Molecular Property Prediction by Combining LSTM and GAT
Abstract
1. Introduction
2. Materials and Methods
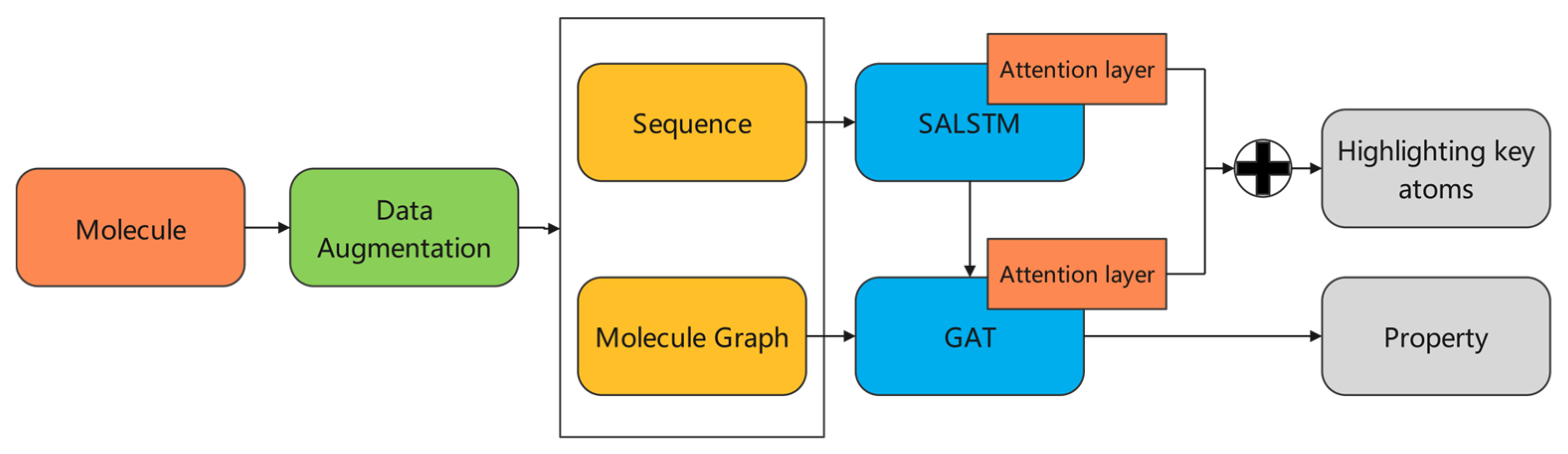
2.1. Overview
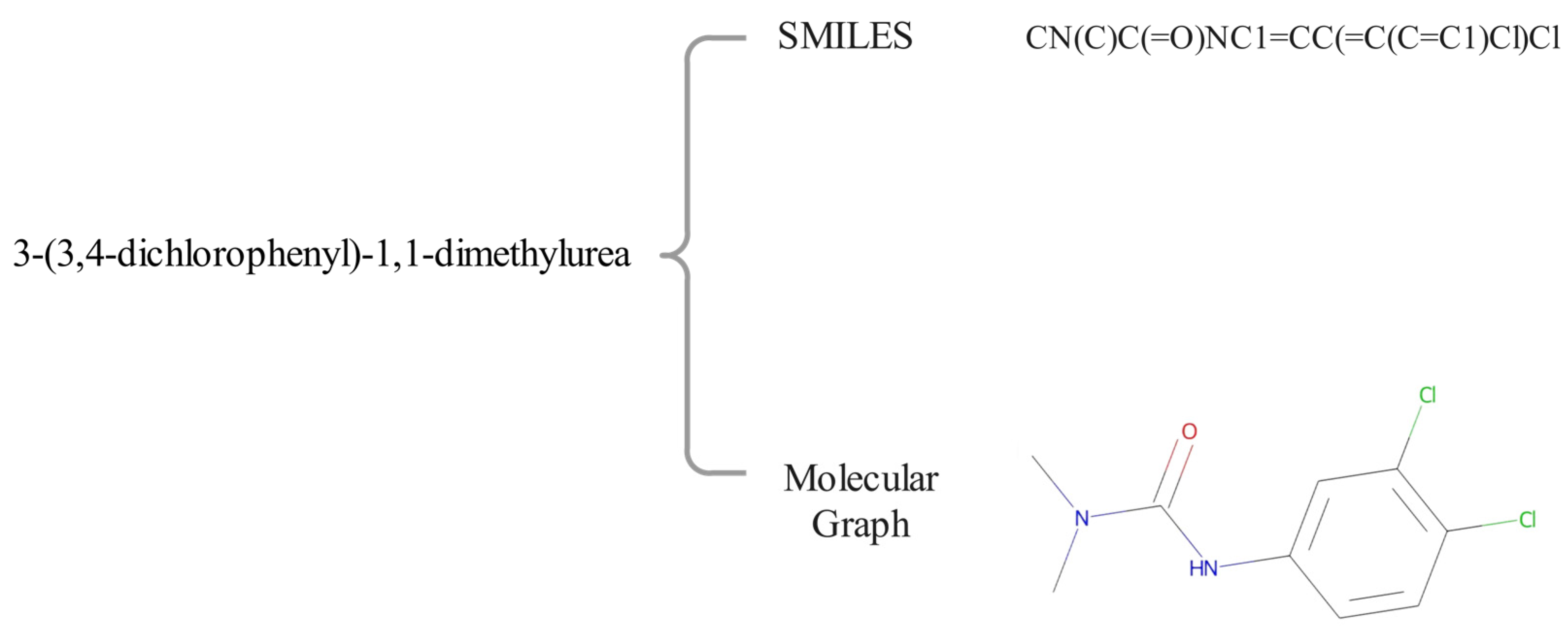
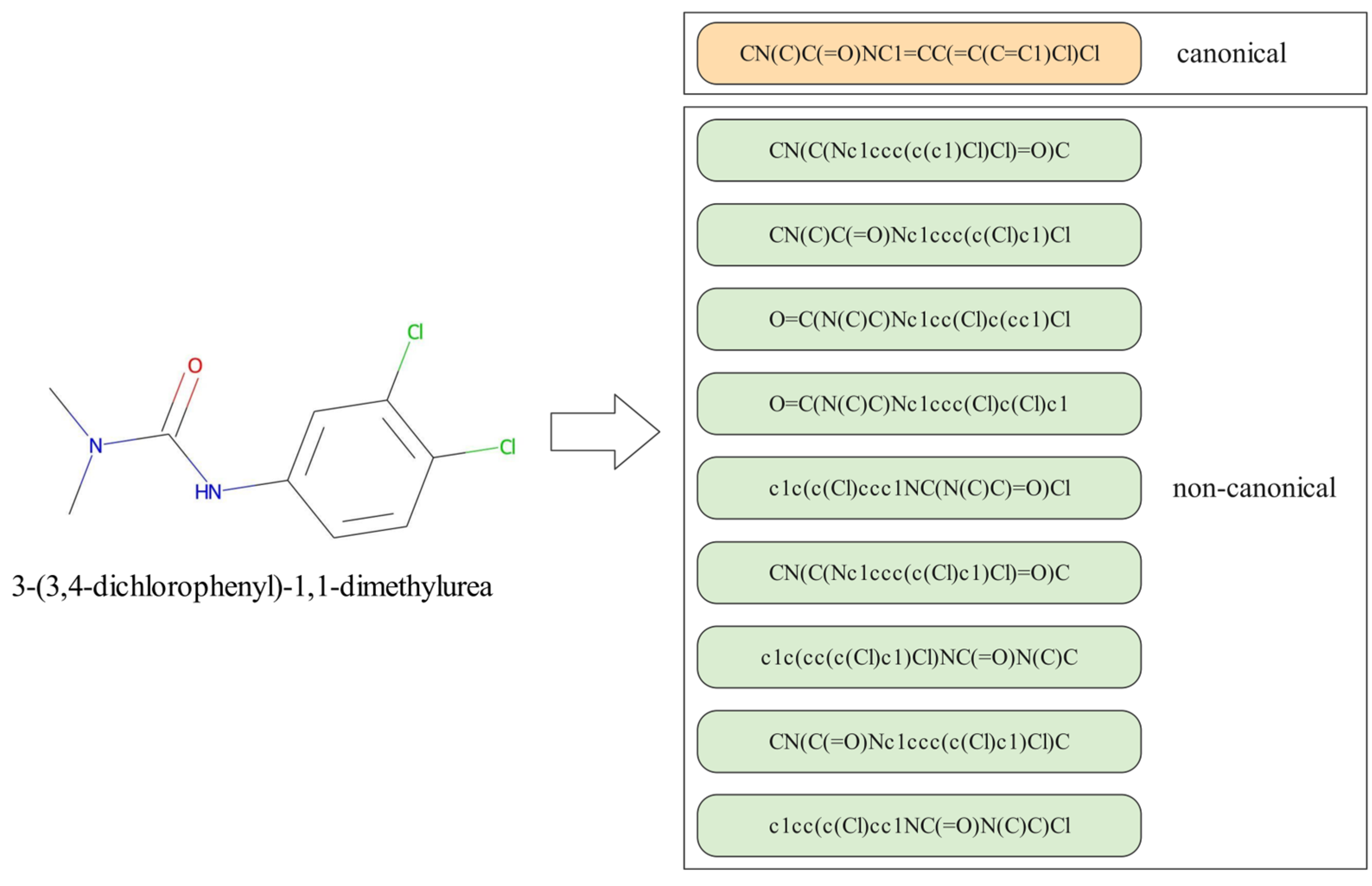
2.2. Data Pre-Processing and Augmentation
2.3. SALSTM Model
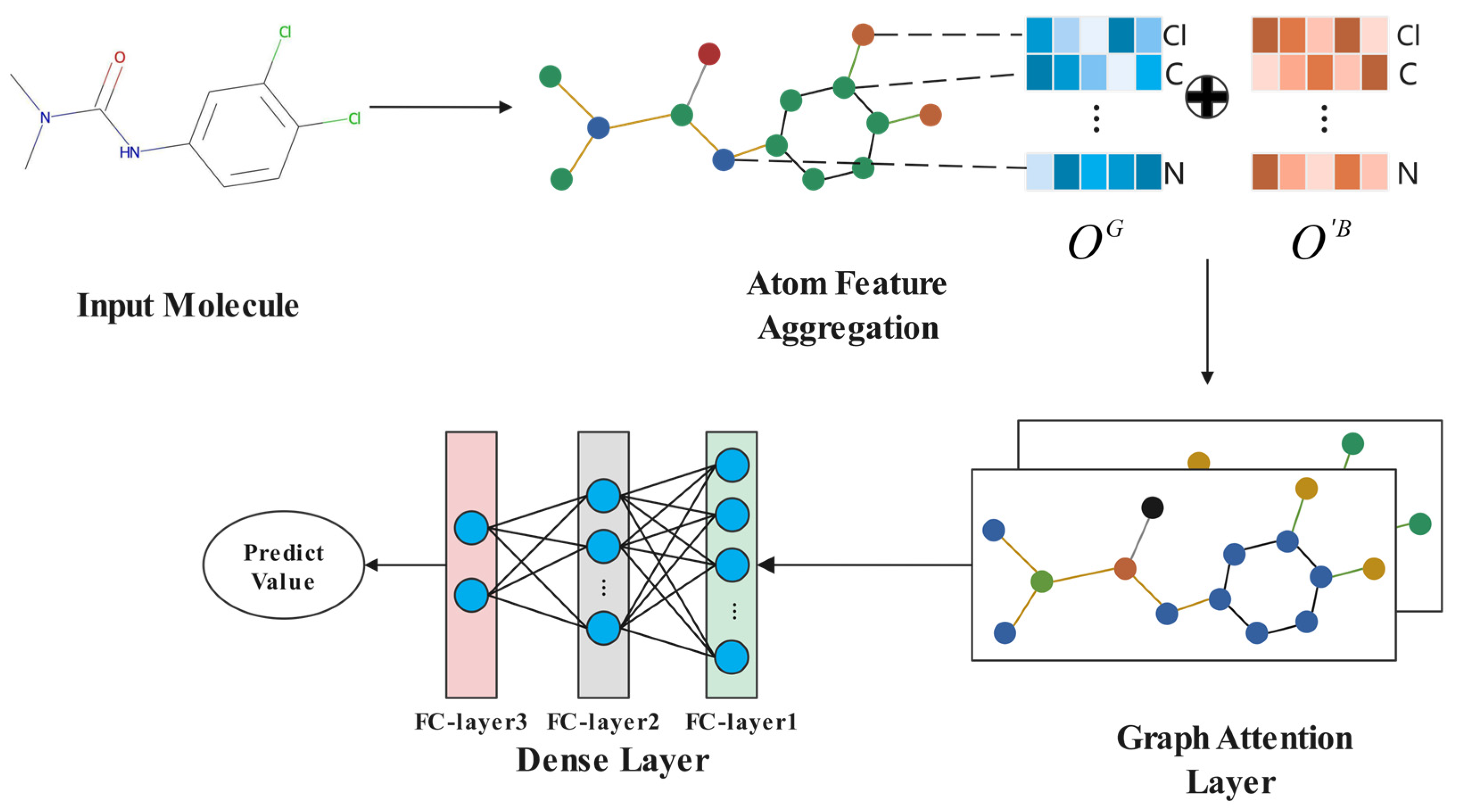
2.4. GAT Model
2.5. Interpretability
3. Results
3.1. Dataset
3.1.1. Regression Task
3.1.2. Classification Task
3.2. Experiment
3.3. Ablation Experiment
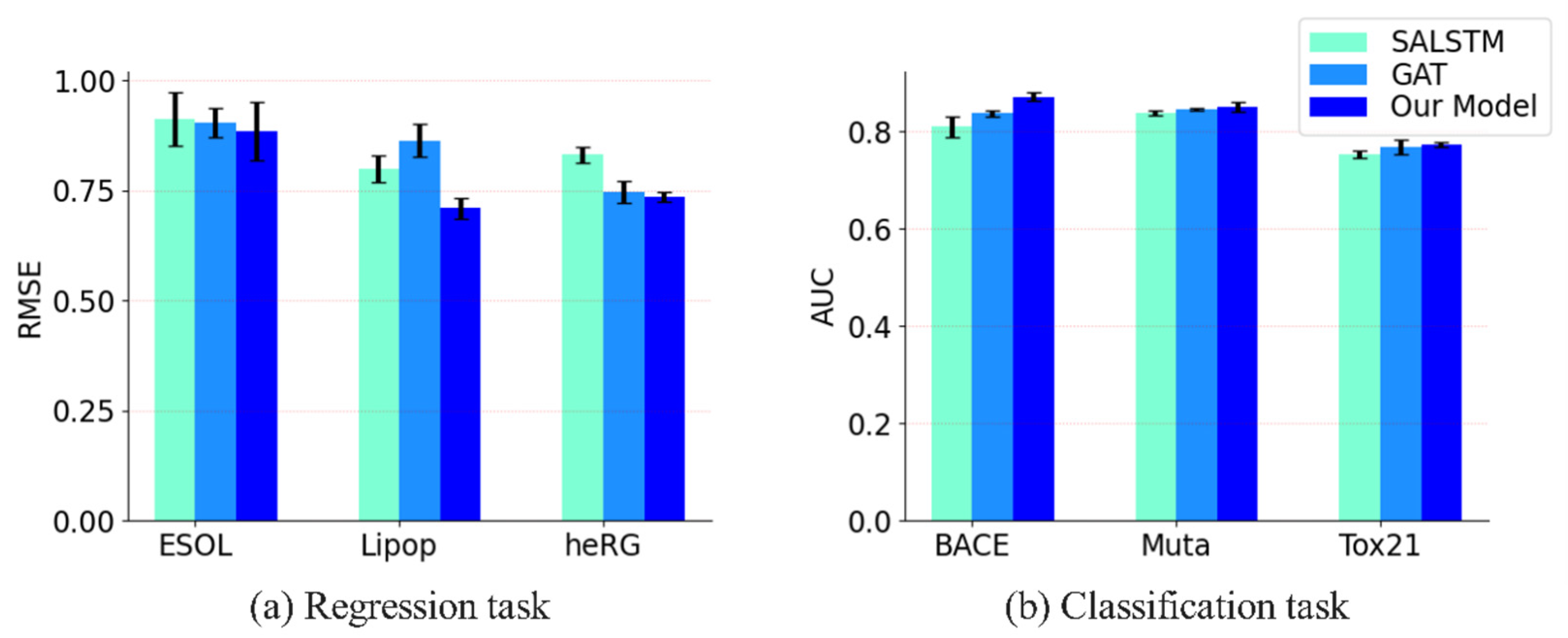
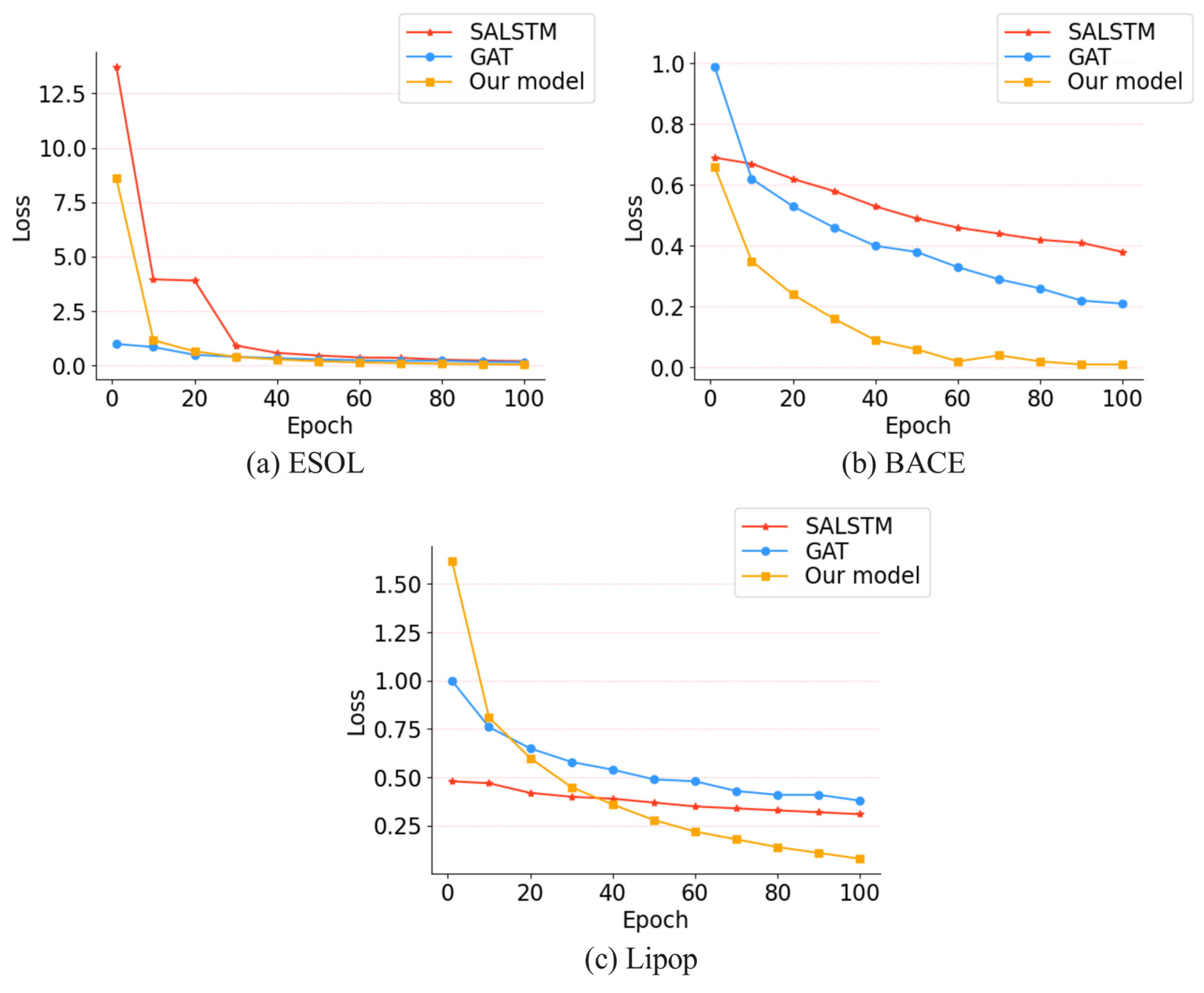
3.3.1. Comparison with SALSTM and GAT
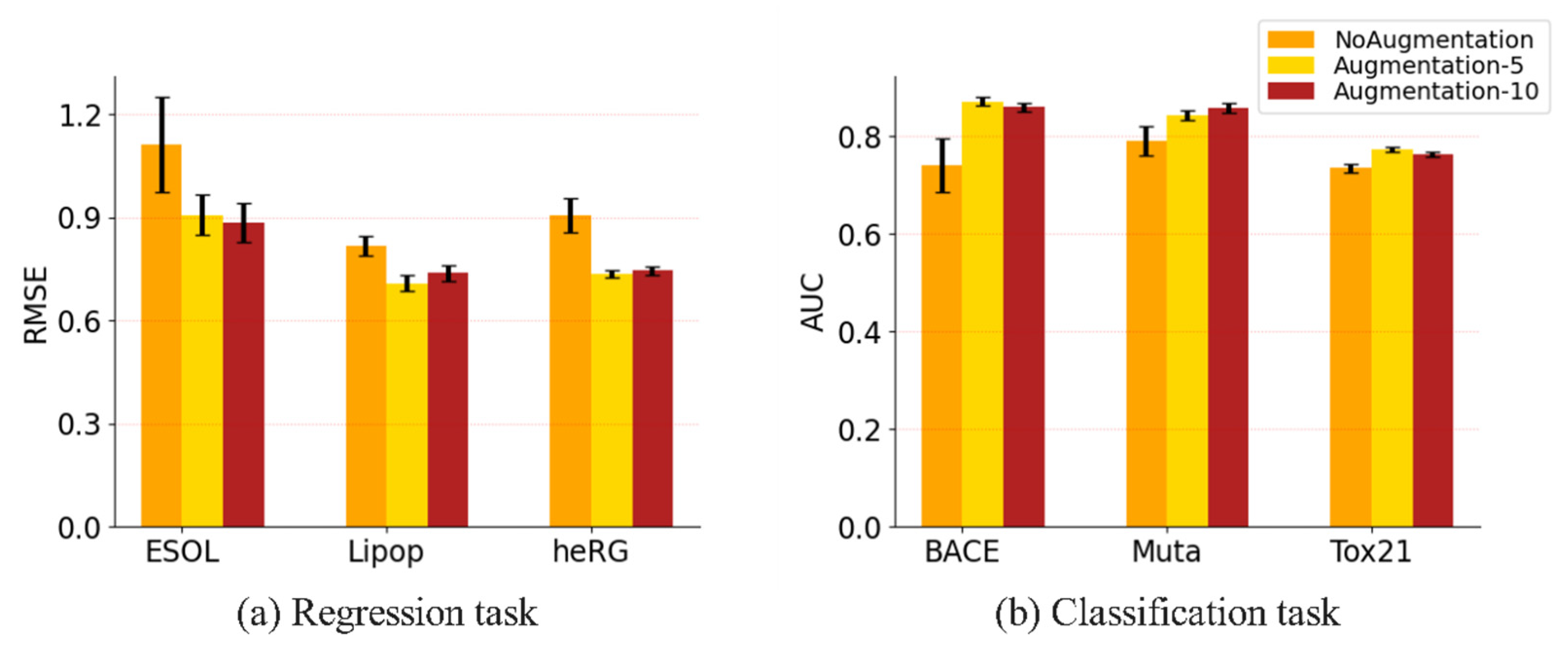
3.3.2. Evaluation on Different Data Augmentation Methods
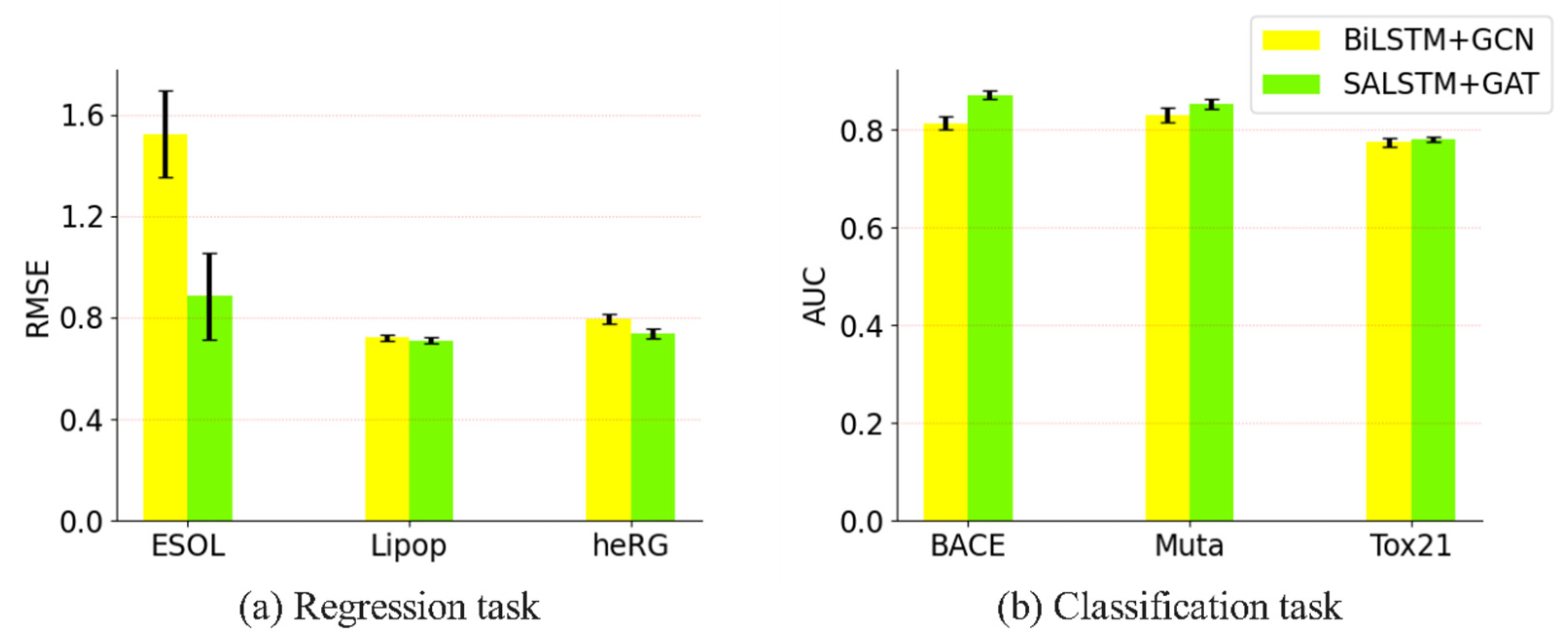
3.3.3. Impact of Adding Attention Mechanism to the Proposed Model
3.4. Interpretability
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mouchlis, V.D.; Afantitis, A.; Serra, A.; Fratello, M.; Papadiamantis, A.G.; Aidinis, V.; Lynch, I.; Greco, D.; Melagraki, G. Advances in de Novo Drug Design: From Conventional to Machine Learning Methods. Int. J. Mol. Sci. 2021, 22, 1676. [Google Scholar] [CrossRef] [PubMed]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. An Updated Review of Computer-Aided Drug Design and Its Application to COVID-19. BioMed Res. Int. 2021, 2021, 8853056. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, M.; Wang, S.; Zhang, S. Deep Learning Methods for Molecular Representation and Property Prediction. Drug Discov. Today 2022, 27, 103373. [Google Scholar] [CrossRef] [PubMed]
- Ndagi, U.; Falaki, A.A.; Abdullahi, M.; Lawal, M.M.; Soliman, M.E. Antibiotic Resistance: Bioinformatics-Based Understanding as a Functional Strategy for Drug Design. RSC Adv. 2020, 10, 18451–18468. [Google Scholar] [CrossRef]
- Raghavachari, K.; Stefanov, B.B.; Curtiss, L.A. Accurate Density Functional Thermochemistry for Larger Molecules. Mol. Phys. 1997, 91, 555–560. [Google Scholar] [CrossRef]
- Jena, B.; Saxena, S.; Nayak, G.K.; Saba, L.; Sharma, N.; Suri, J.S. Artificial Intelligence-Based Hybrid Deep Learning Models for Image Classification: The First Narrative Review. Comput. Biol. Med. 2021, 137, 104803. [Google Scholar] [CrossRef]
- Pradhyumna, P.; Shreya, G.P.; Mohana. Graph Neural Network (GNN) in Image and Video Understanding Using Deep Learning for Computer Vision Applications. In Proceedings of the 2nd International Conference on Electronics and Sustainable Communication Systems, ICESC 2021, Coimbatore, India, 4–6 August 2021; pp. 1183–1189. [Google Scholar]
- Pandey, B.; Kumar Pandey, D.; Pratap Mishra, B.; Rhmann, W. A Comprehensive Survey of Deep Learning in the Field of Medical Imaging and Medical Natural Language Processing: Challenges and Research Directions. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 5083–5099. [Google Scholar] [CrossRef]
- Yi, H.C.; You, Z.H.; Huang, D.S.; Kwoh, C.K. Graph Representation Learning in Bioinformatics: Trends, Methods and Applications. Brief. Bioinform. 2022, 23, bbab340. [Google Scholar] [CrossRef]
- Berrar, D.; Dubitzky, W. Deep Learning in Bioinformatics and Biomedicine. Brief. Bioinform. 2021, 22, 1513–1514. [Google Scholar] [CrossRef]
- Xia, J.; Wang, J.; Niu, S. Research Challenges and Opportunities for Using Big Data in Global Change Biology. Glob. Chang. Biol. 2020, 26, 6040–6061. [Google Scholar] [CrossRef]
- Tetko, I.V.; Engkvist, O. From Big Data to Artificial Intelligence: Chemoinformatics Meets New Challenges. J. Cheminform. 2020, 12, 12–14. [Google Scholar] [CrossRef]
- Awrahman, B.J.; Aziz Fatah, C.; Hamaamin, M.Y. A Review of the Role and Challenges of Big Data in Healthcare Informatics and Analytics. Comput. Intell. Neurosci. 2022, 2022, 5317760. [Google Scholar] [CrossRef] [PubMed]
- Segota, S.B.; Andelic, N.; Lorencin, I.; Musulin, J.; Stifanic, D.; Car, Z. Preparation of Simplified Molecular Input Line Entry System Notation Datasets for Use in Convolutional Neural Networks. In Proceedings of the BIBE 2021—21st IEEE International Conference on BioInformatics and BioEngineering, Kragujevac, Serbia, 25–27 October 2021; Volume 9, pp. 12–17. [Google Scholar]
- Kachalkin, M.N.; Ryazanova, T.K.; Sokolova, I.V.; Voronin, A.V. Prediction of COX-2 Inhibitory Activity Using LSTM-Network. In Proceedings of the 2022 Ural-Siberian Conference on Computational Technologies in Cognitive Science, Genomics and Biomedicine, CSGB 2022, Novosibirsk, Russia, 4–8 July 2022; pp. 160–163. [Google Scholar]
- Segler, M.H.S.; Kogej, T.; Tyrchan, C.; Waller, M.P. Generating Focused Molecule Libraries for Drug Discovery with Recurrent Neural Networks. ACS Cent. Sci. 2018, 4, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient Estimation of Word Representations in Vector Space. In Proceedings of the 1st International Conference on Learning Representations, ICLR 2013—Workshop Track Proceedings, Scottsdale, AZ, USA, 2–4 May 2013; pp. 1–12. [Google Scholar]
- Jaeger, S.; Fulle, S.; Turk, S. Mol2vec: Unsupervised Machine Learning Approach with Chemical Intuition. J. Chem. Inf. Model. 2018, 58, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Chen, G.; Zhao, L.; Zhong, W.; Yu-Chian Chen, C. Mol2Context-Vec: Learning Molecular Representation from Context Awareness for Drug Discovery. Brief. Bioinform. 2021, 22, bbab317. [Google Scholar] [CrossRef]
- Datta, R.; Das, D.; Das, S. Efficient Lipophilicity Prediction of Molecules Employing Deep-Learning Models. Chemom. Intell. Lab. Syst. 2021, 213, 104309. [Google Scholar] [CrossRef]
- Wu, C.K.; Zhang, X.C.; Yang, Z.J.; Lu, A.P.; Hou, T.J.; Cao, D.S. Learning to SMILES: BAN-Based Strategies to Improve Latent Representation Learning from Molecules. Brief. Bioinform. 2021, 22, bbab327. [Google Scholar] [CrossRef]
- Zheng, S.; Yan, X.; Yang, Y.; Xu, J. Identifying Structure-Property Relationships through SMILES Syntax Analysis with Self-Attention Mechanism. J. Chem. Inf. Model. 2019, 59, 914–923. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Zhang, C.; Wang, S. SSGraphCPI: A Novel Model for Predicting Compound-Protein Interactions Based on Deep Learning. Int. J. Mol. Sci. 2022, 23, 3780. [Google Scholar] [CrossRef]
- Oyewola, D.O.; Dada, E.G.; Emebo, O.; Oluwagbemi, O.O. Using Deep 1D Convolutional Grated Recurrent Unit Neural Network to Optimize Quantum Molecular Properties and Predict Intramolecular Coupling Constants of Molecules of Potential Health Medications and Other Generic Molecules. Appl. Sci. 2022, 12, 7228. [Google Scholar] [CrossRef]
- Kimber, T.B.; Gagnebin, M.; Volkamer, A. Maxsmi: Maximizing Molecular Property Prediction Performance with Confidence Estimation Using SMILES Augmentation and Deep Learning. Artif. Intell. Life Sci. 2021, 1, 100014. [Google Scholar] [CrossRef]
- Wang, S.; Jiang, M.; Zhang, S.; Wang, X.; Yuan, Q.; Wei, Z.; Li, Z. Mcn-cpi: Multiscale Convolutional Network for Compound–Protein Interaction Prediction. Biomolecules 2021, 11, 1119. [Google Scholar] [CrossRef]
- Sun, M.; Zhao, S.; Gilvary, C.; Elemento, O.; Zhou, J.; Wang, F. Graph Convolutional Networks for Computational Drug Development and Discovery. Brief. Bioinform. 2020, 21, 919–935. [Google Scholar] [CrossRef]
- Beck, D.; Haffari, G.; Cohn, T. Graph-to-Sequence Learning Using Gated Graph Neural Networks. In Proceedings of the ACL 2018—56th Annual Meeting of the Association for Computational Linguistics, Proceedings of the Conference (Long Papers), Melbourne, Australia, 15–20 July 2018; Volume 1, pp. 273–283. [Google Scholar]
- Ma, H.; Bian, Y.; Rong, Y.; Huang, W.; Xu, T.; Xie, W.; Ye, G.; Huang, J. Dual Message Passing Neural Network for Molecular Property Prediction. arXiv 2020, arXiv:2005.13607. [Google Scholar]
- Deng, D.; Chen, X.; Zhang, R.; Lei, Z.; Wang, X.; Zhou, F. XGraphBoost: Extracting Graph Neural Network-Based Features for a Better Prediction of Molecular Properties. J. Chem. Inf. Model. 2021, 61, 2697–2705. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Z.; Jiang, M.; Wang, S.; Zhang, S.; Wei, Z. Molecule Property Prediction Based on Spatial Graph Embedding. J. Chem. Inf. Model. 2019, 59, 3817–3828. [Google Scholar] [CrossRef]
- Weber, J.K.; Morrone, J.A.; Bagchi, S.; Pabon, J.D.E.; Kang, S.G.; Zhang, L.; Cornell, W.D. Simplified, Interpretable Graph Convolutional Neural Networks for Small Molecule Activity Prediction. J. Comput.-Aided Mol. Des. 2021, 36, 391–404. [Google Scholar] [CrossRef]
- Jiménez-Luna, J.; Skalic, M.; Weskamp, N.; Schneider, G. Coloring Molecules with Explainable Artificial Intelligence for Preclinical Relevance Assessment. J. Chem. Inf. Model. 2021, 61, 1083–1094. [Google Scholar] [CrossRef]
- Guo, Z.; Yu, W.; Zhang, C.; Jiang, M.; Chawla, N.V. GraSeq: Graph and Sequence Fusion Learning for Molecular Property Prediction. In Proceedings of the International Conference on Information and Knowledge Management, Virtual, 19–23 October 2020; pp. 435–443. [Google Scholar]
- Jin, Y.; Lu, J.; Shi, R.; Yang, Y. EmbedDTI: Enhancing the Molecular Representations via Sequence Embedding and Graph Convolutional Network for the Prediction of Drug-Target Interaction. Biomolecules 2021, 11, 1783. [Google Scholar] [CrossRef]
- Shrivastava, A.D.; Swainston, N.; Samanta, S.; Roberts, I.; Muelas, M.W.; Kell, D.B. Massgenie: A Transformer-Based Deep Learning Method for Identifying Small Molecules from Their Mass Spectra. Biomolecules 2021, 11, 1793. [Google Scholar] [CrossRef]
- Li, C.; Feng, J.; Liu, S.; Yao, J. A Novel Molecular Representation Learning for Molecular Property Prediction with a Multiple SMILES-Based Augmentation. Comput. Intell. Neurosci. 2022, 2022, 8464452. [Google Scholar] [CrossRef]
- Sumner, D.; He, J.; Thakkar, A.; Engkvist, O.; Bjerrum, E.J. Levenshtein Augmentation Improves Performance of SMILES Based Deep-Learning Synthesis Prediction. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Arús-Pous, J.; Johansson, S.V.; Prykhodko, O.; Bjerrum, E.J.; Tyrchan, C.; Reymond, J.L.; Chen, H.; Engkvist, O. Randomized SMILES Strings Improve the Quality of Molecular Generative Models. J. Cheminform. 2019, 11, 71. [Google Scholar] [CrossRef]
- Landrum, G. RDKit: Open-Source Cheminformatics. 2006. Available online: http://www.rdkit.org/ (accessed on 10 May 2022).
- Yang, K.K.; Wu, Z.; Bedbrook, C.N.; Arnold, F.H. Learned Protein Embeddings for Machine Learning. Bioinformatics 2018, 34, 2642–2648. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, S.; Zhu, F.; Huang, J. Seq2seq Fingerprint: An Unsupervised Deep Molecular Embedding for Drug Discovery. In Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Boston, MA, USA, 20–23 August 2017; pp. 285–294. [Google Scholar]
- Yang, K.; Swanson, K.; Jin, W.; Coley, C.; Eiden, P.; Gao, H.; Guzman-Perez, A.; Hopper, T.; Kelley, B.; Mathea, M.; et al. Analyzing Learned Molecular Representations for Property Prediction. J. Chem. Inf. Model. 2019, 59, 3370–3388. [Google Scholar] [CrossRef]
- Li, R.; Wang, S.; Zhu, F.; Huang, J. Adaptive Graph Convolutional Neural Networks. In Proceedings of the AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018; Volume 32, pp. 3546–3553. [Google Scholar]
- Veličković, P.; Casanova, A.; Liò, P.; Cucurull, G.; Romero, A.; Bengio, Y. Graph Attention Networks. arXiv 2018, arXiv:1710.10903. [Google Scholar]
- Wu, Z.; Ramsundar, B.; Feinberg, E.N.; Gomes, J.; Geniesse, C.; Pappu, A.S.; Leswing, K.; Pande, V. MoleculeNet: A Benchmark for Molecular Machine Learning. Chem. Sci. 2018, 9, 513–530. [Google Scholar] [CrossRef]
- Delaney, J.S. ESOL: Estimating Aqueous Solubility Directly from Molecular Structure. J. Chem. Inf. Comput. Sci. 2004, 44, 1000–1005. [Google Scholar] [CrossRef]
- Mobley, D.L.; Guthrie, J.P. FreeSolv: A Database of Experimental and Calculated Hydration Free Energies, with Input Files. J. Comput.-Aided Mol. Des. 2014, 28, 711–720. [Google Scholar] [CrossRef]
- Waring, M.J. Lipophilicity in Drug Discovery. Expert Opin. Drug Discov. 2010, 5, 235–248. [Google Scholar] [CrossRef]
- Garrido, A.; Lepailleur, A.; Mignani, S.M.; Dallemagne, P.; Rochais, C. HERG Toxicity Assessment: Useful Guidelines for Drug Design. Eur. J. Med. Chem. 2020, 195, 112290. [Google Scholar] [CrossRef]
- Subramanian, G.; Ramsundar, B.; Pande, V.; Denny, R.A. Computational Modeling of β-Secretase 1 (BACE-1) Inhibitors Using Ligand Based Approaches. J. Chem. Inf. Model. 2016, 56, 1936–1949. [Google Scholar] [CrossRef] [PubMed]
- Boria, I.; Garelli, E.; Gazda, H.T.; Aspesi, A.; Quarello, P.; Pavesi, E.; Ferrante, D.; Meerpohl, J.J.; Kartal, M.; Da Costa, L.; et al. The Ribosomal Basis of Diamond-Blackfan Anemia: Mutation and Database Update. Hum. Mutat. 2010, 31, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Huang, R.; Tetko, I.V.; Xia, Z.; Xu, J.; Tong, W. Trade-off Predictivity and Explainability for Machine-Learning Powered Predictive Toxicology: An in-Depth Investigation with Tox21 Data Sets. Chem. Res. Toxicol. 2021, 34, 541–549. [Google Scholar] [CrossRef]
- Gray, V.E.; Hause, R.J.; Luebeck, J.; Shendure, J.; Fowler, D.M. Quantitative Missense Variant Effect Prediction Using Large-Scale Mutagenesis Data. Cell Syst. 2018, 6, 116–124.e3. [Google Scholar] [CrossRef]
- Zhang, Y.; Lee, J.D.; Wainwright, M.J.; Jordan, M.I. On the Learnability of Fully-Connected Neural Networks. PMLR 2017, 54, 83–91. [Google Scholar]
- Liu, S.; Demirel, M.F.; Liang, Y. N-Gram Graph: Simple Unsupervised Representation for Graphs, with Applications to Molecules. Adv. Neural Inf. Process. Syst. 2019, 32, 1–13. [Google Scholar]
- Honda, S.; Shi, S.; Ueda, H.R. SMILES Transformer: Pre-Trained Molecular Fingerprint for Low Data Drug Discovery. arXiv 2019, arXiv:1911.04738. [Google Scholar]
- Jeon, W.; Kim, D. FP2VEC: A New Molecular Featurizer for Learning Molecular Properties. Bioinformatics 2019, 35, 4979–4985. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, R.; Ma, J.; Liu, Y.; Yang, E.; Du, S.; Zhao, Z.; Yuan, Y. TranGRU: Focusing on Both the Local and Global Information of Molecules for Molecular Property Prediction. Appl. Intell. 2022, 52, 1–15. [Google Scholar] [CrossRef]
- Li, J.; Zhang, T.; Tian, H.; Jin, S.; Fardad, M.; Zafarani, R. SGCN: A Graph Sparsifier Based on Graph Convolutional Networks. In Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining 2020, Singapore, 11–14 May 2020; Volume 12084, pp. 275–287. [Google Scholar]
- Gilmer, J.; Schoenholz, S.S.; Riley, P.F.; Vinyals, O.; Dahl, G.E. Neural Message Passing for Quantum Chemistry. In Proceedings of the 34th International Conference on Machine Learning, ICML 2017, Sydney, Australia, 6–11 August 2017; Volume 3, pp. 2053–2070. [Google Scholar]
- Lu, C.; Liu, Q.; Wang, C.; Huang, Z.; Lin, P.; He, L. Molecular Property Prediction: A Multilevel Quantum Interactions Modeling Perspective. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 17 July 2019; Volume 33, pp. 1052–1060. [Google Scholar]
- Xiong, Z.; Wang, D.; Liu, X.; Zhong, F.; Wan, X.; Li, X.; Li, Z.; Luo, X.; Chen, K.; Jiang, H.; et al. Pushing the Boundaries of Molecular Representation for Drug Discovery with the Graph Attention Mechanism. J. Med. Chem. 2020, 63, 8749–8760. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Liu, B.; Gomes, J.; Zitnik, M.; Liang, P.; Pande, V.; Leskovec, J. Strategies for Pre-Training Graph Neural Networks. arXiv 2019, arXiv:1905.12265. [Google Scholar]
- Chiriano, G.; De Simone, A.; Mancini, F.; Perez, D.I.; Cavalli, A.; Bolognesi, M.L.; Legname, G.; Martinez, A.; Andrisano, V.; Carloni, P.; et al. A Small Chemical Library of 2-Aminoimidazole Derivatives as BACE-1 Inhibitors: Structure-Based Design, Synthesis, and Biological Evaluation. Eur. J. Med. Chem. 2012, 48, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Mureddu, L.G.; Vuister, G.W. Fragment-Based Drug Discovery by NMR. Where Are the Successes and Where Can It Be Improved? Front. Mol. Biosci. 2022, 9, 834453. [Google Scholar] [CrossRef] [PubMed]

| Feature | Description |
|---|---|
| Atomic number | Atomic number |
| Degree | Number of directly bonded neighbors (one-hot) |
| Formal charge | Integer electronic charge (one-hot) |
| Chiral tag | Chirality information of atoms (one-hot) |
| Hs num | Number of hydrogen atoms (one-hot) |
| Hybridization | SP,SP2,SP3,SP3D,SP3D2 (one-hot) |
| Aromaticity | Whether the atom is in an aromatic hydrocarbon |
| Mass | Atomic mass |
| Dataset | Task | Task Type | #Molecule | Splits | Metric |
|---|---|---|---|---|---|
| ESOL | 1 | Regression | 1128 | Random | RMSE |
| FreeSolv | 1 | Regression | 642 | Random | RMSE |
| Lipophilicity | 1 | Regression | 4200 | Random | RMSE |
| heRG | 1 | Regression | 4813 | Random | RMSE |
| BACE | 1 | Classification | 1513 | Random | ROC-AUC |
| Mutagenesis | 1 | Classification | 6506 | Random | ROC-ACU |
| ClinTox | 2 | Classification | 1478 | Random | ROC-AUC |
| Tox21 | 12 | Classification | 7831 | Random | ROC-AUC |
| FreeSolv | ESOL | Lipophilicity | ||
|---|---|---|---|---|
| Sequence-based | FCNN | 1.87 ± 0.07 | 1.12 ± 0.15 | 0.86 ± 0.01 |
| N-GRAM | 2.512 ± 0.190 | 1.100 ± 0.160 | 0.876 ± 0.033 | |
| RNNS2S | 2.987 | 1.317 | 1.219 | |
| SMILES Transformers | 2.246 | 1.144 | 1.169 | |
| FP2VEC | 2.512 ± 0.190 | 1.100 ± 0.160 | 0.876 ± 0.033 | |
| Graph-based | SGCN | 2.158 ± 0.049 | 1.345 ± 0.019 | 1.074 ± 0.007 |
| MPNN | 1.327 ± 0.279 | 0.700 ± 0.073 | 0.673 ± 0.038 | |
| DMPNN | 2.177 | 0.980 | / | |
| MGCN | 3.349 ± 0.097 | 1.266 ± 0.147 | 1.113 ± 0.041 | |
| AttentionFP | 2.030 ± 0.420 | 0.853 ± 0.060 | 0.650 ± 0.030 | |
| our method | 1.211 ± 0.192 | 0.885 ± 0.067 | 0.709 ± 0.023 | |
| BACE | ClinTox | Tox21 | ||
|---|---|---|---|---|
| Sequence-based | N-GRAM | 0.876 ± 0.035 | 0.855 ± 0.037 | 0.769 ± 0.027 |
| RNNS2S | 0.717 | \ | 0.702 | |
| SMILES Transformers | 0.719 | \ | 0.706 | |
| TranGRU | 0.790 | \ | 0.813 | |
| Graph-based | SGCN | \ | 0.820 ± 0.009 | 0.766 ± 0.002 |
| MPNN | 0.793 ± 0.031 | 0.879 ± 0.054 | 0.809 ± 0.017 | |
| MGCN | 0.734 ± 0.030 | 0.634 ± 0.042 | 0.707 ± 0.016 | |
| AttentionFP | 0.863 ± 0.015 | 0.796 ± 0.005 | 0.807 ± 0.020 | |
| PreGNN | 0.845 | \ | 0.781 | |
| GraSeq | 0.838 | \ | 0.820 | |
| our method | 0.880 ± 0.009 | 0.883 ± 0.025 | 0.774 ± 0.005 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Pan, S.; Xia, L.; Li, Z. Molecular Property Prediction by Combining LSTM and GAT. Biomolecules 2023, 13, 503. https://doi.org/10.3390/biom13030503
Xu L, Pan S, Xia L, Li Z. Molecular Property Prediction by Combining LSTM and GAT. Biomolecules. 2023; 13(3):503. https://doi.org/10.3390/biom13030503
Chicago/Turabian StyleXu, Lei, Shourun Pan, Leiming Xia, and Zhen Li. 2023. "Molecular Property Prediction by Combining LSTM and GAT" Biomolecules 13, no. 3: 503. https://doi.org/10.3390/biom13030503
APA StyleXu, L., Pan, S., Xia, L., & Li, Z. (2023). Molecular Property Prediction by Combining LSTM and GAT. Biomolecules, 13(3), 503. https://doi.org/10.3390/biom13030503







