Screening of EWI-2-Derived Peptides for Targeting Tetraspanin CD81 and Their Effect on Cancer Cell Migration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Preparation of the EWI-2 Peptide Array
2.3. Preparation of Fluorescently Labeled CD81 Protein
2.4. Binding Assay between EWI-2 Peptide Array and Fluorescently Labeled CD81 Protein
2.5. Characterizations of Candidate Peptides
2.6. Preparation of Candidate Peptide Powders
2.7. Boyden Chamber Assay
2.8. Statistical Analysis
3. Results and Discussion
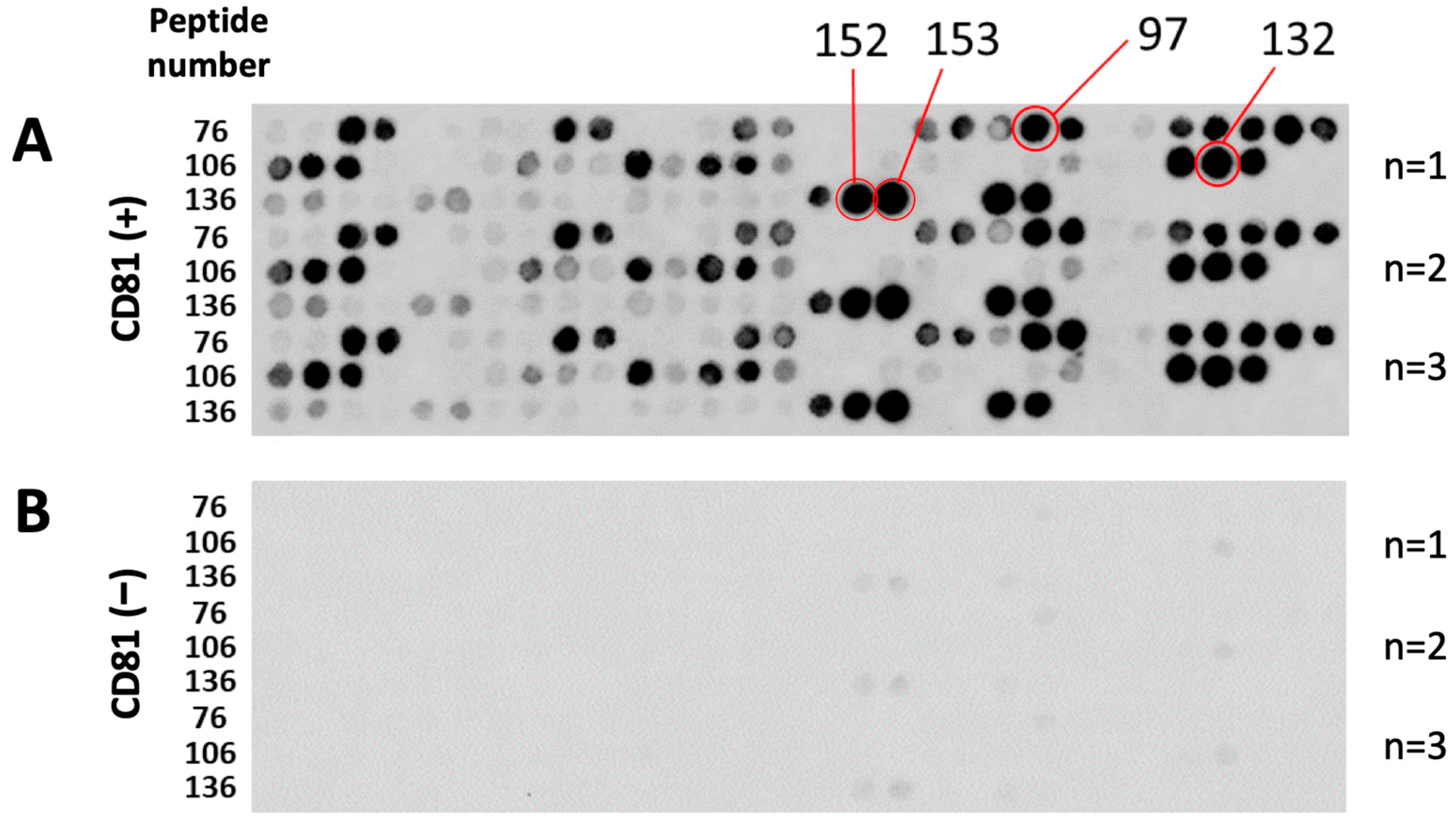
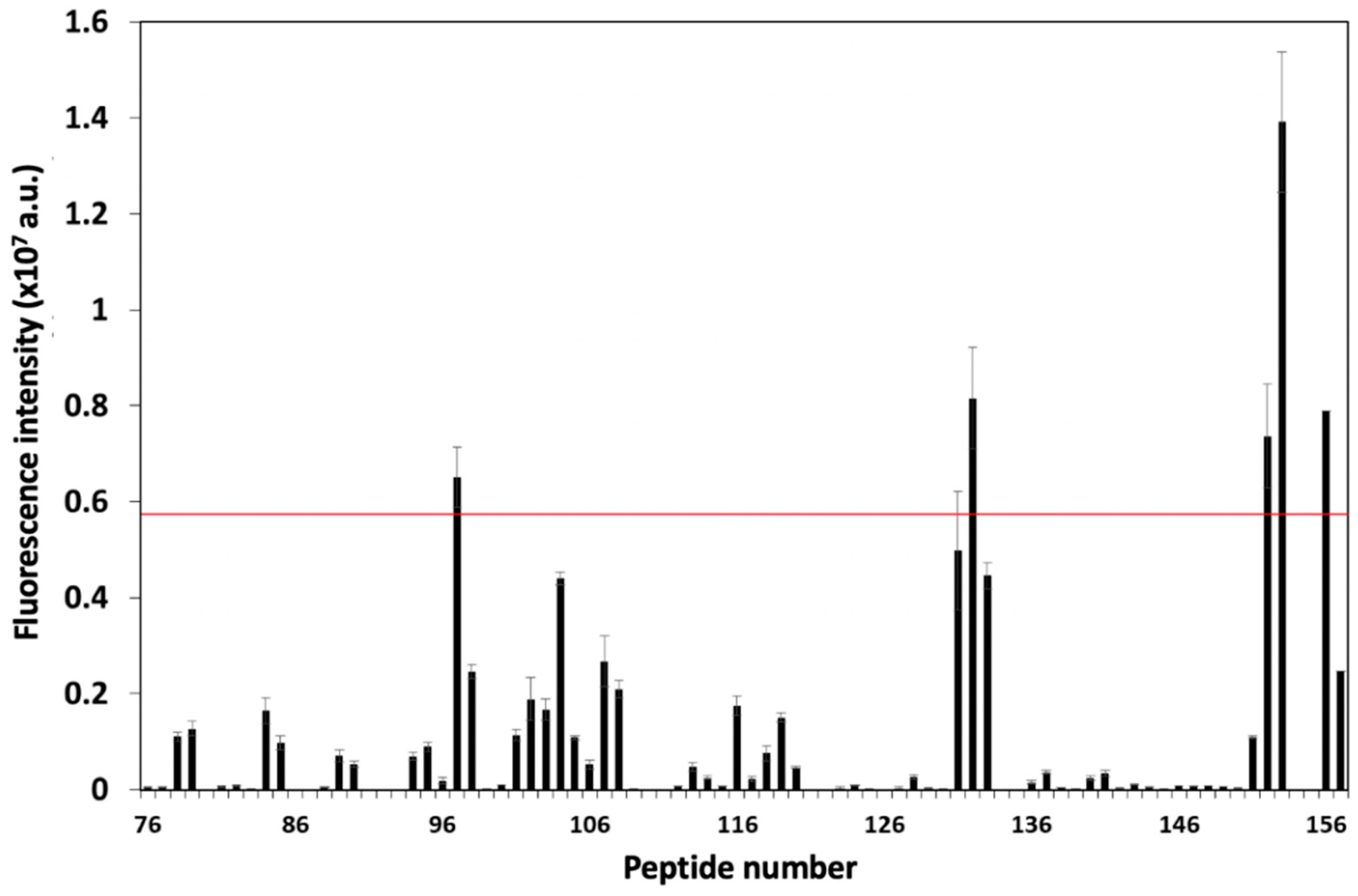
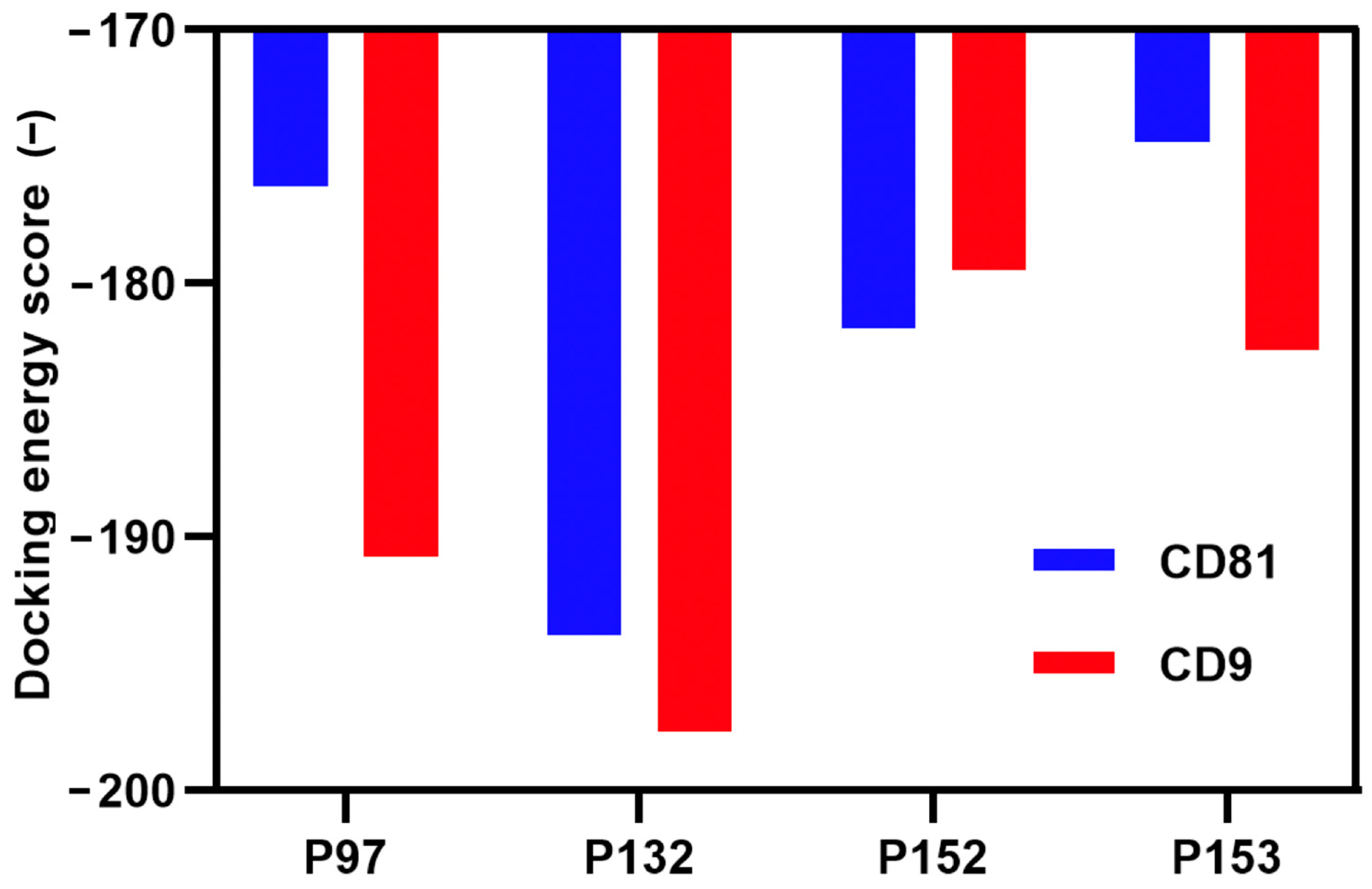
3.1. Screening and Characterization of CD81-BP Candidates
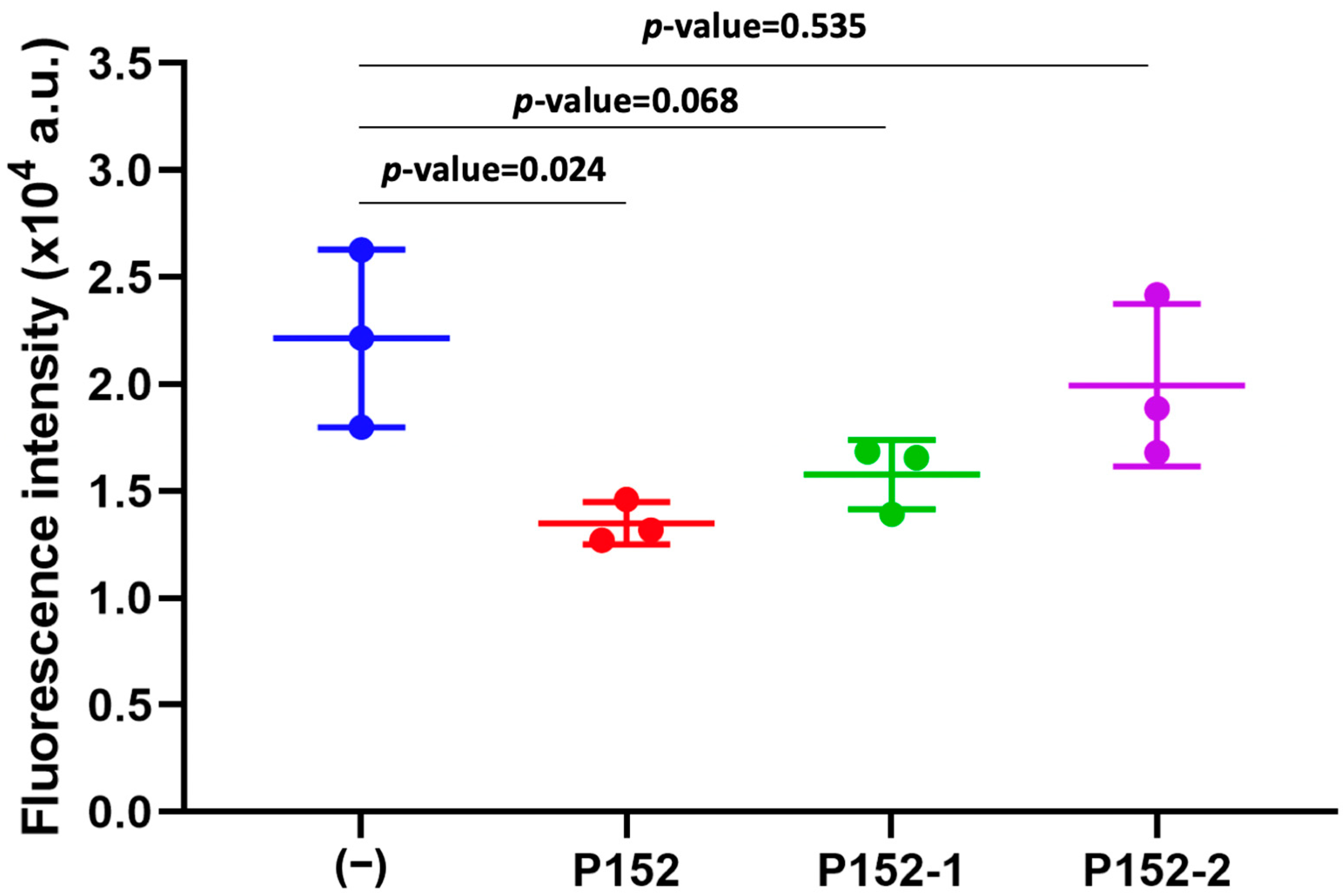
3.2. Effect of CD81-BP Candidates on Cancer Cell Migration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deng, Y.; Cai, S.; Shen, J.; Peng, H. Tetraspanins: Novel Molecular Regulators of Gastric Cancer. Front. Oncol. 2021, 11, 702510. [Google Scholar] [CrossRef]
- Hemler, M.E. Targeting of tetraspanin proteins—Potential benefits and strategies. Nat. Rev. Drug Discov. 2008, 7, 747–758. [Google Scholar] [CrossRef] [Green Version]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005, 6, 801–811. [Google Scholar] [CrossRef]
- Hochheimer, N.; Sies, R.; Aschenbrenner, A.C.; Schneider, D.; Lang, T. Classes of non-conventional tetraspanins defined by alternative splicing. Sci. Rep. 2019, 9, 14075. [Google Scholar] [CrossRef] [Green Version]
- Levy, S.; Shoham, T. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 2005, 5, 136–148. [Google Scholar] [CrossRef]
- Pileri, P.; Uematsu, Y.; Campagnoli, S.; Galli, G.; Falugi, F.; Petracca, R.; Weiner, A.J.; Houghton, M.; Rosa, D.; Grandi, G.; et al. Binding of Hepatitis C Virus to CD81. Science 1998, 282, 938–941. [Google Scholar] [CrossRef]
- Kong, L.; Giang, E.; Nieusma, T.; Kadam, R.U.; Cogburn, K.E.; Hua, Y.; Dai, X.; Stanfield, R.L.; Burton, D.R.; Ward, A.B.; et al. Hepatitis C Virus E2 Envelope Glycoprotein Core Structure. Science 2013, 342, 1090–1094. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.G.; Whidby, J.; Miller, M.T.; Scarborough, H.; Zatorski, A.V.; Cygan, A.; Price, A.A.; Yost, S.A.; Bohannon, C.D.; Jacob, J.; et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 2014, 509, 381–384. [Google Scholar] [CrossRef] [Green Version]
- Fénéant, L.; Cocquerel, L. SRFBP1, an Additional Player in HCV Entry. Trends Microbiol. 2015, 23, 590–593. [Google Scholar] [CrossRef]
- Vences-Catalán, F.; Duault, C.; Kuo, C.-C.; Rajapaksa, R.; Levy, R.; Levy, S. CD81 as a tumor target. Biochem. Soc. Trans. 2017, 45, 531–535. [Google Scholar] [CrossRef]
- Zhang, N.; Zuo, L.; Zheng, H.; Li, G.; Hu, X. Increased Expression of CD81 in Breast Cancer Tissue is Associated with Reduced Patient Prognosis and Increased Cell Migration and Proliferation in MDA-MB-231 and MDA-MB-435S Human Breast Cancer Cell Lines In Vitro. Med. Sci. Monit. 2018, 24, 5739–5747. [Google Scholar] [CrossRef]
- Hong, I.-K.; Byun, H.-J.; Lee, J.; Jin, Y.-J.; Wang, S.-J.; Jeoung, D.-I.; Kim, Y.-M.; Lee, H. The Tetraspanin CD81 Protein Increases Melanoma Cell Motility by Up-regulating Metalloproteinase MT1-MMP Expression through the Pro-oncogenic Akt-dependent Sp1 Activation Signaling Pathways. J. Biol. Chem. 2014, 289, 15691–15704. [Google Scholar] [CrossRef] [Green Version]
- Vexler, V.; Yu, L.; Pamulapati, C.; Garrido, R.; Grimm, H.P.; Sriraman, P.; Bohini, S.; Schraeml, M.; Singh, U.; Brandt, M.; et al. Target-mediated drug disposition and prolonged liver accumulation of a novel humanized anti-CD81 monoclonal antibody in cynomolgus monkeys. Mabs 2013, 5, 776–786. [Google Scholar] [CrossRef] [Green Version]
- Vences-Catalán, F.; Rajapaksa, R.; Kuo, C.-C.; Miller, C.L.; Lee, A.; Ramani, V.C.; Jeffrey, S.S.; Levy, R.; Levy, S. Targeting the tetraspanin CD81 reduces cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 2021, 118, e2018961118. [Google Scholar] [CrossRef]
- Lorico, A.; Lorico-Rappa, M.; Karbanová, J.; Corbeil, D.; Pizzorno, G. CD9, a tetraspanin target for cancer therapy? Exp. Biol. Med. 2021, 246, 1121–1138. [Google Scholar] [CrossRef]
- Liu, D.; Guo, P.; McCarthy, C.; Wang, B.; Tao, Y.; Auguste, D. Peptide density targets and impedes triple negative breast cancer metastasis. Nat. Commun. 2018, 9, 2612. [Google Scholar] [CrossRef] [Green Version]
- Suwatthanarak, T.; Tanaka, M.; Minamide, T.; Harvie, A.J.; Tamang, A.; Critchley, K.; Evans, S.D.; Okochi, M. Screening and characterisation of CdTe/CdS quantum dot-binding peptides for material surface functionalisation. RSC Adv. 2020, 10, 8218–8223. [Google Scholar] [CrossRef]
- Suwatthanarak, T.; Tanaka, M.; Miyamoto, Y.; Miyado, K.; Okochi, M. Inhibition of cancer-cell migration by tetraspanin CD9-binding peptide. Chem. Commun. 2021, 57, 4906–4909. [Google Scholar] [CrossRef]
- Frank, R. Spot-synthesis: An easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 1992, 48, 9217–9232. [Google Scholar] [CrossRef]
- Suwatthanarak, T.; Thiodorus, I.A.; Tanaka, M.; Shimada, T.; Takeshita, D.; Yasui, T.; Baba, Y.; Okochi, M. Microfluidic-based capture and release of cancer-derived exosomes via peptide–nanowire hybrid interface. Lab Chip 2020, 21, 597–607. [Google Scholar] [CrossRef]
- Stipp, C.S.; Kolesnikova, T.V.; Hemler, M.E. EWI-2 Is a Major CD9 and CD81 Partner and Member of a Novel Ig Protein Subfamily. J. Biol. Chem. 2001, 276, 40545–40554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Yan, C.; Zou, X. MDockPeP: An ab-initio protein–peptide docking server. J. Comput. Chem. 2018, 39, 2409–2413. [Google Scholar] [CrossRef]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.-Y. HPEPDOCK: A web server for blind peptide–protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef]
- Clark, K.L.; Zeng, Z.; Langford, A.L.; Bowen, S.M.; Todd, S.C. PGRL Is a Major CD81-Associated Protein on Lymphocytes and Distinguishes a New Family of Cell Surface Proteins. J. Immunol. 2001, 167, 5115–5121. [Google Scholar] [CrossRef] [Green Version]
- Montpellier, C.; Tews, B.A.; Poitrimole, J.; Rocha-Perugini, V.; D’Arienzo, V.; Potel, J.; Zhang, X.A.; Rubinstein, E.; Dubuisson, J.; Cocquerel, L. Interacting Regions of CD81 and Two of Its Partners, EWI-2 and EWI-2wint, and Their Effect on Hepatitis C Virus Infection. J. Biol. Chem. 2011, 286, 13954–13965. [Google Scholar] [CrossRef] [Green Version]
- Watson, J.D.; Milner-White, E. A novel main-chain anion-binding site in proteins: The nest. A particular combination of φ,ψ values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J. Mol. Biol. 2002, 315, 171–182. [Google Scholar] [CrossRef]
- Milner-White, E.J.; Nissink, J.W.M.; Allen, F.H.; Duddy, W.J. Recurring main-chain anion-binding motifs in short polypeptides: Nests. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 1935–1942. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Wang, K.; Liu, Q.; Zhang, X. Discovery of monoamine oxidase A inhibitory peptides from hairtail (Trichiurus japonicus) using in vitro simulated gastrointestinal digestion and in silico studies. Bioorg. Chem. 2020, 101, 104032. [Google Scholar] [CrossRef]
- Suwatthanarak, T.; Ito, K.; Tanaka, M.; Sugiura, K.; Hoshino, A.; Miyamoto, Y.; Miyado, K.; Okochi, M. A peptide binding to the tetraspanin CD9 reduces cancer metastasis. Biomater. Adv. 2023, 146, 213283. [Google Scholar] [CrossRef]
- Sowińska, M.; Szeliga, M.; Morawiak, M.; Zabłocka, B.; Urbanczyk-Lipkowska, Z. Design, Synthesis and Activity of New N1-Alkyl Tryptophan Functionalized Dendrimeric Peptides against Glioblastoma. Biomolecules 2022, 12, 1116. [Google Scholar] [CrossRef]

| Peptide a | Sequence | pI b | Charge c | GRAVY d | Estimated KD (µM) e |
|---|---|---|---|---|---|
| P153 | FMKRLRKR | 12.3 | +4.5 | −1.6 | 12.6 |
| P132 | RSHRLRLH | 12.3 | +2.6 | −1.6 | 1.63 |
| P152 | CFMKRLRK | 11.0 | +3.5 | −0.72 | 0.91 |
| P97 | ASRTYRLR | 11.7 | +2.6 | −1.3 | 1.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suwatthanarak, T.; Usuba, K.; Kuroha, K.; Tanaka, M.; Okochi, M. Screening of EWI-2-Derived Peptides for Targeting Tetraspanin CD81 and Their Effect on Cancer Cell Migration. Biomolecules 2023, 13, 510. https://doi.org/10.3390/biom13030510
Suwatthanarak T, Usuba K, Kuroha K, Tanaka M, Okochi M. Screening of EWI-2-Derived Peptides for Targeting Tetraspanin CD81 and Their Effect on Cancer Cell Migration. Biomolecules. 2023; 13(3):510. https://doi.org/10.3390/biom13030510
Chicago/Turabian StyleSuwatthanarak, Thanawat, Kei Usuba, Kotomi Kuroha, Masayoshi Tanaka, and Mina Okochi. 2023. "Screening of EWI-2-Derived Peptides for Targeting Tetraspanin CD81 and Their Effect on Cancer Cell Migration" Biomolecules 13, no. 3: 510. https://doi.org/10.3390/biom13030510





