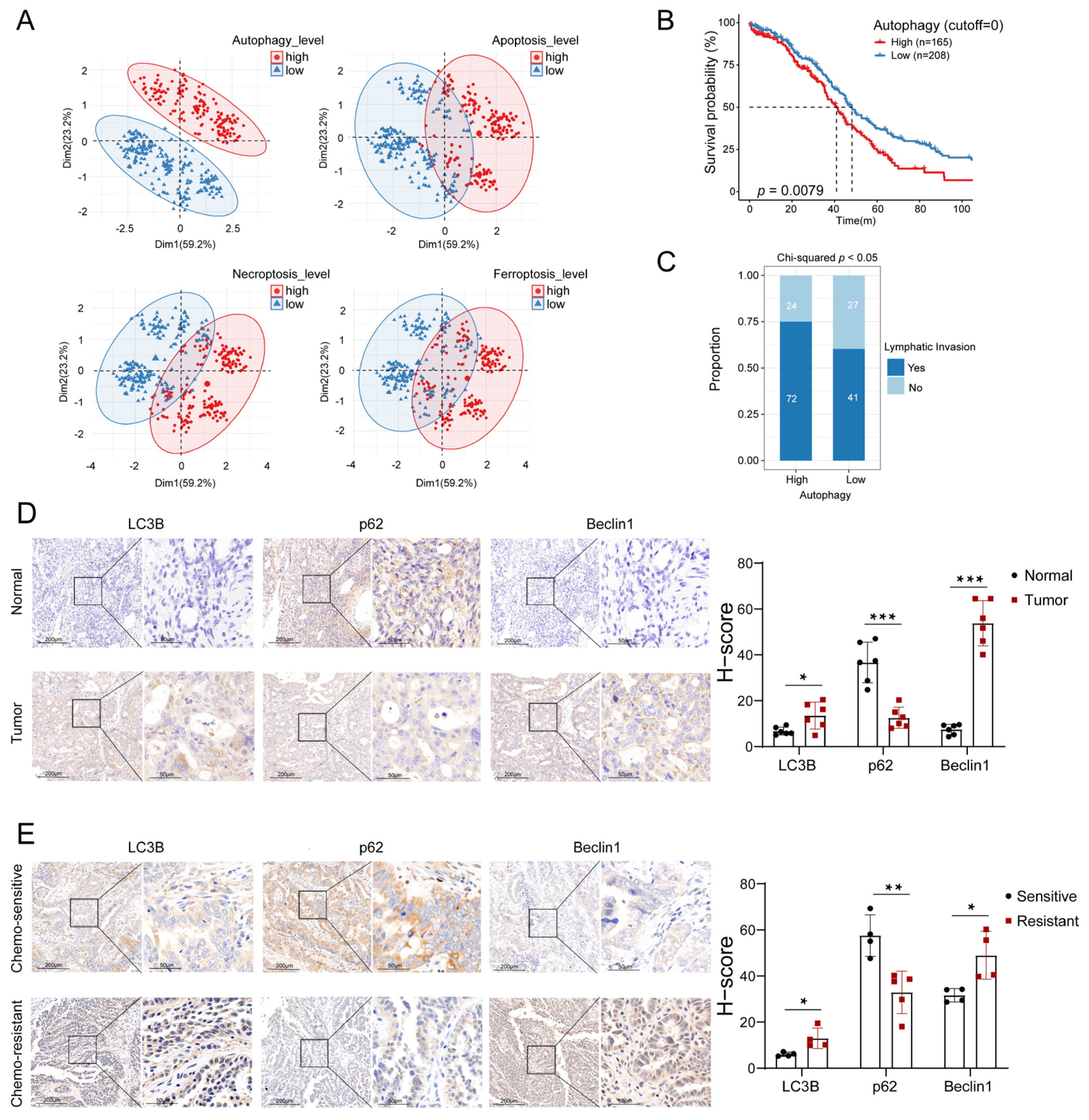
3.1. Autophagy Levels Were Highly Relevant to the Prognosis of OC Patients
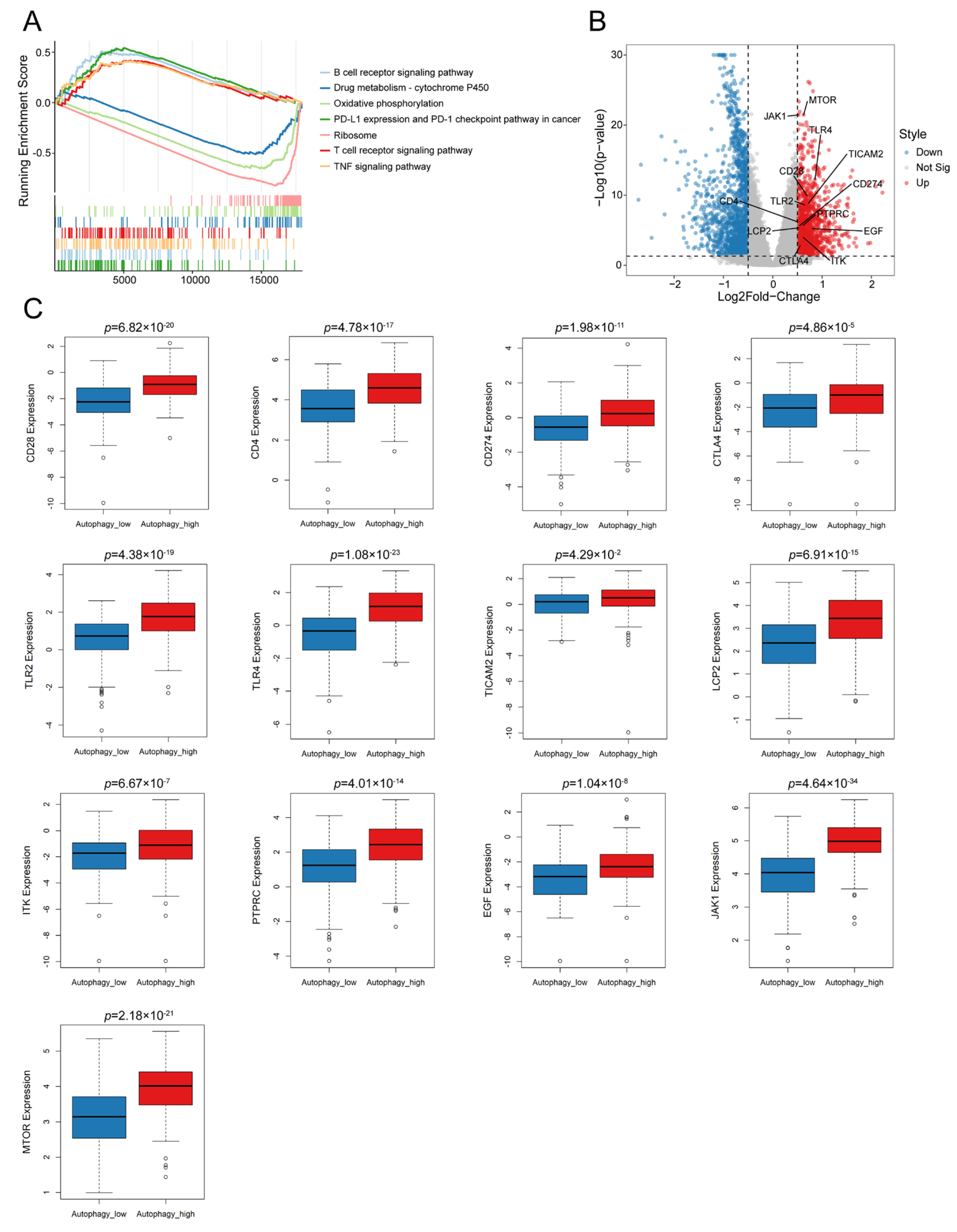
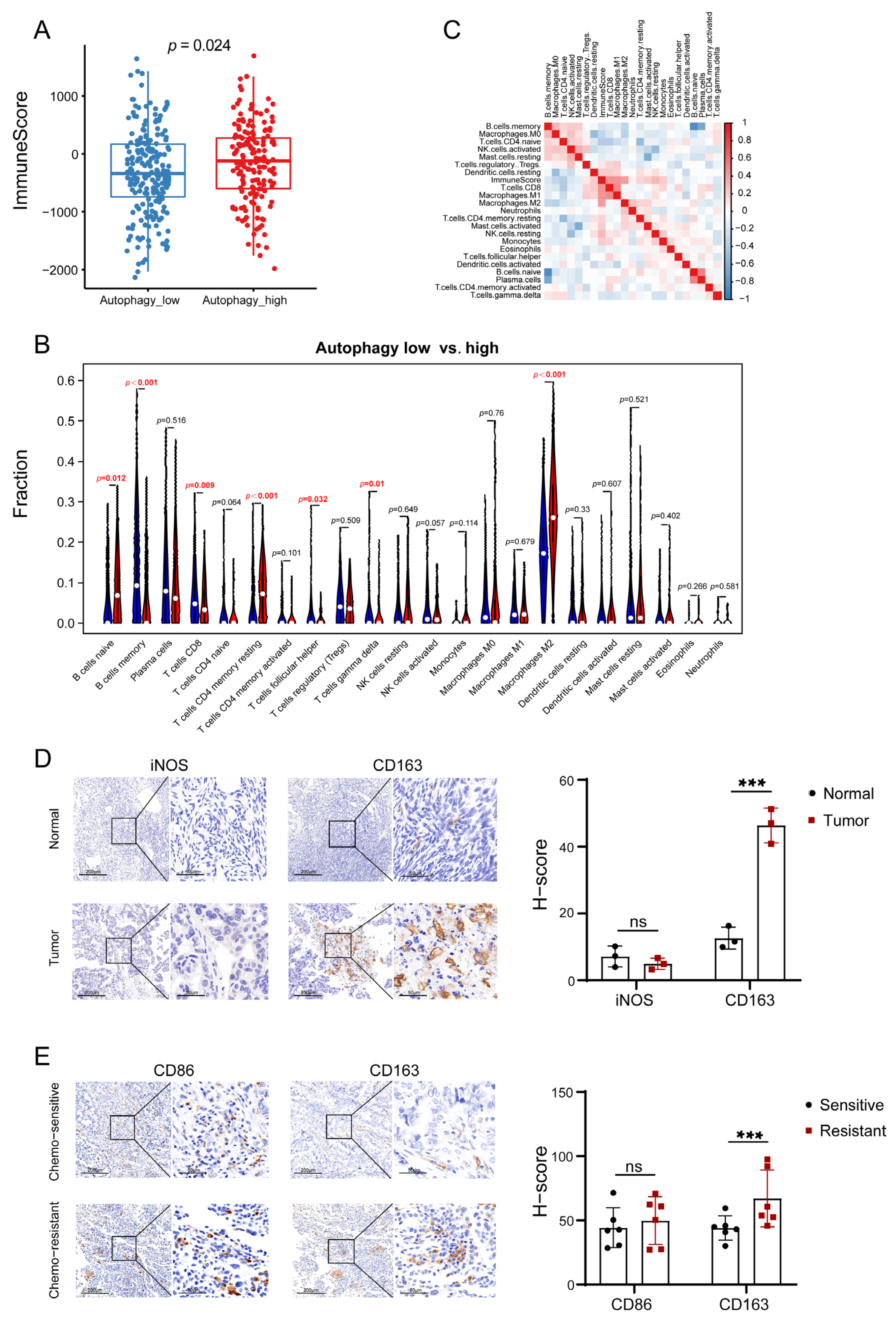
3.2. Autophagy Was Significantly Correlated with the Immune Microenvironment in OC
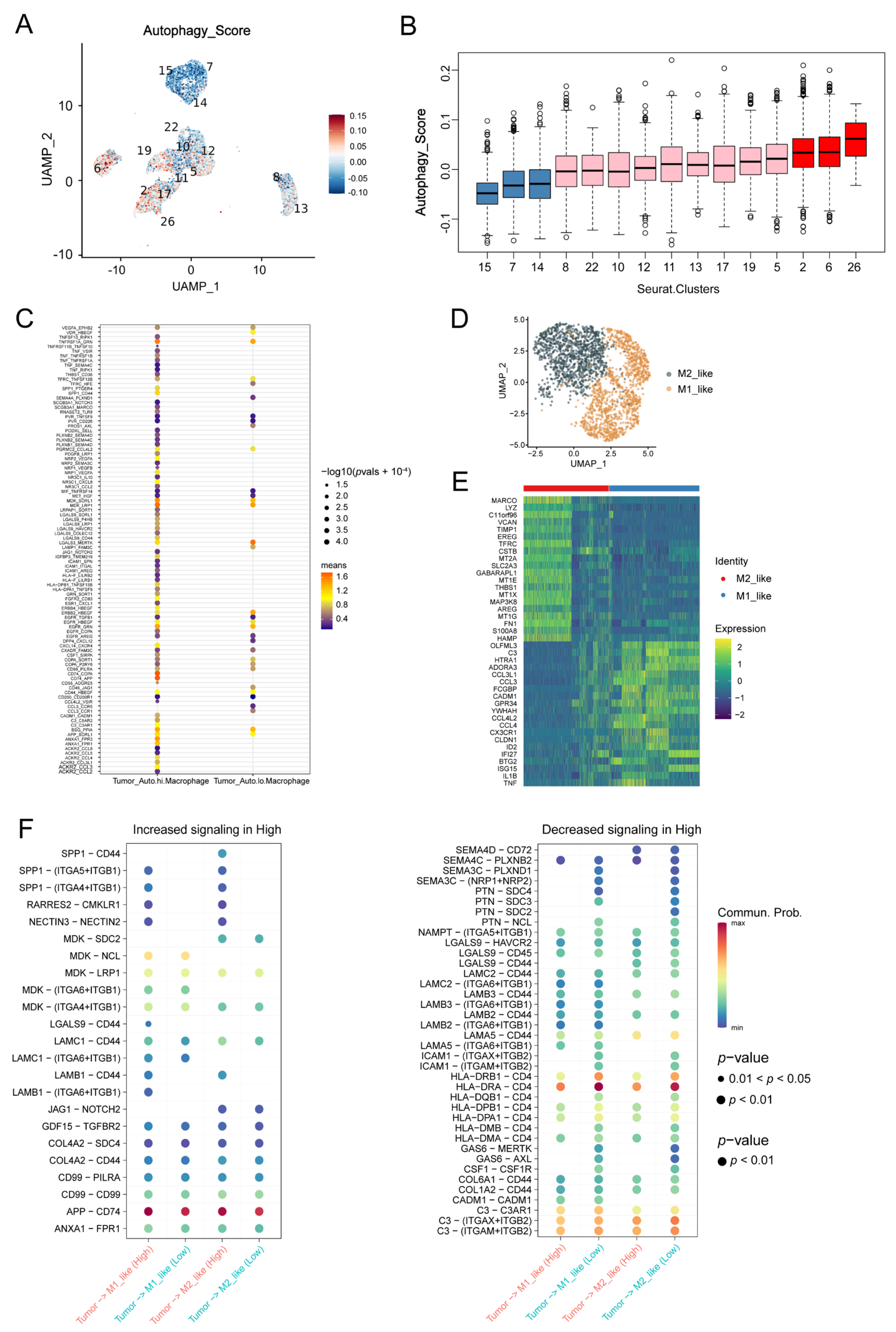
3.3. Macrophages Contributed to the Autophagy-Involved Immune Microenvironment of OC
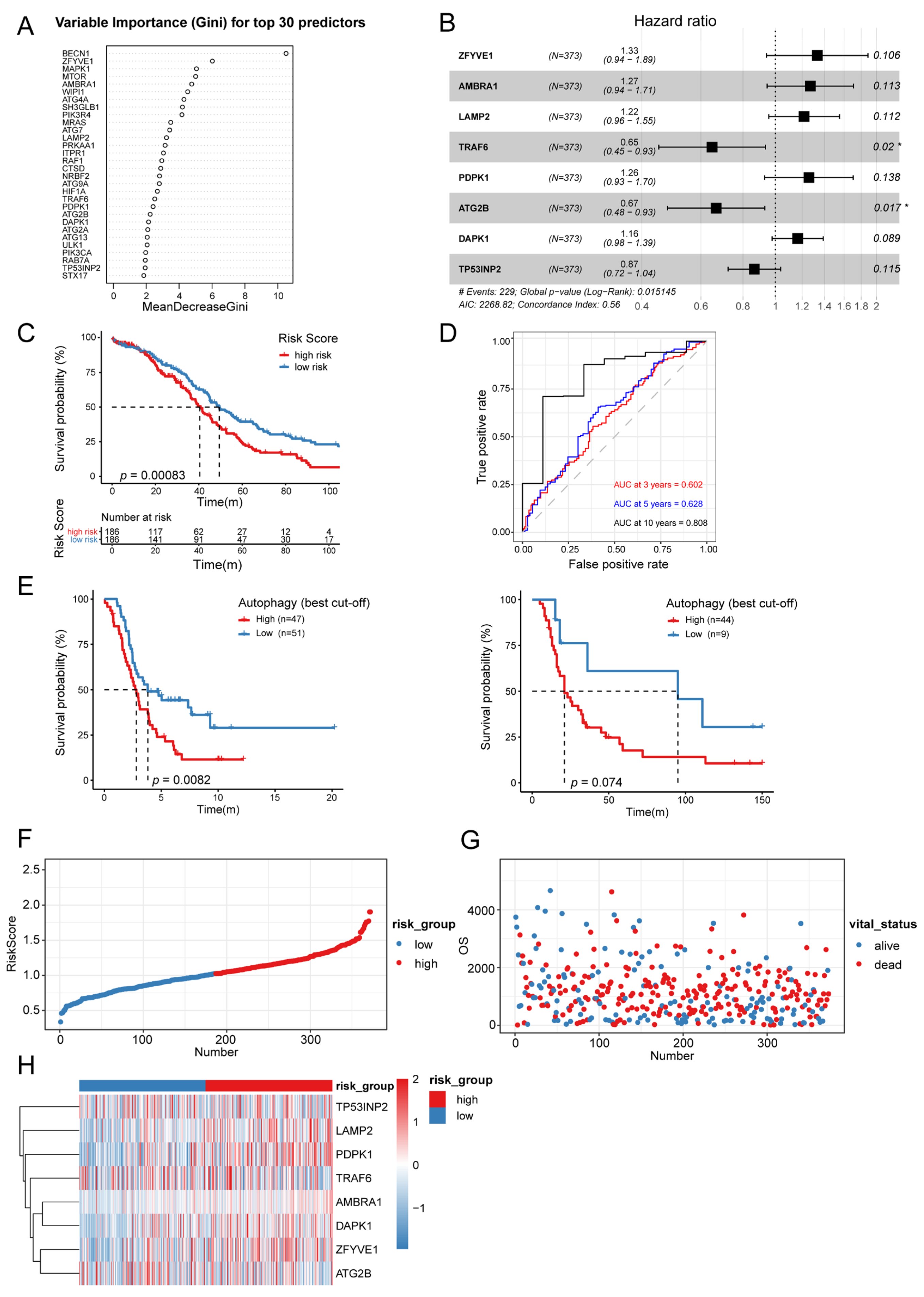
3.4. Establishment of Autophagy-Related Signature
3.5. Autophagy-Related Signature Was an Independent Prognostic Predictor
3.6. Autophagy-Related Signature Predicted ICIs Therapy Efficiency of OC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kandalaft, L.E.; Odunsi, K.; Coukos, G. Immunotherapy in Ovarian Cancer: Are We There Yet? J. Clin. Oncol. 2019, 37, 2460–2471. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Sikora, M.J.; Boisen, M.M.; Ma, T.; Christie, A.; Tseng, G.; Park, Y.; Luthra, S.; Chandran, U.; Haluska, P.; et al. Active Estrogen Receptor-alpha Signaling in Ovarian Cancer Models and Clinical Specimens. Clin. Cancer Res. 2017, 23, 3802–3812. [Google Scholar] [CrossRef] [PubMed]
- McMullen, M.; Karakasis, K.; Madariaga, A.; Oza, A.M. Overcoming Platinum and PARP-Inhibitor Resistance in Ovarian Cancer. Cancers 2020, 12, 1607. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.L.; Cummings, M.; Thangavelu, A.; Theophilou, G.; de Jong, D.; Orsi, N.M. Barriers to Immunotherapy in Ovarian Cancer: Metabolic, Genomic, and Immune Perturbations in the Tumour Microenvironment. Cancers 2021, 13, 6231. [Google Scholar] [CrossRef]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef]
- Galluzzi, L.; Green, D.R. Autophagy-Independent Functions of the Autophagy Machinery. Cell 2019, 177, 1682–1699. [Google Scholar] [CrossRef]
- Clarke, A.J.; Simon, A.K. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat. Rev. Immunol. 2019, 19, 170–183. [Google Scholar] [CrossRef]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef]
- Hu, W.-H.; Yang, W.-C.; Liu, P.-F.; Liu, T.-T.; Morgan, P.; Tsai, W.-L.; Pan, H.-W.; Lee, C.-H.; Shu, C.-W. Clinicopathological Association of Autophagy Related 5 Protein with Prognosis of Colorectal Cancer. Diagnostics 2021, 11, 782. [Google Scholar] [CrossRef]
- Yang, Y.-H.; Liu, J.-B.; Gui, Y.; Lei, L.-L.; Zhang, S.-J. Relationship between autophagy and perineural invasion, clinicopathological features, and prognosis in pancreatic cancer. World J. Gastroenterol. 2017, 23, 7232–7241. [Google Scholar] [CrossRef]
- Hu, D.; Jiang, L.; Luo, S.; Zhao, X.; Hu, H.; Zhao, G.; Tang, W. Development of an autophagy-related gene expression signature for prognosis prediction in prostate cancer patients. J. Transl. Med. 2020, 18, 160. [Google Scholar] [CrossRef]
- Ma, J.-Y.; Liu, Q.; Liu, G.; Peng, S.; Wu, G. Identification and validation of a robust autophagy-related molecular model for predicting the prognosis of breast cancer patients. Aging 2021, 13, 16684–16695. [Google Scholar] [CrossRef]
- Meng, D.; Jin, H.; Zhang, X.; Yan, W.; Xia, Q.; Shen, S.; Xie, S.; Cui, M.; Ding, B.; Gu, Y.; et al. Identification of autophagy-related risk signatures for the prognosis, diagnosis, and targeted therapy in cervical cancer. Cancer Cell Int. 2021, 21, 362. [Google Scholar] [CrossRef]
- Levine, B. Unraveling the role of autophagy in cancer. Autophagy 2006, 2, 65–66. [Google Scholar] [CrossRef]
- Mathew, R.; Karantza-Wadsworth, V.; White, E. Role of autophagy in cancer. Nat. Rev. Cancer 2007, 7, 961–967. [Google Scholar] [CrossRef]
- Yang, Z.J.; Chee, C.E.; Huang, S.; Sinicrope, F.A. The role of autophagy in cancer: Therapeutic implications. Mol. Cancer Ther. 2011, 10, 1533–1541. [Google Scholar] [CrossRef]
- White, E. The role for autophagy in cancer. J. Clin. Investig. 2015, 125, 42–46. [Google Scholar] [CrossRef]
- Mowers, E.E.; Sharifi, M.N.; Macleod, K.F. Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J. 2018, 285, 1751–1766. [Google Scholar] [CrossRef]
- Ma, F.; Ding, M.-G.; Lei, Y.-Y.; Luo, L.-H.; Jiang, S.; Feng, Y.-H.; Liu, X.-L. SKIL facilitates tumorigenesis and immune escape of NSCLC via upregulating TAZ/autophagy axis. Cell Death Dis. 2020, 11, 1028. [Google Scholar] [CrossRef]
- Yamamoto, K.; Venida, A.; Yano, J.; Biancur, D.E.; Kakiuchi, M.; Gupta, S.; Sohn, A.S.W.; Mukhopadhyay, S.; Lin, E.Y.; Parker, S.J.; et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020, 581, 100–105. [Google Scholar] [CrossRef]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E.; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Shiuan, E.; Reddy, A.; Dudzinski, S.O.; Lim, A.R.; Sugiura, A.; Hongo, R.; Young, K.; Liu, X.D.; Smith, C.C.; O’Neal, J.; et al. Clinical Features and Multiplatform Molecular Analysis Assist in Understanding Patient Response to Anti-PD-1/PD-L1 in Renal Cell Carcinoma. Cancers 2021, 13, 1475. [Google Scholar] [CrossRef] [PubMed]
- Notter, T.; Panzanelli, P.; Pfister, S.; Mircsof, D.; Fritschy, J.-M. A protocol for concurrent high-quality immunohistochemical and biochemical analyses in adult mouse central nervous system. Eur. J. Neurosci. 2014, 39, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Detre, S.; Saclani Jotti, G.; Dowsett, M. A “quickscore” method for immunohistochemical semiquantitation: Validation for oestrogen receptor in breast carcinomas. J. Clin. Pathol. 1995, 48, 876–878. [Google Scholar] [CrossRef]
- Derangère, V.; Lecuelle, J.; Lepage, C.; Aoulad-Ben Salem, O.; Allatessem, B.M.; Ilie, A.; Bouché, O.; Phelip, J.-M.; Baconnier, M.; Pezet, D.; et al. Combination of CDX2 H-score quantitative analysis with CD3 AI-guided analysis identifies patients with a good prognosis only in stage III colon cancer. Eur. J. Cancer 2022, 172, 221–230. [Google Scholar] [CrossRef]
- Mazières, J.; Brugger, W.; Cappuzzo, F.; Middel, P.; Frosch, A.; Bara, I.; Klingelschmitt, G.; Klughammer, B. Evaluation of EGFR protein expression by immunohistochemistry using H-score and the magnification rule: Re-analysis of the SATURN study. Lung Cancer 2013, 82, 231–237. [Google Scholar] [CrossRef]
- Vento-Tormo, R.; Efremova, M.; Botting, R.A.; Turco, M.Y.; Vento-Tormo, M.; Meyer, K.B.; Park, J.-E.; Stephenson, E.; Polański, K.; Goncalves, A.; et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018, 563, 347–353. [Google Scholar] [CrossRef]
- Liu, J.; Hong, M.; Li, Y.; Chen, D.; Wu, Y.; Hu, Y. Programmed Cell Death Tunes Tumor Immunity. Front. Immunol. 2022, 13, 847345. [Google Scholar] [CrossRef]
- Jiang, G.-M.; Tan, Y.; Wang, H.; Peng, L.; Chen, H.-T.; Meng, X.-J.; Li, L.-L.; Liu, Y.; Li, W.-F.; Shan, H. The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol. Cancer 2019, 18, 17. [Google Scholar] [CrossRef]
- Lisi, L.; Ciotti, G.M.P.; Braun, D.; Kalinin, S.; Currò, D.; Dello Russo, C.; Coli, A.; Mangiola, A.; Anile, C.; Feinstein, D.L.; et al. Expression of iNOS, CD163 and ARG-1 taken as M1 and M2 markers of microglial polarization in human glioblastoma and the surrounding normal parenchyma. Neurosci. Lett. 2017, 645, 106–112. [Google Scholar] [CrossRef]
- Dong, P.; Ma, L.; Liu, L.; Zhao, G.; Zhang, S.; Dong, L.; Xue, R.; Chen, S. CD86⁺/CD206⁺, Diametrically Polarized Tumor-Associated Macrophages, Predict Hepatocellular Carcinoma Patient Prognosis. Int. J. Mol. Sci. 2016, 17, 320. [Google Scholar] [CrossRef]
- Ferrisse, T.M.; de Oliveira, A.B.; Palaçon, M.P.; Silva, E.V.; Massucato, E.M.S.; de Almeida, L.Y.; Léon, J.E.; Bufalino, A. The role of CD68+ and CD163+ macrophages in immunopathogenesis of oral lichen planus and oral lichenoid lesions. Immunobiology 2021, 226, 152072. [Google Scholar] [CrossRef]
- Liu, J.; Geng, X.; Hou, J.; Wu, G. New insights into M1/M2 macrophages: Key modulators in cancer progression. Cancer Cell Int. 2021, 21, 389. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, W.; Chen, Z.; Xiang, C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp. Cell Res. 2017, 359, 449–457. [Google Scholar] [CrossRef]
- Qi, J.; Sun, H.; Zhang, Y.; Wang, Z.; Xun, Z.; Li, Z.; Ding, X.; Bao, R.; Hong, L.; Jia, W.; et al. Single-cell and spatial analysis reveal interaction of FAP fibroblasts and SPP1 macrophages in colorectal cancer. Nat. Commun. 2022, 13, 1742. [Google Scholar] [CrossRef]
- Ho, D.W.-H.; Tsui, Y.-M.; Chan, L.-K.; Sze, K.M.-F.; Zhang, X.; Cheu, J.W.-S.; Chiu, Y.-T.; Lee, J.M.-F.; Chan, A.C.-Y.; Cheung, E.T.-Y.; et al. Single-cell RNA sequencing shows the immunosuppressive landscape and tumor heterogeneity of HBV-associated hepatocellular carcinoma. Nat. Commun. 2021, 12, 3684. [Google Scholar] [CrossRef]
- Dai, J.-C.; Yang, J.-Y.; Chang, R.-Q.; Liang, Y.; Hu, X.-Y.; Li, H.; You, S.; He, F.; Hu, L.-N. GAS6-mediated dialogue between decidual stromal cells and macrophages is essential for early pregnancy maintenance by inducing M2-like polarization and cell proliferation of decidual macrophages. Mol. Hum. Reprod. 2022, 28. [Google Scholar] [CrossRef]
- Ran, T.; Chen, Z.; Zhao, L.; Ran, W.; Fan, J.; Hong, S.; Yang, Z. LAMB1 Is Related to the T Stage and Indicates Poor Prognosis in Gastric Cancer. Technol. Cancer Res. Treat. 2021, 20, 15330338211004944. [Google Scholar] [CrossRef]
- Ma, D.; Zhan, D.; Fu, Y.; Wei, S.; Lal, B.; Wang, J.; Li, Y.; Lopez-Bertoni, H.; Yalcin, F.; Dzaye, O.; et al. Mutant IDH1 promotes phagocytic function of microglia/macrophages in gliomas by downregulating ICAM1. Cancer Lett. 2021, 517, 35–45. [Google Scholar] [CrossRef]
- Fujiwara, T.; Yakoub, M.A.; Chandler, A.; Christ, A.B.; Yang, G.; Ouerfelli, O.; Rajasekhar, V.K.; Yoshida, A.; Kondo, H.; Hata, T.; et al. CSF1/CSF1R Signaling Inhibitor Pexidartinib (PLX3397) Reprograms Tumor-Associated Macrophages and Stimulates T-cell Infiltration in the Sarcoma Microenvironment. Mol. Cancer Ther. 2021, 20, 1388–1399. [Google Scholar] [CrossRef]
- Shi, Y.; Ping, Y.-F.; Zhou, W.; He, Z.-C.; Chen, C.; Bian, B.-S.-J.; Zhang, L.; Chen, L.; Lan, X.; Zhang, X.-C.; et al. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat. Commun. 2017, 8, 15080. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Beltran, A.; Cheng, L. Histologic variants of urothelial carcinoma: Differential diagnosis and clinical implications. Hum. Pathol. 2006, 37, 1371–1388. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, Y.; Wang, C. Beyond Tumor Mutation Burden: Tumor Neoantigen Burden as a Biomarker for Immunotherapy and Other Types of Therapy. Front. Oncol. 2021, 11, 672677. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018, 362. [Google Scholar] [CrossRef]
- Tan, W.-X.; Xu, T.-M.; Zhou, Z.-L.; Lv, X.-J.; Liu, J.; Zhang, W.-J.; Cui, M.-H. TRP14 promotes resistance to cisplatin by inducing autophagy in ovarian cancer. Oncol. Rep. 2019, 42, 1343–1354. [Google Scholar] [CrossRef]
- Zhou, F.; Yang, X.; Zhao, H.; Liu, Y.; Feng, Y.; An, R.; Lv, X.; Li, J.; Chen, B. Down-regulation of OGT promotes cisplatin resistance by inducing autophagy in ovarian cancer. Theranostics 2018, 8, 5200–5212. [Google Scholar] [CrossRef]
- Sutton, E.C.; DeRose, V.J. Early nucleolar responses differentiate mechanisms of cell death induced by oxaliplatin and cisplatin. J. Biol. Chem. 2021, 296, 100633. [Google Scholar] [CrossRef]
- Pecoraro, A.; Pagano, M.; Russo, G.; Russo, A. Role of Autophagy in Cancer Cell Response to Nucleolar and Endoplasmic Reticulum Stress. Int. J. Mol. Sci. 2020, 21, 7334. [Google Scholar] [CrossRef]
- Pecoraro, A.; Carotenuto, P.; Franco, B.; De Cegli, R.; Russo, G.; Russo, A. Role of uL3 in the Crosstalk between Nucleolar Stress and Autophagy in Colon Cancer Cells. Int. J. Mol. Sci. 2020, 21, 2143. [Google Scholar] [CrossRef]
- Siva Sankar, D.; Dengjel, J. Protein complexes and neighborhoods driving autophagy. Autophagy 2021, 17, 2689–2705. [Google Scholar] [CrossRef]
- Zhong, X.; Feng, L.; Xu, W.-H.; Wu, X.; Ding, Y.-D.; Zhou, Y.; Lei, C.-Q.; Shu, H.-B. The zinc-finger protein ZFYVE1 modulates TLR3-mediated signaling by facilitating TLR3 ligand binding. Cell. Mol. Immunol. 2020, 17, 741–752. [Google Scholar] [CrossRef]
- Di Leo, L.; De Zio, D. AMBRA1 has an impact on melanoma development beyond autophagy. Autophagy 2021, 17, 1802–1803. [Google Scholar] [CrossRef]
- Eskelinen, E.-L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Asp. Med. 2006, 27, 495–502. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Sousa, L.G.; Perrod, C.; Maceiras, A.R.; Ferreirinha, P.; Pombinho, R.; Romera-Cárdenas, G.; Gomez-Lazaro, M.; Senkara, M.; Pistolic, J.; et al. LAMP2 regulates autophagy in the thymic epithelium and thymic stroma-dependent CD4 T cell development. Autophagy 2023, 19, 426–439. [Google Scholar] [CrossRef]
- Min, Y.; Kim, M.-J.; Lee, S.; Chun, E.; Lee, K.-Y. Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy 2018, 14, 1347–1358. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, B.; Li, Y.; Deng, T.; Xu, Y.; Lei, J.; Zhou, J. Binding of VP3 to the PIK3C3-PDPK1 complex inhibits autophagy by activating the AKT-MTOR pathway. Autophagy 2020, 16, 1697–1710. [Google Scholar] [CrossRef]
- Velikkakath, A.K.G.; Nishimura, T.; Oita, E.; Ishihara, N.; Mizushima, N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell 2012, 23, 896–909. [Google Scholar] [CrossRef]
- Singh, P.; Ravanan, P.; Talwar, P. Death Associated Protein Kinase 1 (DAPK1): A Regulator of Apoptosis and Autophagy. Front. Mol. Neurosci. 2016, 9, 46. [Google Scholar] [CrossRef]
- You, Z.; Xu, Y.; Wan, W.; Zhou, L.; Li, J.; Zhou, T.; Shi, Y.; Liu, W. TP53INP2 contributes to autophagosome formation by promoting LC3-ATG7 interaction. Autophagy 2019, 15, 1309–1321. [Google Scholar] [CrossRef]
- Hessmann, E.; Buchholz, S.M.; Demir, I.E.; Singh, S.K.; Gress, T.M.; Ellenrieder, V.; Neesse, A. Microenvironmental Determinants of Pancreatic Cancer. Physiol. Rev. 2020, 100, 1707–1751. [Google Scholar] [CrossRef]
- Liu, T.; Wang, L.; Liang, P.; Wang, X.; Liu, Y.; Cai, J.; She, Y.; Wang, D.; Wang, Z.; Guo, Z.; et al. USP19 suppresses inflammation and promotes M2-like macrophage polarization by manipulating NLRP3 function via autophagy. Cell. Mol. Immunol. 2021, 18, 2431–2442. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Green, D.R.; Zou, W. Autophagy in tumour immunity and therapy. Nat. Rev. Cancer 2021, 21, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Poillet-Perez, L.; Sharp, D.W.; Yang, Y.; Laddha, S.V.; Ibrahim, M.; Bommareddy, P.K.; Hu, Z.S.; Vieth, J.; Haas, M.; Bosenberg, M.W.; et al. Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response. Nat. Cancer 2020, 1, 923–934. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.P.B.; Rose, C.J.; Waterton, J.C.; Carano, R.A.D.; Parker, G.J.M.; Jackson, A. Imaging intratumor heterogeneity: Role in therapy response, resistance, and clinical outcome. Clin. Cancer Res. 2015, 21, 249–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, J.; Wang, C.; Sun, Y.; Guo, J.; Liu, S.; Cheng, Z. Identification of an Autophagy-Related Signature for Prognosis and Immunotherapy Response Prediction in Ovarian Cancer. Biomolecules 2023, 13, 339. https://doi.org/10.3390/biom13020339
Ding J, Wang C, Sun Y, Guo J, Liu S, Cheng Z. Identification of an Autophagy-Related Signature for Prognosis and Immunotherapy Response Prediction in Ovarian Cancer. Biomolecules. 2023; 13(2):339. https://doi.org/10.3390/biom13020339
Chicago/Turabian StyleDing, Jinye, Chunyan Wang, Yaoqi Sun, Jing Guo, Shupeng Liu, and Zhongping Cheng. 2023. "Identification of an Autophagy-Related Signature for Prognosis and Immunotherapy Response Prediction in Ovarian Cancer" Biomolecules 13, no. 2: 339. https://doi.org/10.3390/biom13020339




