Importance of ACE2 for SARS-CoV-2 Infection of Kidney Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Samples
2.2. Protein Quantification by LC-MS/MS
2.3. Immunofluorescence Staining
2.4. Immunohistochemistry
2.5. Generation of Stable Cell Lines
2.6. Real-Time PCR Analysis
2.7. Infection Assay
2.8. Plaque Assay
2.9. SARS-CoV-2 Pseudovirus Preparation and Infection
2.10. Statistical Analysis
3. Results
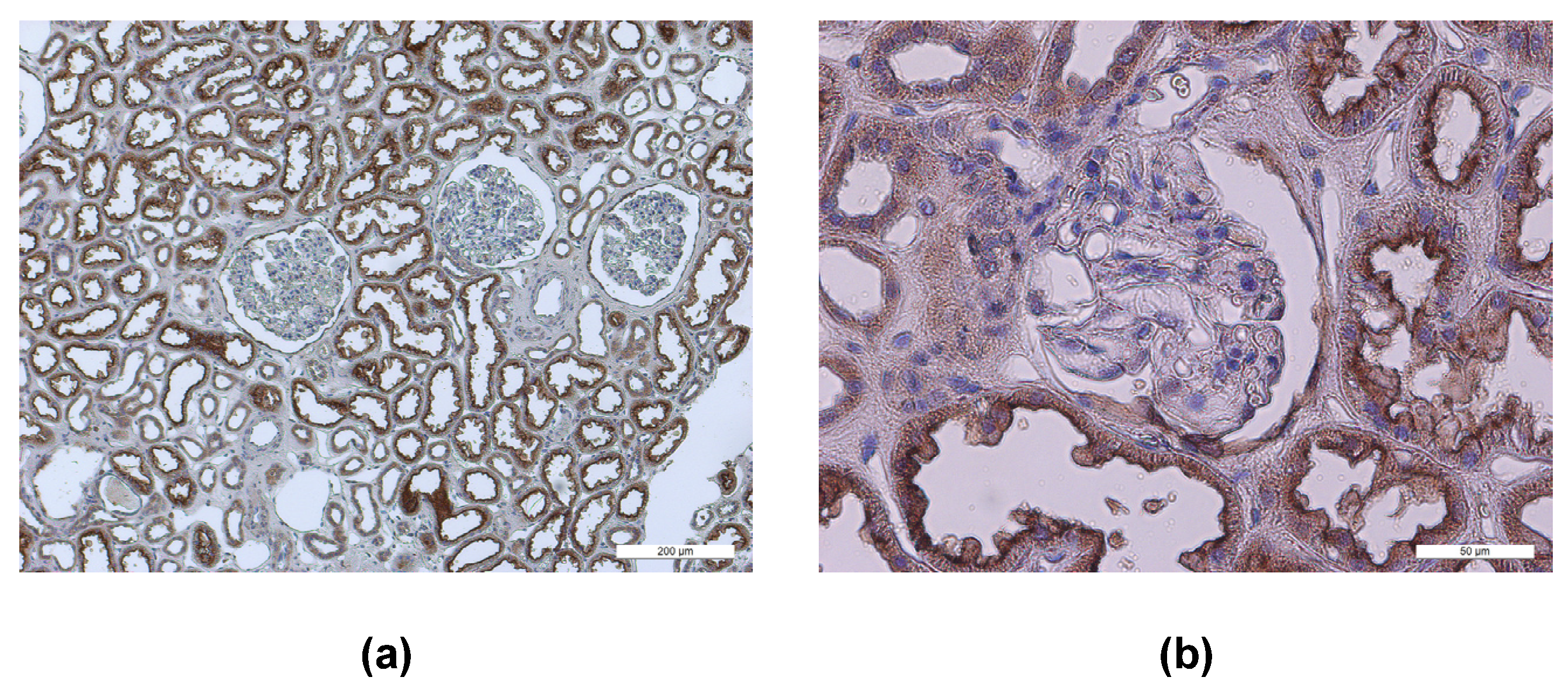
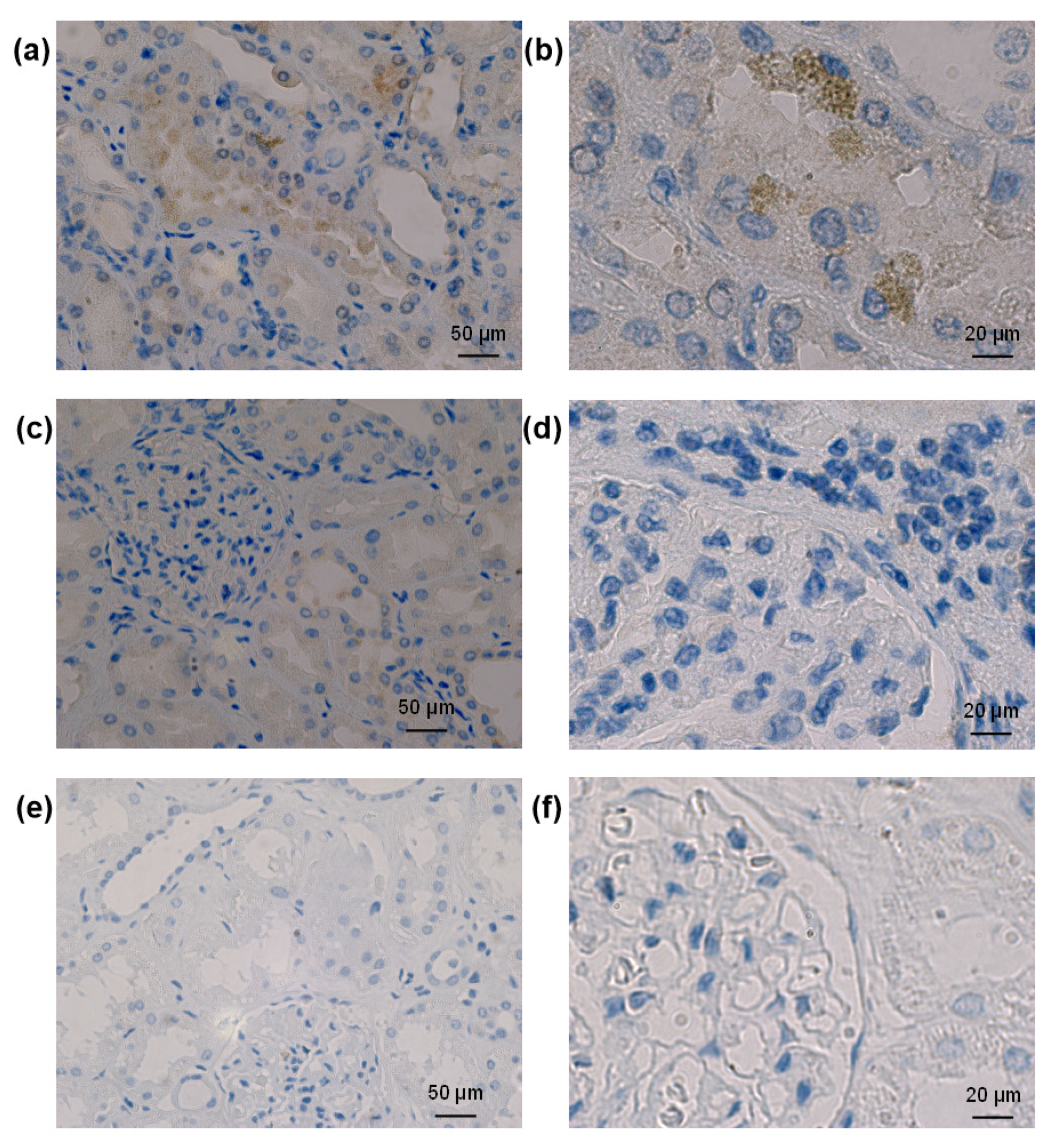
3.1. Localization of Entry Receptors in Human Kidneys
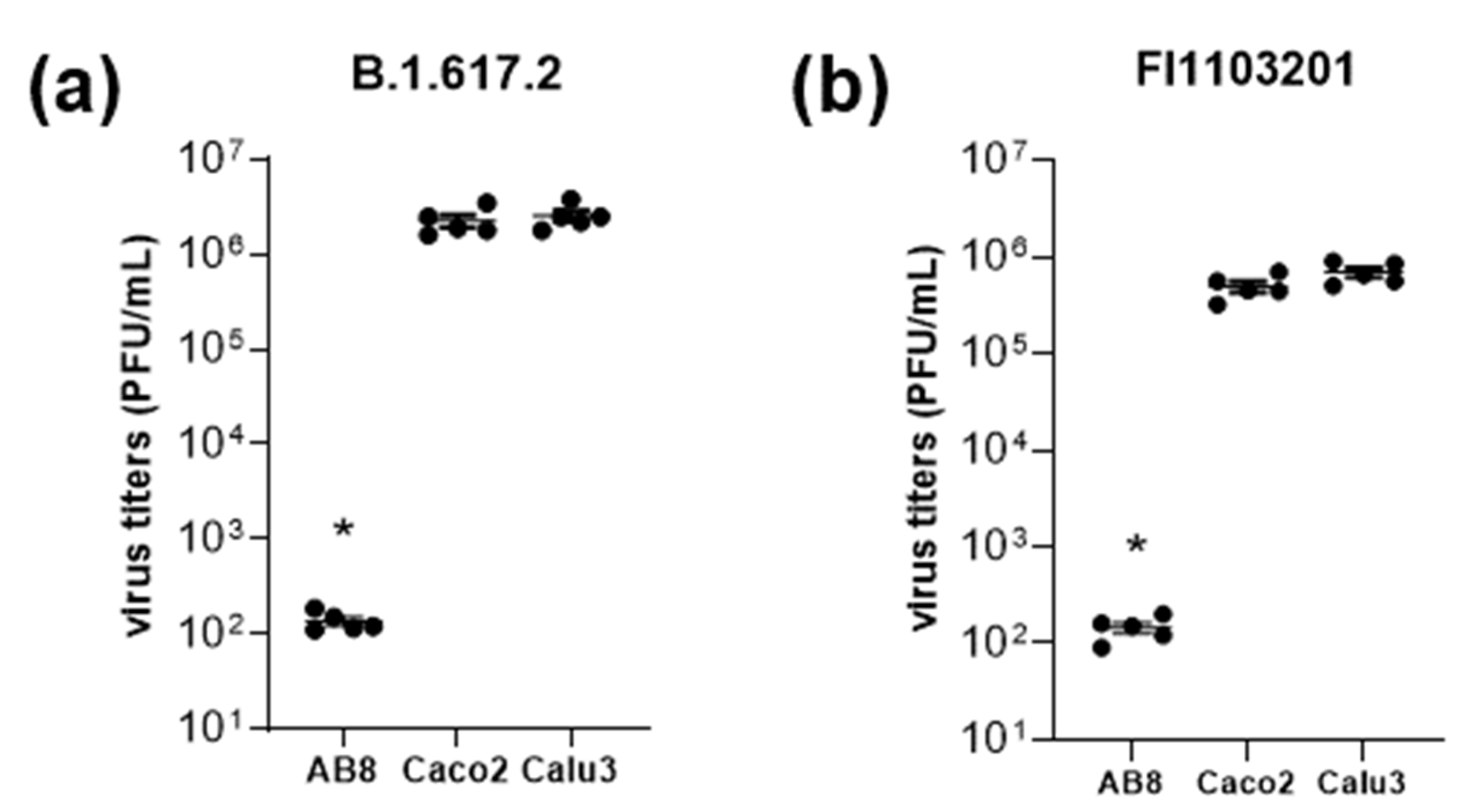
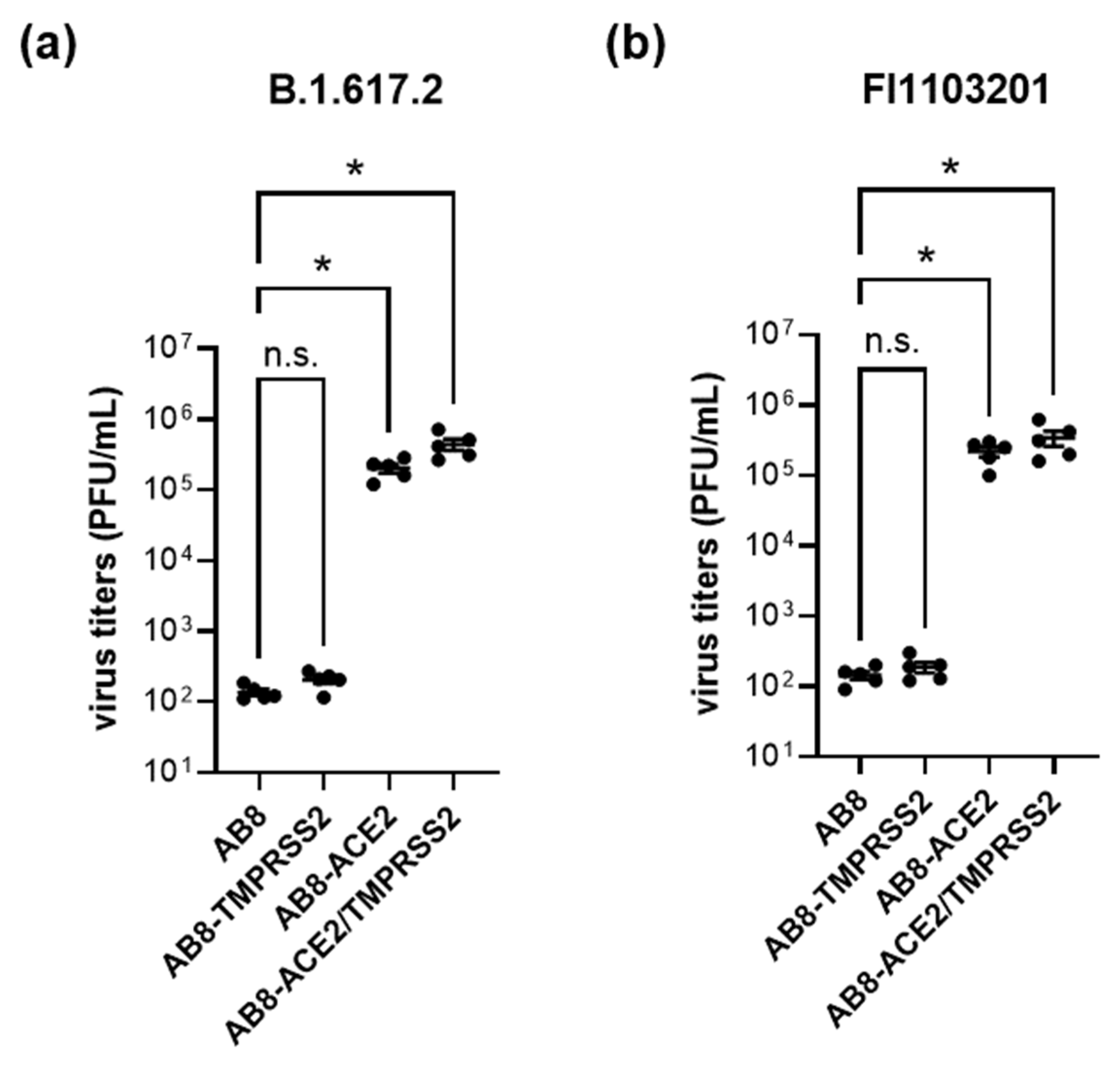
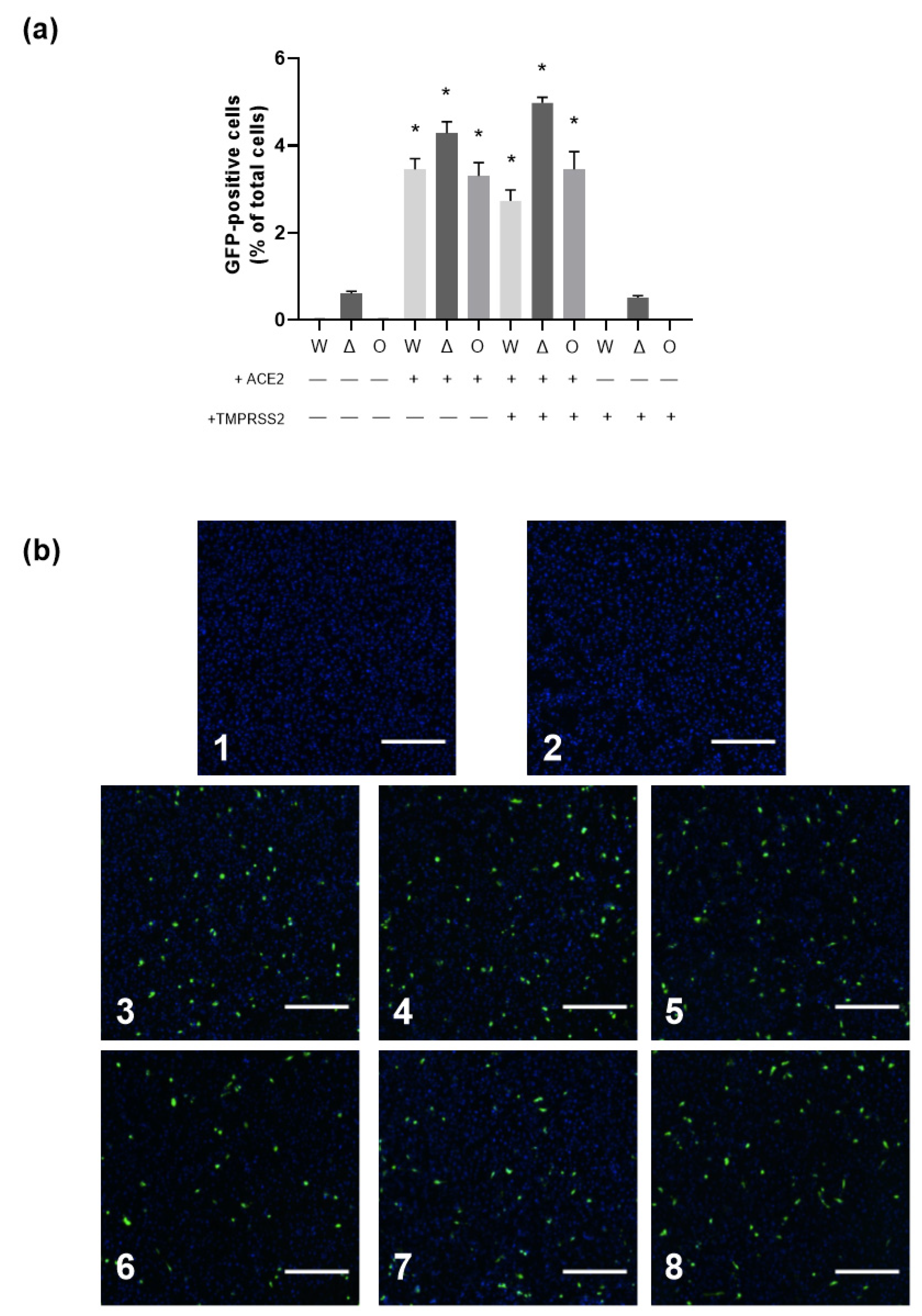
3.2. Role of Virus Entry Receptors for Infection of AB8 Podocytes
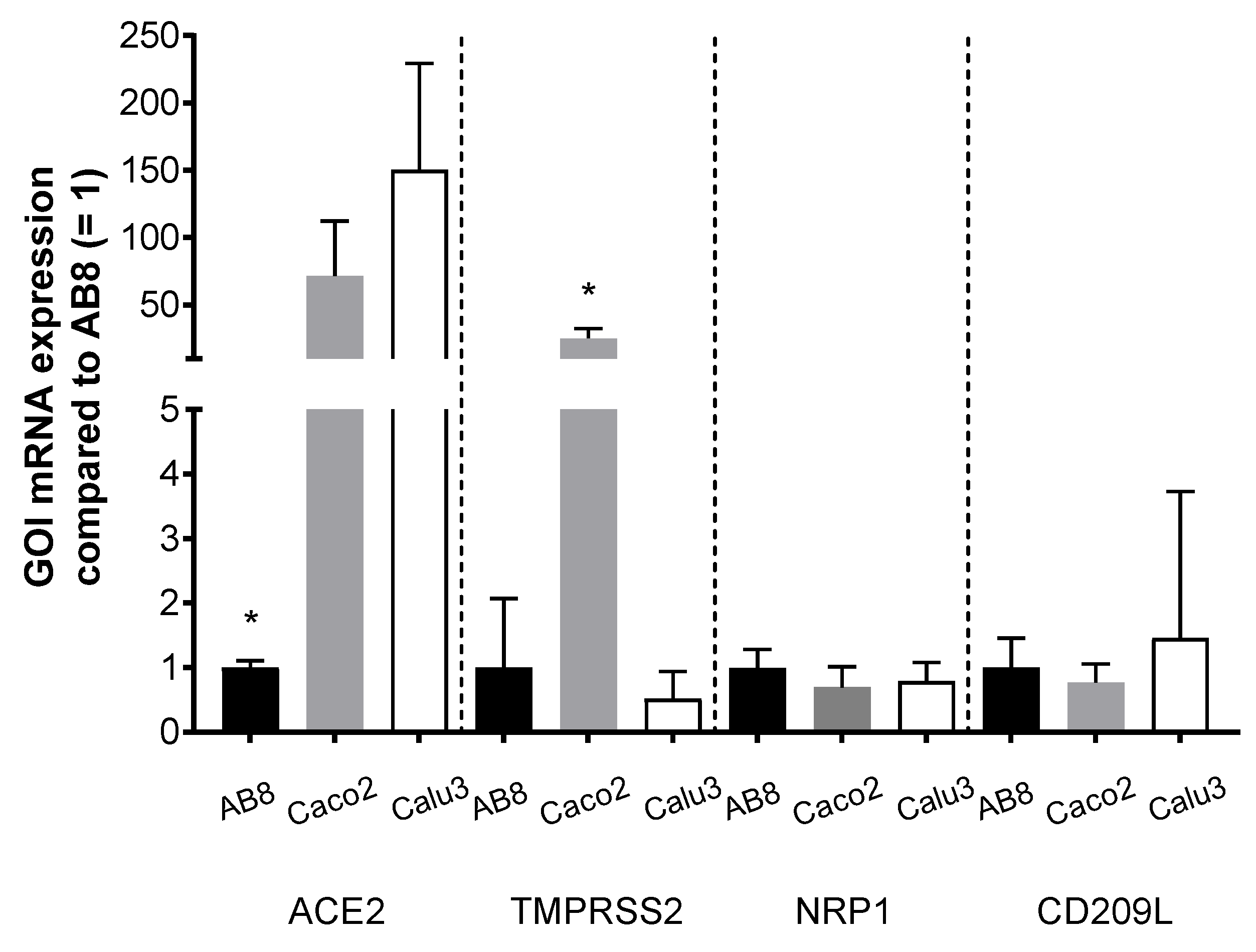
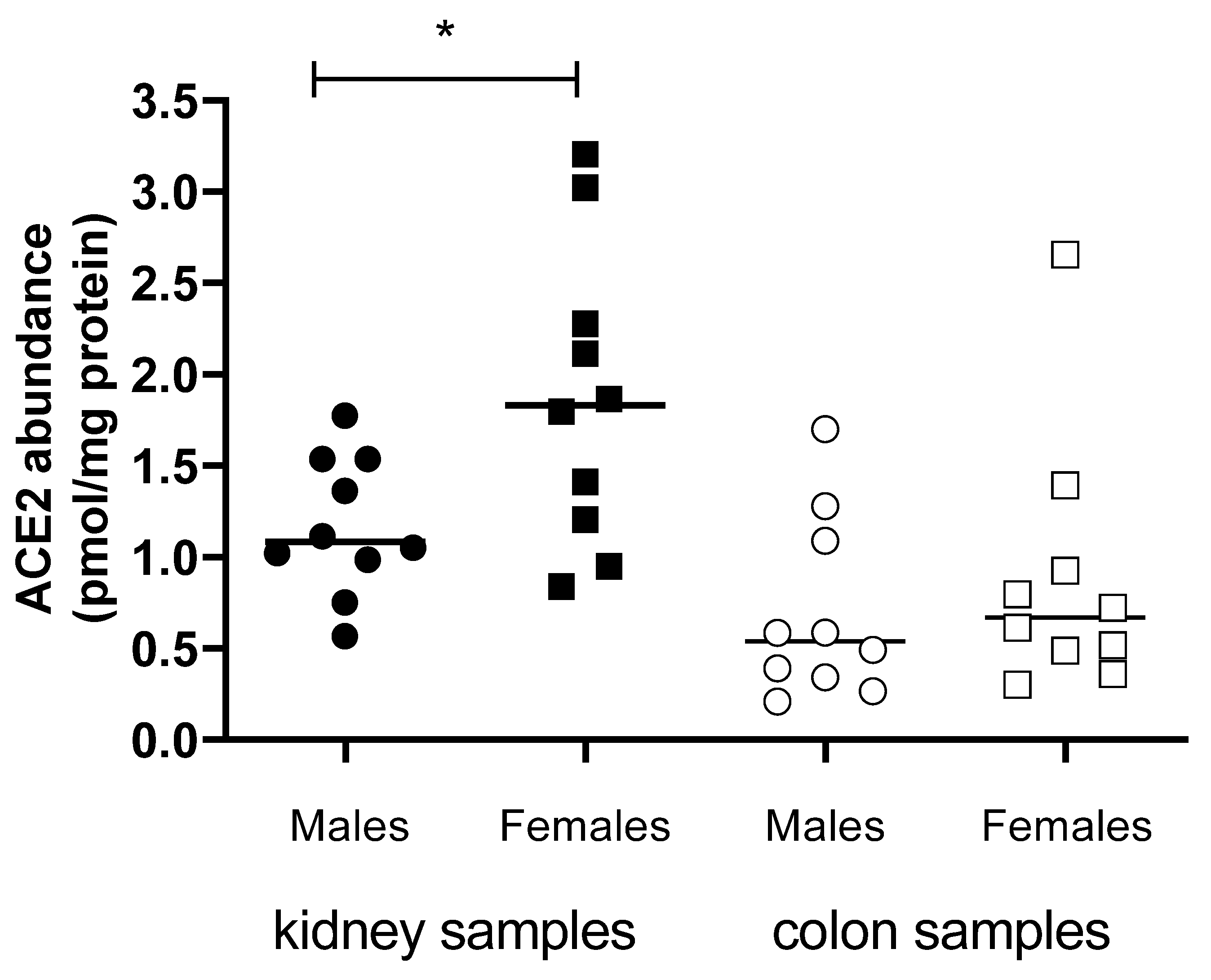
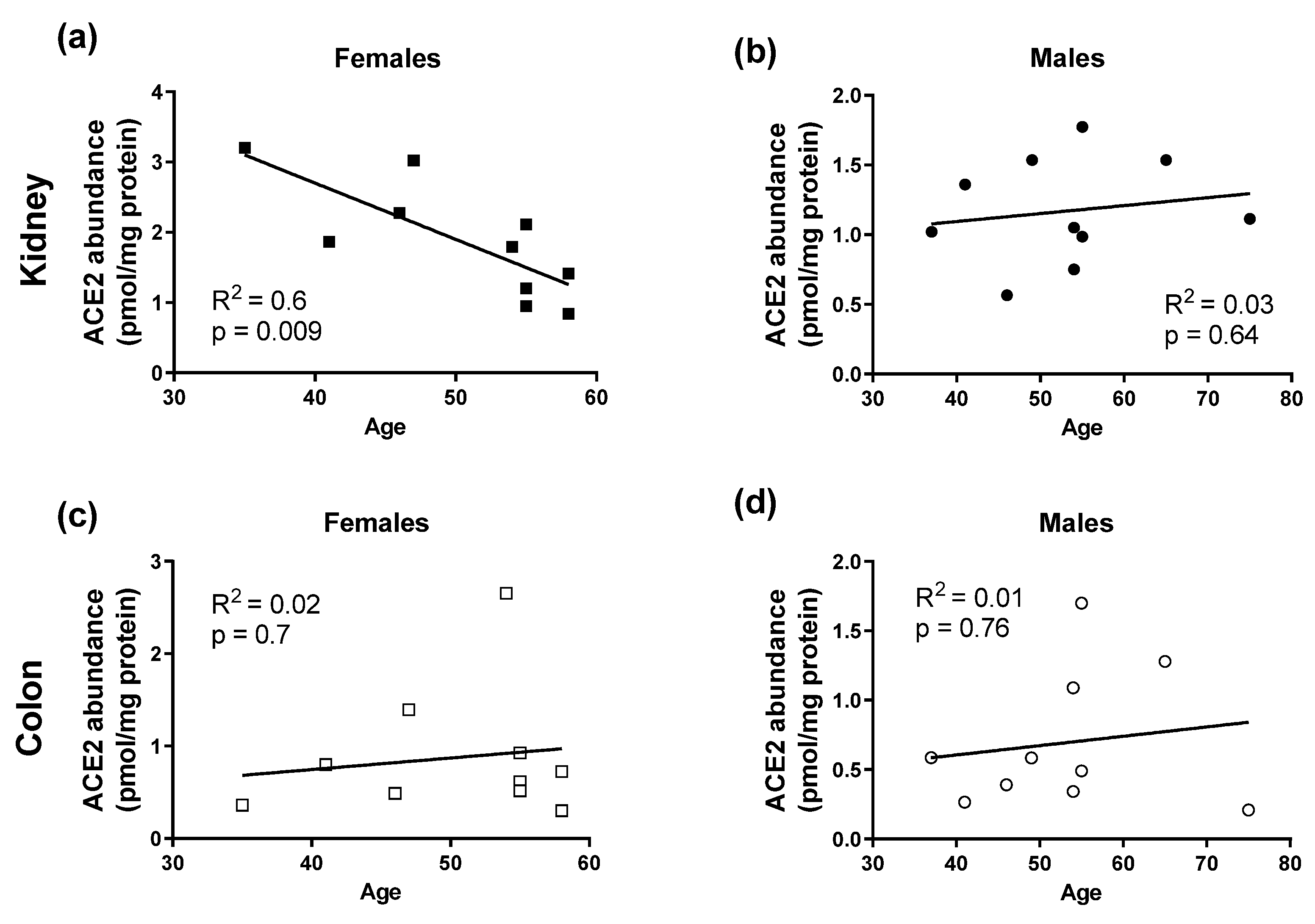
3.3. ACE2 and TMPRSS2 Protein Abundance in Human Kidneys and Colon
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 81, 18–19. [Google Scholar] [CrossRef]
- Vennema, H.; Godeke, G.J.; Rossen, J.W.A.; Voorhout, W.F.; Horzinek, M.C.; Opstelten, D.J.E.; Rottier, P.J.M. Nucleocapsid-Independent Assembly of Coronavirus-like Particles by Co-Expression of Viral Envelope Protein Genes. EMBO J. 1996, 15, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Holmes, K.V. CORONAVIRUSES (CORONAVIRIDAE). In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 1999; pp. 291–298. [Google Scholar]
- Snijder, E.J.; Decroly, E.; Ziebuhr, J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016, 96, 59–126. [Google Scholar] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. In Coronaviruses: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–23. [Google Scholar]
- Lai, M.M.; Cavanagh, D. The Molecular Biology of Coronaviruses. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 1997; pp. 1–100. [Google Scholar]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 Facilitates SARS-CoV-2 Cell Entry and Infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Amraei, R.; Yin, W.; Napoleon, M.A.; Suder, E.L.; Berrigan, J.; Zhao, Q.; Olejnik, J.; Chandler, K.B.; Xia, C.; Feldman, J.; et al. CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2. ACS Cent. Sci. 2021, 7, 1156–1165. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061. [Google Scholar] [CrossRef]
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Grasselli, G.; Zangrillo, A.; Zanella, A.; Antonelli, M.; Cabrini, L.; Castelli, A.; Cereda, D.; Coluccello, A.; Foti, G.; Fumagalli, R.; et al. Baseline Characteristics and Outcomes of 1591 Patients Infected with SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA 2020, 323, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Chen, S.; Li, X.; Liu, S.; Wang, F. Prevalence of Venous Thromboembolism in Patients with Severe Novel Coronavirus Pneumonia. J. Thromb. Haemost. 2020, 18, 1412–1424. [Google Scholar] [CrossRef]
- Tang, N.; Li, D.; Wang, X.; Sun, Z. Abnormal Coagulation Parameters Are Associated with Poor Prognosis in Patients with Novel Coronavirus Pneumonia. J. Thromb. Haemost. 2020, 18, 844–847. [Google Scholar] [CrossRef] [PubMed]
- Driggin, E.; Madhavan, M.V.; Bikdeli, B.; Chuich, T.; Laracy, J.; Biondi-Zoccai, G.; Brown, T.S.; Der Nigoghossian, C.; Zidar, D.A.; Haythe, J.; et al. Cardiovascular Considerations for Patients, Health Care Workers, and Health Systems During the COVID-19 Pandemic. J. Am. Coll. Cardiol. 2020, 75, 2352–2371. [Google Scholar] [CrossRef] [PubMed]
- Oxley, T.J.; Mocco, J.; Majidi, S.; Kellner, C.P.; Shoirah, H.; Singh, I.P.; De Leacy, R.A.; Shigematsu, T.; Ladner, T.R.; Yaeger, K.A.; et al. Large-Vessel Stroke as a Presenting Feature of COVID-19 in the Young. N. Engl. J. Med. 2020, 382, e60. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wu, D.; Chen, H.; Yan, W.; Yang, D.; Chen, G.; Ma, K.; Xu, D.; Yu, H.; Wang, H.; et al. Clinical Characteristics of 113 Deceased Patients with Coronavirus Disease 2019: Retrospective Study. BMJ 2020, 368, m1091. [Google Scholar] [CrossRef]
- Cheng, Y.; Luo, R.; Wang, K.; Zhang, M.; Wang, Z.; Dong, L.; Li, J.; Yao, Y.; Ge, S.; Xu, G. Kidney Disease Is Associated with In-Hospital Death of Patients with COVID-19. Kidney Int. 2020, 97, 829–838. [Google Scholar] [CrossRef]
- Faour, W.H.; Choaib, A.; Issa, E.; El Choueiry, F.; Shbaklo, K.; Alhajj, M.; Sawaya, R.T.; Harhous, Z.; Alefishat, E.; Nader, M. Mechanisms of COVID-19-Induced Kidney Injury and Current Pharmacotherapies. Inflamm. Res. 2022, 71, 39–56. [Google Scholar] [CrossRef]
- Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A Human Homolog of Angiotensin-Converting Enzyme: Cloning and Functional Expression as a Captopril-Insensitive Carboxypeptidase. J. Biol. Chem. 2000, 275, 33238–33243. [Google Scholar] [CrossRef] [PubMed]
- D’Ardes, D.; Boccatonda, A.; Rossi, I.; Guagnano, M.T.; Santilli, F.; Cipollone, F.; Bucci, M. COVID-19 and RAS: Unravelling an Unclear Relationship. Int. J. Mol. Sci. 2020, 21, 3003. [Google Scholar] [CrossRef] [PubMed]
- Reid, I.A.; Morris, B.J.; Ganong, W.F. The Renin-Angiotensin System. Annu. Rev. Physiol. 1978, 40, 377–410. [Google Scholar] [CrossRef] [PubMed]
- Fountain, J.H.; Lappin, S.L. Physiology, Renin Angiotensin System; StatPearls Publishing: Treasure Island, FL, USA, 2018. [Google Scholar]
- Shenoy, V.; Ferreira, A.J.; Qi, Y.; Fraga-Silva, R.A.; Díez-Freire, C.; Dooies, A.; Jun, J.Y.; Sriramula, S.; Mariappan, N.; Pourang, D.; et al. The Angiotensin-Converting Enzyme 2/Angiogenesis-(1-7)/Mas Axis Confers Cardiopulmonary Protection against Lung Fibrosis and Pulmonary Hypertension. Am. J. Respir Crit. Care Med. 2010, 182, 1065–1072. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, Y.; Huang, Y.; Pan, C.; Liu, L.; Qiu, H. Angiotensin-(1-7) Attenuates Lung Fibrosis by Way of Mas Receptor in Acute Lung Injury. J. Surg. Res. 2013, 185, 740–747. [Google Scholar] [CrossRef]
- Reindl-Schwaighofer, R.; Hödlmoser, S.; Domenig, O.; Krenn, K.; Eskandary, F.; Krenn, S.; Schörgenhofer, C.; Rumpf, B.; Karolyi, M.; Traugott, M.T.; et al. The Systemic Renin-Angiotensin System in COVID-19. Sci. Rep. 2022, 12, 20117. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk Factors Associated with Acute Respiratory Distress Syndrome and Death in Patients with Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef]
- Karagiannidis, C.; Mostert, C.; Hentschker, C.; Voshaar, T.; Malzahn, J.; Schillinger, G.; Klauber, J.; Janssens, U.; Marx, G.; Weber-Carstens, S.; et al. Case Characteristics, Resource Use, and Outcomes of 10 021 Patients with COVID-19 Admitted to 920 German Hospitals: An Observational Study. Lancet Respir Med. 2020, 8, 853–862. [Google Scholar] [CrossRef]
- Ortolan, A.; Lorenzin, M.; Felicetti, M.; Doria, A.; Ramonda, R. Does Gender Influence Clinical Expression and Disease Outcomes in COVID-19? A Systematic Review and Meta-Analysis. Int. J. Infect. Dis. 2020, 99, 496–504. [Google Scholar] [CrossRef]
- Naicker, S.; Yang, C.W.; Hwang, S.J.; Liu, B.C.; Chen, J.H.; Jha, V. The Novel Coronavirus 2019 Epidemic and Kidneys. Kidney Int. 2020, 97, 824–828. [Google Scholar] [CrossRef]
- Sinha, I.P.; Harwood, R.; Semple, M.G.; Hawcutt, D.B.; Thursfield, R.; Narayan, O.; Kenny, S.E.; Viner, R.; Hewer, S.L.; Southern, K.W.; et al. COVID-19 Infection in Children. Lancet Respir Med. 2020, 8, 446–447. [Google Scholar] [CrossRef]
- Khemiri, H.; Ayouni, K.; Triki, H.; Haddad-Boubaker, S. SARS-CoV-2 Infection in Pediatric Population before and during the Delta (B.1.617.2) and Omicron (B.1.1.529) Variants Era. Virol. J. 2022, 19, 144. [Google Scholar] [CrossRef] [PubMed]
- Pietri, L.; Giorgi, R.; Bégu, A.; Lojou, M.; Koubi, M.; Cauchois, R.; Grangeot, R.; Dubois, N.; Kaplanski, G.; Valéro, R.; et al. Excess Body Weight Is an Independent Risk Factor for Severe Forms of COVID-19. Metabolism 2021, 117, 154703. [Google Scholar] [CrossRef] [PubMed]
- Shimabukuro-Vornhagen, A.; Gödel, P.; Subklewe, M.; Stemmler, H.J.; Schlößer, H.A.; Schlaak, M.; Kochanek, M.; Böll, B.; von Bergwelt-Baildon, M.S. Cytokine Release Syndrome. J. Immunother. Cancer 2018, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, C.; Drozdzik, M.; Oswald, S. Mass Spectrometry-Based Targeted Proteomics Method for the Quantification of Clinically Relevant Drug Metabolizing Enzymes in Human Specimens. J. Chromatogr. B 2021, 1180, 122891. [Google Scholar] [CrossRef]
- Oswald, S.; Gröer, C.; Drozdzik, M.; Siegmund, W. Mass Spectrometry-Based Targeted Proteomics as a Tool to Elucidate the Expression and Function of Intestinal Drug Transporters. AAPS J. 2013, 15, 1128–1140. [Google Scholar] [CrossRef]
- Engel, U.; Breborowicz, D.; Bog-Hansen, T.; Francis, D. Lectin Staining of Renal Tubules in Normal Kidney. APMIS 1997, 105, 31–34. [Google Scholar] [CrossRef]
- Duning, K.; Schurek, E.-M.; Schlüter, M.; Bayer, M.; Reinhardt, H.-C.; Schwab, A.; Schaefer, L.; Benzing, T.; Schermer, B.; Saleem, M.A.; et al. KIBRA Modulates Directional Migration of Podocytes. J. Am. Soc. Nephrol. 2008, 19, 1891–1903. [Google Scholar] [CrossRef]
- Saleem, M.A.; O’Hare, M.J.; Reiser, J.; Coward, R.J.; Inward, C.D.; Farren, T.; Xing, C.Y.; Ni, L.; Mathieson, P.W.; Mundel, P. A Conditionally Immortalized Human Podocyte Cell Line Demonstrating Nephrin and Podocin Expression. J. Am. Soc. Nephrol. 2002, 13, 630–638. [Google Scholar] [CrossRef]
- Lepa, C.; Möller-Kerutt, A.; Stölting, M.; Picciotto, C.; Eddy, M.; Butt, E.; Kerjaschki, D.; Korb-Pap, A.; Vollenbröker, B.; Weide, T.; et al. LIM and SH3 Protein 1 (LASP-1): A Novel Link between the Slit Membrane and Actin Cytoskeleton Dynamics in Podocytes. FASEB J. 2020, 34, 5453–5464. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schloer, S.; Brunotte, L.; Goretzko, J.; Mecate-Zambrano, A.; Korthals, N.; Gerke, V.; Ludwig, S.; Rescher, U. Targeting the Endolysosomal Host-SARS-CoV-2 Interface by Clinically Licensed Functional Inhibitors of Acid Sphingomyelinase (FIASMA) Including the Antidepressant Fluoxetine. Emerg. Microbes Infect 2020, 9, 2245–2255. [Google Scholar] [CrossRef] [PubMed]
- Schloer, S.; Brunotte, L.; Mecate-Zambrano, A.; Zheng, S.; Tang, J.; Ludwig, S.; Rescher, U. Drug Synergy of Combinatory Treatment with Remdesivir and the Repurposed Drugs Fluoxetine and Itraconazole Effectively Impairs SARS-CoV-2 Infection in Vitro. Br. J. Pharm. 2021, 178, 2339–2350. [Google Scholar] [CrossRef]
- Mautner, L.; Hoyos, M.; Dangel, A.; Berger, C.; Ehrhardt, A.; Baiker, A. Replication Kinetics and Infectivity of SARS-CoV-2 Variants of Concern in Common Cell Culture Models. Virol. J. 2022, 19, 76. [Google Scholar] [CrossRef] [PubMed]
- Saccon, E.; Chen, X.; Mikaeloff, F.; Rodriguez, J.E.; Szekely, L.; Vinhas, B.S.; Krishnan, S.; Byrareddy, S.N.; Frisan, T.; Végvári, Á.; et al. Cell-Type-Resolved Quantitative Proteomics Map of Interferon Response against SARS-CoV-2. iScience 2021, 24, 102420. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wu, M.; Yao, J.; Guo, J.; Liao, X.; Song, S.; Li, J.; Duan, G.; Zhou, Y.; Wu, X.; et al. Caution on Kidney Dysfunctions of COVID-19 Patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Su, H.; Yang, M.; Wan, C.; Yi, L.X.; Tang, F.; Zhu, H.Y.; Yi, F.; Yang, H.C.; Fogo, A.B.; Nie, X.; et al. Renal Histopathological Analysis of 26 Postmortem Findings of Patients with COVID-19 in China. Kidney Int. 2020, 98, 219–227. [Google Scholar] [CrossRef]
- Puelles, V.G.; Lütgehetmann, M.; Lindenmeyer, M.T.; Sperhake, J.P.; Wong, M.N.; Allweiss, L.; Chilla, S.; Heinemann, A.; Wanner, N.; Liu, S.; et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 590–592. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Wang, R.; Feng, Z.; Zhang, J.; Yang, H.; Tan, Y.; Wang, H.; Wang, C.; Liu, L.; et al. Human Kidney Is a Target for Novel Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Nat. Commun. 2021, 12, 2506. [Google Scholar] [CrossRef]
- Sharma, P.; Uppal, N.N.; Wanchoo, R.; Shah, H.H.; Yang, Y.; Parikh, R.; Khanin, Y.; Madireddy, V.; Larsen, C.P.; Jhaveri, K.D.; et al. COVID-19–Associated Kidney Injury: A Case Series of Kidney Biopsy Findings. J. Am. Soc. Nephrol. 2020, 31, 1948–1958. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, C.S.; Miller, S.E.; Martines, R.B.; Bullock, H.A.; Zaki, S.R. Electron Microscopy of SARS-CoV-2: A Challenging Task. Lancet 2020, 395, e99. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial Cell Infection and Endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.E.; Brealey, J.K. Visualization of Putative Coronavirus in Kidney. Kidney Int. 2020, 98, 231–232. [Google Scholar] [CrossRef]
- Coate, K.C.; Cha, J.; Shrestha, S.; Wang, W.; Goncalves, L.M.; Almaca, J.; Knapp, M.E.; Fasolino, M.; Morgan, A.; Dai, C.; et al. SARS-CoV-2 Cell Entry Factors ACE2 and TMPRSS2 Are Expressed in the Microvasculature and Ducts of Human Pancreas but Are Not Enriched in β Cells. Cell Metab. 2020, 32, 1028–1040. [Google Scholar] [CrossRef]
- Ziegler, C.; Allon, S.J.; Nyguist, S.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Yuming, C.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035. [Google Scholar] [CrossRef]
- Haraldsson, B.; Nyström, J.; Deen, W.M. Properties of the Glomerular Barrier and Mechanisms of Proteinuria. Physiol. Rev. 2008, 88, 451–487. [Google Scholar] [CrossRef]
- Tukiainen, T.; Villani, A.-C.; Yen, A.; Rivas, M.A.; Marshall, J.L.; Satija, R.; Aguirre, M.; Gauthier, L.; Fleharty, M.; Kirby, A.; et al. Landscape of X Chromosome Inactivation across Human Tissues. Nature 2017, 550, 244–248. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, Q.; Xia, X.; Liu, K.; Yu, Z.; Tao, W.; Gong, W.; Han, J.D.J. Individual Variation of the SARS-CoV-2 Receptor ACE2 Gene Expression and Regulation. Aging Cell 2020, 19, e13168. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fett, C.; Mack, M.; Ten Eyck, P.P.; Meyerholz, D.K.; Perlman, S. Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection. J. Immunol. 2017, 198, 4046–4053. [Google Scholar] [CrossRef]
- Klein, S.L.; Flanagan, K.L. Sex Differences in Immune Responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Canney, M.; Atiquzzaman, M.; Cunningham, A.M.; Zheng, Y.; Er, L.; Hawken, S.; Zhao, Y.; Barbour, S.J. A Population-Based Analysis of the Risk of Glomerular Disease Relapse after COVID-19 Vaccination. J. Am. Soc. Nephrol. 2022, 33, 2247–2257. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Kuba, K.; Rao, S.; Huan, Y.; Guo, F.; Guan, B.; Yang, P.; Sarao, R.; Wada, T.; Leong-Poi, H.; et al. Angiotensin-Converting Enzyme 2 Protects from Severe Acute Lung Failure. Nature 2005, 436, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Batlle, D.; Wysocki, J.; Satchell, K. Soluble Angiotensin-Converting Enzyme 2: A Potential Approach for Coronavirus Infection Therapy? Clin. Sci. 2020, 134, 543–545. [Google Scholar] [CrossRef]
- Roshanravana, N.; Ghaffaria, S.; Hedayat, M. Angiotensin Converting Enzyme-2 as Therapeutic Target in COVID-19. Diabetes Metab. Syndr. 2020, 14, 637–639. [Google Scholar] [CrossRef]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913. [Google Scholar] [CrossRef]
- Li, M.Y.; Li, L.; Zhang, Y.; Wang, X.S. Expression of the SARS-CoV-2 Cell Receptor Gene ACE2 in a Wide Variety of Human Tissues. Infect. Dis. Poverty 2020, 9, 45. [Google Scholar] [CrossRef]
- Bertram, S.; Glowacka, I.; Blazejewska, P.; Soilleux, E.; Allen, P.; Danisch, S.; Steffen, I.; Choi, S.-Y.; Park, Y.; Schneider, H.; et al. TMPRSS2 and TMPRSS4 Facilitate Trypsin-Independent Spread of Influenza Virus in Caco-2 Cells. J. Virol. 2010, 84, 10016–10025. [Google Scholar] [CrossRef]
- Kühn, N.; Bergmann, S.; Kösterke, N.; Lambertz, R.L.O.; Keppner, A.; van den Brand, J.M.A.; Pöhlmann, S.; Weiß, S.; Hummler, E.; Hatesuer, B.; et al. The Proteolytic Activation of (H3N2) Influenza A Virus Hemagglutinin Is Facilitated by Different Type II Transmembrane Serine Proteases. J. Virol. 2016, 90, 4298–4307. [Google Scholar] [CrossRef]
- Böttcher, E.; Matrosovich, T.; Beyerle, M.; Klenk, H.-D.; Garten, W.; Matrosovich, M. Proteolytic Activation of Influenza Viruses by Serine Proteases TMPRSS2 and HAT from Human Airway Epithelium. J. Virol. 2006, 80, 9896–9898. [Google Scholar] [CrossRef]


| Primer | Sequence 5′–3′ |
|---|---|
| ACE2 sense | CATTGGTCTTCTGTCACCCGA |
| ACE2 antisense | CCCCAACTATCTCTCGCTTCATCT |
| TMPRSS2 sense | AAATCCCCATCCGGGACAGT |
| TMPRSS2 antisense | GAGCACTTGCTGCCCATGAA |
| CD209 1 sense | TTCCAGAAGTAACCGCTTCACC |
| CD209 antisense | CTGCTTGAAGCTGGGCAACA |
| CD209L 2 sense | AGTGGCTGGAACGACAATCG |
| CD209L antisense | AGAGAACCGTCTCAGGGCAT |
| NRP1 3 sense | AGGATCACACAGGAGATGGC |
| NRP1 antisense | CTGGTAGCGCAGTTTGACCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kroll, M.-K.; Schloer, S.; Candan, P.; Korthals, N.; Wenzel, C.; Ihle, H.; Gilhaus, K.; Liedtke, K.R.; Schöfbänker, M.; Surmann, B.; et al. Importance of ACE2 for SARS-CoV-2 Infection of Kidney Cells. Biomolecules 2023, 13, 472. https://doi.org/10.3390/biom13030472
Kroll M-K, Schloer S, Candan P, Korthals N, Wenzel C, Ihle H, Gilhaus K, Liedtke KR, Schöfbänker M, Surmann B, et al. Importance of ACE2 for SARS-CoV-2 Infection of Kidney Cells. Biomolecules. 2023; 13(3):472. https://doi.org/10.3390/biom13030472
Chicago/Turabian StyleKroll, Marie-Kristin, Sebastian Schloer, Peynaz Candan, Nadia Korthals, Christoph Wenzel, Hannah Ihle, Kevin Gilhaus, Kim Rouven Liedtke, Michael Schöfbänker, Beate Surmann, and et al. 2023. "Importance of ACE2 for SARS-CoV-2 Infection of Kidney Cells" Biomolecules 13, no. 3: 472. https://doi.org/10.3390/biom13030472
APA StyleKroll, M.-K., Schloer, S., Candan, P., Korthals, N., Wenzel, C., Ihle, H., Gilhaus, K., Liedtke, K. R., Schöfbänker, M., Surmann, B., Schröter, R., Neugebauer, U., Mall, G., Oswald, S., Ludwig, S., Rescher, U., Vollenbröker, B., & Ciarimboli, G. (2023). Importance of ACE2 for SARS-CoV-2 Infection of Kidney Cells. Biomolecules, 13(3), 472. https://doi.org/10.3390/biom13030472









