Structural Characterization of Nanobodies during Germline Maturation
Abstract
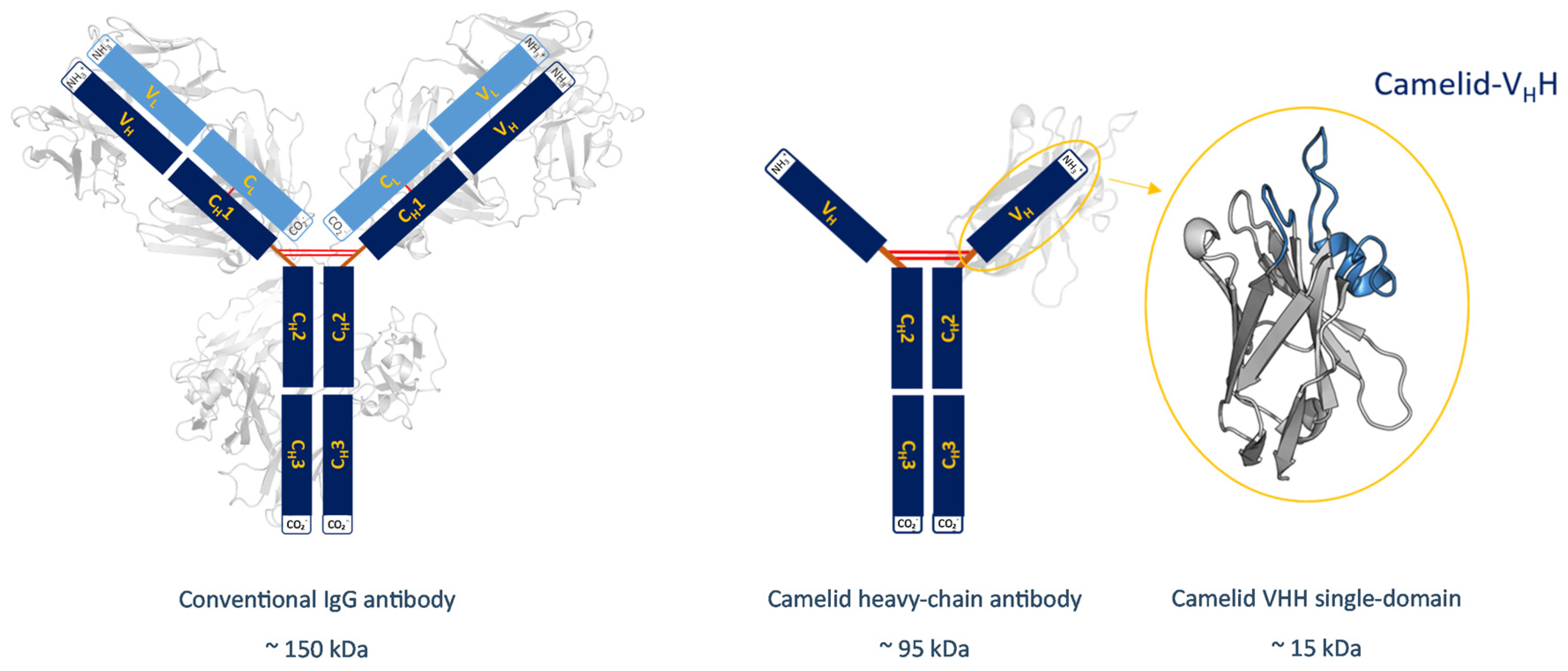
1. Introduction
2. Materials and Methods
2.1. Metadynamics Simulations
2.2. Molecular Dynamics Simulations
2.3. Analysis of the Simulation Trajectories
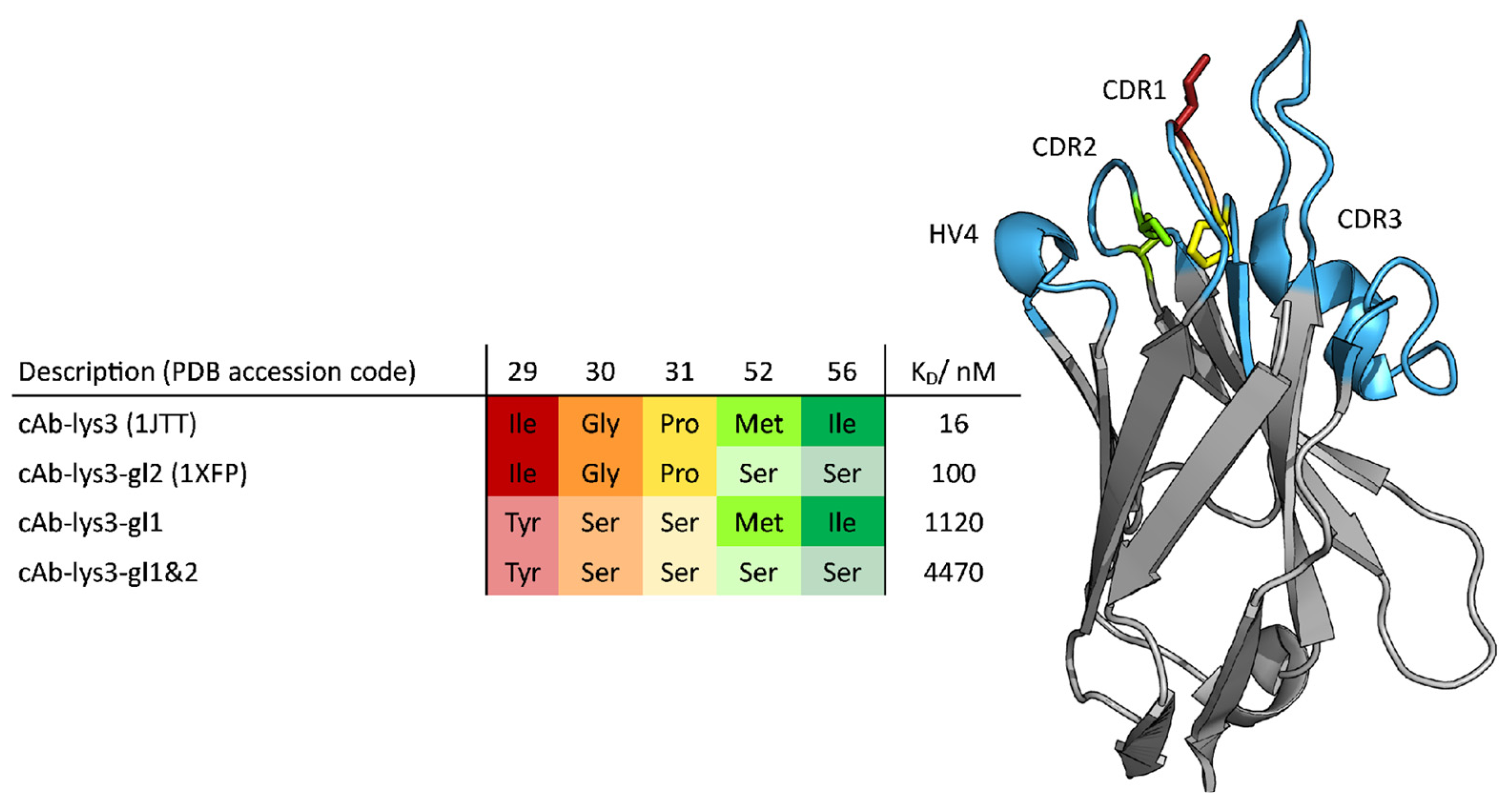
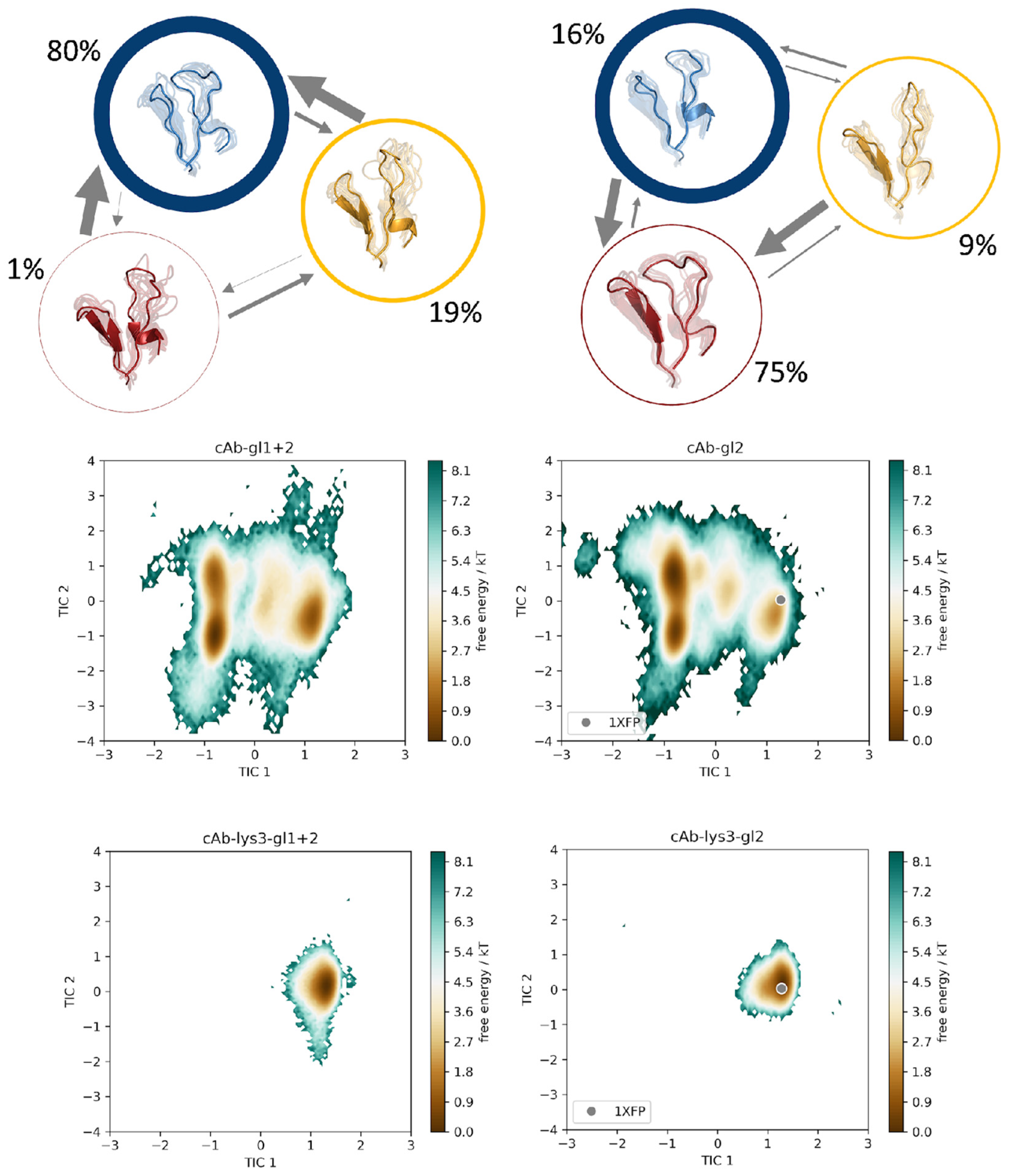
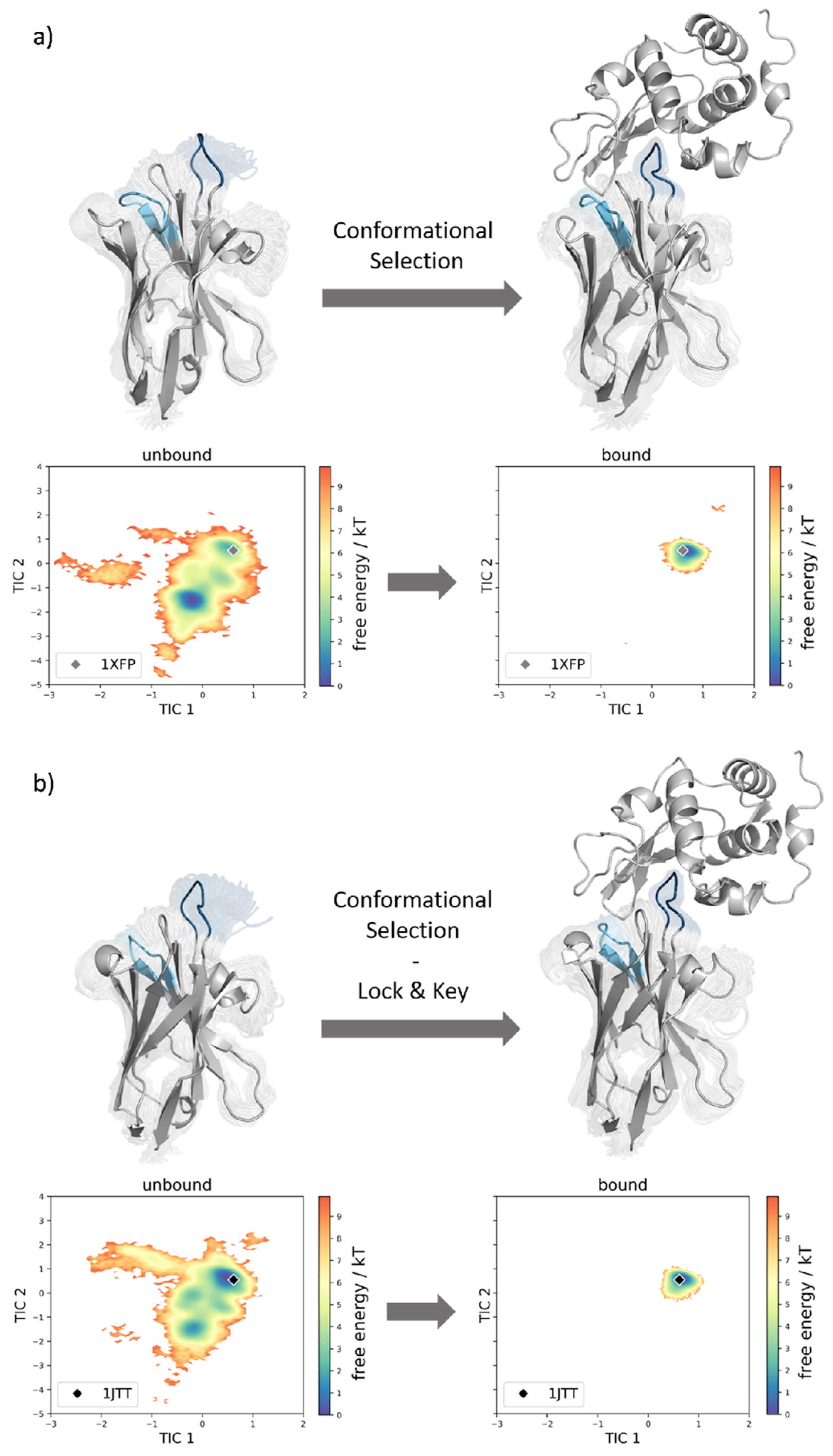
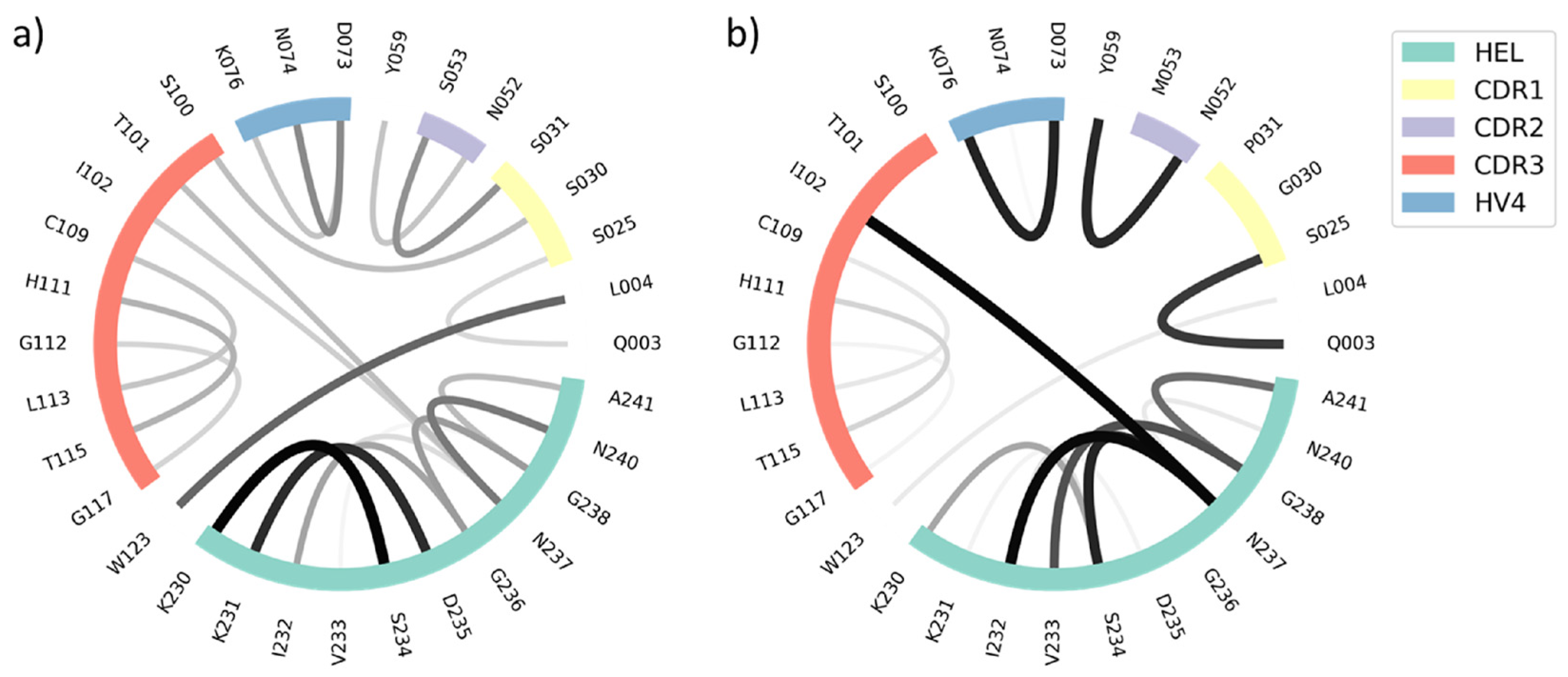
3. Results
Time-Lagged Independent Component Analysis (tICA) and Markov-State Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Genst, E.; Saerens, D.; Muyldermans, S.; Conrath, K. Antibody Repertoire Development in Camelids. Dev. Comp. Immunol. 2006, 30, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Dooley, H.; Flajnik, M.F. Antibody Repertoire Development in Cartilaginous Fish. Dev. Comp. Immunol. 2006, 30, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S.; Baral, T.N.; Retamozzo, V.C.; De Baetselier, P.; De Genst, E.; Kinne, J.; Leonhardt, H.; Magez, S.; Nguyen, V.K.; Revets, H. Camelid Immunoglobulins and Nanobody Technology. Vet. Immunol. Immunopathol. 2009, 128, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F.; Deschacht, N.; Muyldermans, S. A Case of Convergence: Why Did a Simple Alternative to Canonical Antibodies Arise in Sharks and Camels? PLoS Biol. 2011, 9, e1001120. [Google Scholar] [CrossRef]
- Chiu, M.L.; Goulet, D.R.; Teplyakov, A.; Gilliland, G.L. Antibody Structure and Function: The Basis for Engineering Therapeutics. Antibodies 2019, 8, 55. [Google Scholar] [CrossRef]
- Kabat, E.A.; Te Wu, T.; Perry, H.M.; Foeller, C.; Gottesman, K.S. Sequences of Proteins of Immunological Interest; DIANE Publishing: Darby, PA, USA, 1992; ISBN 0-941375-65-X. [Google Scholar]
- Nguyen, V.K.; Muyldermans, S.; Hamers, R. The Specific Variable Domain of Camel Heavy-Chain Antibodies Is Encoded in the Germline. J. Mol. Biol. 1998, 275, 413–418. [Google Scholar] [CrossRef]
- Harmsen, M.M.; De Haard, H.J. Properties, Production, and Applications of Camelid Single-Domain Antibody Fragments. Appl. Microbiol. Biotechnol. 2007, 77, 13–22. [Google Scholar] [CrossRef]
- Hoey, R.J.; Eom, H.; Horn, J.R. Structure and Development of Single Domain Antibodies as Modules for Therapeutics and Diagnostics. Exp. Biol. Med. 2019, 244, 1568–1576. [Google Scholar] [CrossRef]
- Rahbarizadeh, F.; Ahmadvand, D.; Sharifzadeh, Z. Nanobody; an Old Concept and New Vehicle for Immunotargeting. Immunol. Investig. 2011, 40, 299–338. [Google Scholar] [CrossRef]
- Salvador, J.-P.; Vilaplana, L.; Marco, M.-P. Nanobody: Outstanding Features for Diagnostic and Therapeutic Applications. Anal. Bioanal. Chem. 2019, 411, 1703–1713. [Google Scholar] [CrossRef]
- Regep, C.; Georges, G.; Shi, J.; Popovic, B.; Deane, C.M. The H3 Loop of Antibodies Shows Unique Structural Characteristics. Proteins Struct. Funct. Bioinforma. 2017, 85, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Jeliazkov, J.R.; Sljoka, A.; Kuroda, D.; Tsuchimura, N.; Katoh, N.; Tsumoto, K.; Gray, J.J. Repertoire Analysis of Antibody CDR-H3 Loops Suggests Affinity Maturation Does Not Typically Result in Rigidification. Front. Immunol. 2018, 9, 413. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Quintero, M.L.; Loeffler, J.R.; Kraml, J.; Kahler, U.; Kamenik, A.S.; Liedl, K.R. Characterizing the Diversity of the CDR-H3 Loop Conformational Ensembles in Relationship to Antibody Binding Properties. Front. Immunol. 2019, 9, 3065. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.T.; Kelm, S.; Liu, X.; Taylor, R.D.; Baker, T.; Essex, J.W. Higher Affinity Antibodies Bind With Lower Hydration and Flexibility in Large Scale Simulations. Front. Immunol. 2022, 13, 884110. [Google Scholar] [CrossRef] [PubMed]
- Zavrtanik, U.; Lukan, J.; Loris, R.; Lah, J.; Hadži, S. Structural Basis of Epitope Recognition by Heavy-Chain Camelid Antibodies. J. Mol. Biol. 2018, 430, 4369–4386. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, L.; Muyldermans, S. Single Domain Antibodies: Comparison of Camel VH and Camelised Human VH Domains. J. Immunol. Methods 1999, 231, 25–38. [Google Scholar] [CrossRef]
- Ewert, S.; Cambillau, C.; Conrath, K.; Plückthun, A. Biophysical Properties of Camelid VHH Domains Compared to Those of Human VH3 Domains. Biochemistry 2002, 41, 3628–3636. [Google Scholar] [CrossRef]
- Akiba, H.; Tamura, H.; Kiyoshi, M.; Yanaka, S.; Sugase, K.; Caaveiro, J.M.; Tsumoto, K. Structural and Thermodynamic Basis for the Recognition of the Substrate-Binding Cleft on Hen Egg Lysozyme by a Single-Domain Antibody. Sci. Rep. 2019, 9, 15481. [Google Scholar] [CrossRef]
- Cegielska-Radziejewska, R.; Lesnierowski, G.; Kijowski, J. Properties and Application of Egg White Lysozyme and Its Modified Preparations-a Review. Pol. J. Food Nutr. Sci. 2008, 58, 5–10. [Google Scholar]
- Ragland, S.A.; Criss, A.K. From Bacterial Killing to Immune Modulation: Recent Insights into the Functions of Lysozyme. PLoS Pathog. 2017, 13, e1006512. [Google Scholar] [CrossRef]
- De Genst, E.; Handelberg, F.; Van Meirhaeghe, A.; Vynck, S.; Loris, R.; Wyns, L.; Muyldermans, S. Chemical Basis for the Affinity Maturation of a Camel Single Domain Antibody. J. Biol. Chem. 2004, 279, 53593–53601. [Google Scholar] [CrossRef] [PubMed]
- Yau, K.Y.; Dubuc, G.; Li, S.; Hirama, T.; MacKenzie, C.R.; Jermutus, L.; Hall, J.C.; Tanha, J. Affinity Maturation of a VHH by Mutational Hotspot Randomization. J. Immunol. Methods 2005, 297, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Quintero, M.L.; Loeffler, J.R.; Bacher, L.M.; Waibl, F.; Seidler, C.A.; Liedl, K.R. Local and Global Rigidification upon Antibody Affinity Maturation. Front. Mol. Biosci. 2020, 7, 182. [Google Scholar] [CrossRef] [PubMed]
- Chemical Computing Group. Molecular Operating Environment (MOE). 1010 Sherbrooke St. West, Suite #910; H3A. 2R7; Chemical Computing Group: Montreal, QC, Canada, 2020. [Google Scholar]
- Labute, P. Protonate3D: Assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins 2009, 75, 187–205. [Google Scholar] [CrossRef] [PubMed]
- Hub, J.S.; de Groot, B.L.; Grubmüller, H.; Groenhof, G. Quantifying Artifacts in Ewald Simulations of Inhomogeneous Systems with a Net Charge. J. Chem. Theory Comput. 2014, 10, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- El Hage, K.; Hedin, F.; Gupta, P.K.; Meuwly, M.; Karplus, M. Valid Molecular Dynamics Simulations of Human Hemoglobin Require a Surprisingly Large Box Size. Elife 2018, 7, e35560. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Wallnoefer, H.G.; Liedl, K.R.; Fox, T. A Challenging System: Free Energy Prediction for Factor Xa. J. Comput. Chem. 2011, 32, 1743–1752. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Barducci, A.; Bussi, G.; Parrinello, M. Well-Tempered Metadynamics: A Smoothly Converging and Tunable Free-Energy Method. Phys. Rev. Lett. 2008, 100, 020603. [Google Scholar] [CrossRef]
- Barducci, A.; Bonomi, M.; Parrinello, M. Linking Well-Tempered Metadynamics Simulations with Experiments. Biophys. J. 2010, 98, L44–L46. [Google Scholar] [CrossRef]
- Ilott, A.J.; Palucha, S.; Hodgkinson, P.; Wilson, M.R. Well-Tempered Metadynamics as a Tool for Characterizing Multi-Component, Crystalline Molecular Machines. J. Phys. Chem. B 2013, 117, 12286–12295. [Google Scholar] [CrossRef]
- Biswas, M.; Lickert, B.; Stock, G. Metadynamics Enhanced Markov Modeling of Protein Dynamics. J. Phys. Chem. B 2018, 122, 5508–5514. [Google Scholar] [CrossRef] [PubMed]
- Barducci, A.; Bonomi, M.; Parrinello, M. Metadynamics. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011, 1, 826–843. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; Van Der Spoel, D. GROMACS 4.5: A High-Throughput and Highly Parallel Open Source Molecular Simulation Toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Tribello, G.A.; Bonomi, M.; Branduardi, D.; Camilloni, C.; Bussi, G. PLUMED 2: New Feathers for an Old Bird. Comput. Phys. Commun. 2014, 185, 604–613. [Google Scholar] [CrossRef]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of Polypeptide Chain Configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Laio, A.; Gervasio, F.L. Metadynamics: A Method to Simulate Rare Events and Reconstruct the Free Energy in Biophysics, Chemistry and Material Science. Rep. Prog. Phys. 2008, 71, 126601. [Google Scholar] [CrossRef]
- Adelman, S.A.; Doll, J.D. Generalized Langevin Equation Approach for Atom/Solid-surface Scattering: General Formulation for Classical Scattering off Harmonic Solids. J. Chem. Phys. 1976, 64, 2375–2388. [Google Scholar] [CrossRef]
- Berendsen, H.J.; van Postma, J.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Gotz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham III, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.; Brozell, S.R.; Cerutti, D.; Cheatham, T.; Cruzeiro, V.W.D.; Darden, T.; Duke, R.E.; Giambasu, G. Amber 2020; University of California: San Francisco, CA, USA, 2020. [Google Scholar]
- Scherer, M.K.; Trendelkamp-Schroer, B.; Paul, F.; Pérez-Hernández, G.; Hoffmann, M.; Plattner, N.; Wehmeyer, C.; Prinz, J.-H.; Noé, F. PyEMMA 2: A Software Package for Estimation, Validation, and Analysis of Markov Models. J. Chem. Theory Comput. 2015, 11, 5525–5542. [Google Scholar] [CrossRef]
- Pérez-Hernández, G.; Noé, F. Hierarchical Time-Lagged Independent Component Analysis: Computing Slow Modes and Reaction Coordinates for Large Molecular Systems. J. Chem. Theory Comput. 2016, 12, 6118–6129. [Google Scholar] [CrossRef]
- Pande, V.S.; Beauchamp, K.; Bowman, G.R. Everything You Wanted to Know about Markov State Models but Were Afraid to Ask. Methods 2010, 52, 99–105. [Google Scholar] [CrossRef]
- Likas, A.; Vlassis, N.; Verbeek, J.J. The Global K-Means Clustering Algorithm. Pattern Recognit. 2003, 36, 451–461. [Google Scholar] [CrossRef]
- Röblitz, S.; Weber, M. Fuzzy Spectral Clustering by PCCA+: Application to Markov State Models and Data Classification. Adv. Data Anal. Classif. 2013, 7, 147–179. [Google Scholar] [CrossRef]
- Karush, J. On the Chapman-Kolmogorov Equation. Ann. Math. Stat. 1961, 32, 1333–1337. [Google Scholar] [CrossRef]
- Miroshin, R.N. Special Solutions of the Chapman–Kolmogorov Equation for Multidimensional-State Markov Processes with Continuous Time. Vestn. St Petersburg Univ. Math. 2016, 49, 122–129. [Google Scholar] [CrossRef]
- GetContacts. Available online: https://getcontacts.github.io/ (accessed on 21 October 2022).
- Chothia, C.; Lesk, A.M. Canonical Structures for the Hypervariable Regions of Immunoglobulins. J. Mol. Biol. 1987, 196, 901–917. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Quintero, M.L.; Seidler, C.A.; Liedl, K.R. T-Cell Receptor Variable β Domains Rigidify during Affinity Maturation. Sci. Rep. 2020, 10, 4472. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Kumar, S.; Tsai, C.-J.; Nussinov, R. Folding Funnels and Binding Mechanisms. Protein Eng. 1999, 12, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Elgundi, Z.; Reslan, M.; Cruz, E.; Sifniotis, V.; Kayser, V. The State-of-Play and Future of Antibody Therapeutics. Adv. Drug Deliv. Rev. 2017, 122, 2–19. [Google Scholar] [CrossRef]
- Strohl, W.R.; Strohl, L.M. Therapeutic Antibody Engineering: Current and Future Advances Driving the Strongest Growth Area in the Pharmaceutical Industry; Elsevier: Amsterdam, The Netherlands, 2012; ISBN 1-908818-09-3. [Google Scholar]
- Van Bockstaele, F.; Holz, J.-B.; Revets, H. The Development of Nanobodies for Therapeutic Applications. Curr. Opin. Investig. Drugs Lond. Engl. 2000 2009, 10, 1212–1224. [Google Scholar]
- Stocki, P.; Wicher, K.B.; Jacobsen, C.L.; Szary, J.M.; Demydchuk, M.; Northall, L.; Moos, T.; Walsh, F.S.; Rutkowski, J.L. High Efficiency Blood-Brain Barrier Transport Using a VNAR Targeting the Transferrin Receptor 1 (TfR1). BioRxiv 2019, 816900. [Google Scholar]
- Raybould, M.I.; Marks, C.; Krawczyk, K.; Taddese, B.; Nowak, J.; Lewis, A.P.; Bujotzek, A.; Shi, J.; Deane, C.M. Five Computational Developability Guidelines for Therapeutic Antibody Profiling. Proc. Natl. Acad. Sci. USA 2019, 116, 4025–4030. [Google Scholar] [CrossRef]
- Colman, P.M. Structure of Antibody-Antigen Complexes: Implications for Immune Recognition. Adv. Immunol. 1988, 43, 99–132. [Google Scholar]
- Makabe, K.; Nakanishi, T.; Tsumoto, K.; Tanaka, Y.; Kondo, H.; Umetsu, M.; Sone, Y.; Asano, R.; Kumagai, I. Thermodynamic Consequences of Mutations in Vernier Zone Residues of a Humanized Anti-Human Epidermal Growth Factor Receptor Murine Antibody, 528. J. Biol. Chem. 2008, 283, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Arslan, M.; Karadag, D.; Kalyoncu, S. Conformational Changes in a Vernier Zone Region: Implications for Antibody Dual Specificity. Proteins Struct. Funct. Bioinforma. 2020, 88, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Quintero, M.L.; Kroell, K.B.; Hofer, F.; Riccabona, J.R.; Liedl, K.R. Mutation of Framework Residue H71 Results in Different Antibody Paratope States in Solution. Front. Immunol. 2021, 12, 243. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, F.; Corbeil, C.R.; Purisima, E.O.; Sulea, T. Coevolved Canonical Loops Conformations of Single-Domain Antibodies: A Tale of Three Pockets Playing Musical Chairs. Front. Immunol. 2022, 13, 884132. [Google Scholar] [CrossRef]
- Löhr, T.; Sormanni, P.; Vendruscolo, M. Conformational Entropy as a Potential Liability of Computationally Designed Antibodies. Biomolecules 2022, 12, 718. [Google Scholar] [CrossRef]
- Fernández-Quintero, M.L.; Kraml, J.; Georges, G.; Liedl, K.R. CDR-H3 Loop Ensemble in Solution–Conformational Selection upon Antibody Binding. MAbs 2019, 11, 1077–1088. [Google Scholar] [CrossRef]
- Bork, P.; Holm, L.; Sander, C. The Immunoglobulin Fold: Structural Classification, Sequence Patterns and Common Core. J. Mol. Biol. 1994, 242, 309–320. [Google Scholar] [CrossRef]
- Frederick, K.K.; Marlow, M.S.; Valentine, K.G.; Wand, A.J. Conformational Entropy in Molecular Recognition by Proteins. Nature 2007, 448, 325–329. [Google Scholar] [CrossRef]
- Kelow, S.P.; Adolf-Bryfogle, J.; Dunbrack, R.L. Hiding in Plain Sight: Structure and Sequence Analysis Reveals the Importance of the Antibody DE Loop for Antibody-Antigen Binding. MAbs 2020, 12, 1840005. [Google Scholar]


| Description | Bound | Unbound |
|---|---|---|
| Number of Clusters/Simulation Time (μs) | ||
| cAb-lys3 | 179/35.8 | 240/48.0 |
| cAb-lys3-gl2 | 145/29.0 | 321/64.2 |
| cAb-lys3-gl1 | 156/31.2 | 223/44.6 |
| cAb-lys3-gl1&2 | 177/35.4 | 213/42.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seidler, C.A.; Kokot, J.; Fernández-Quintero, M.L.; Liedl, K.R. Structural Characterization of Nanobodies during Germline Maturation. Biomolecules 2023, 13, 380. https://doi.org/10.3390/biom13020380
Seidler CA, Kokot J, Fernández-Quintero ML, Liedl KR. Structural Characterization of Nanobodies during Germline Maturation. Biomolecules. 2023; 13(2):380. https://doi.org/10.3390/biom13020380
Chicago/Turabian StyleSeidler, Clarissa A., Janik Kokot, Monica L. Fernández-Quintero, and Klaus R. Liedl. 2023. "Structural Characterization of Nanobodies during Germline Maturation" Biomolecules 13, no. 2: 380. https://doi.org/10.3390/biom13020380
APA StyleSeidler, C. A., Kokot, J., Fernández-Quintero, M. L., & Liedl, K. R. (2023). Structural Characterization of Nanobodies during Germline Maturation. Biomolecules, 13(2), 380. https://doi.org/10.3390/biom13020380






