LncRNA and circRNA in Patients with Non-Alcoholic Fatty Liver Disease: A Systematic Review
Abstract
1. Introduction
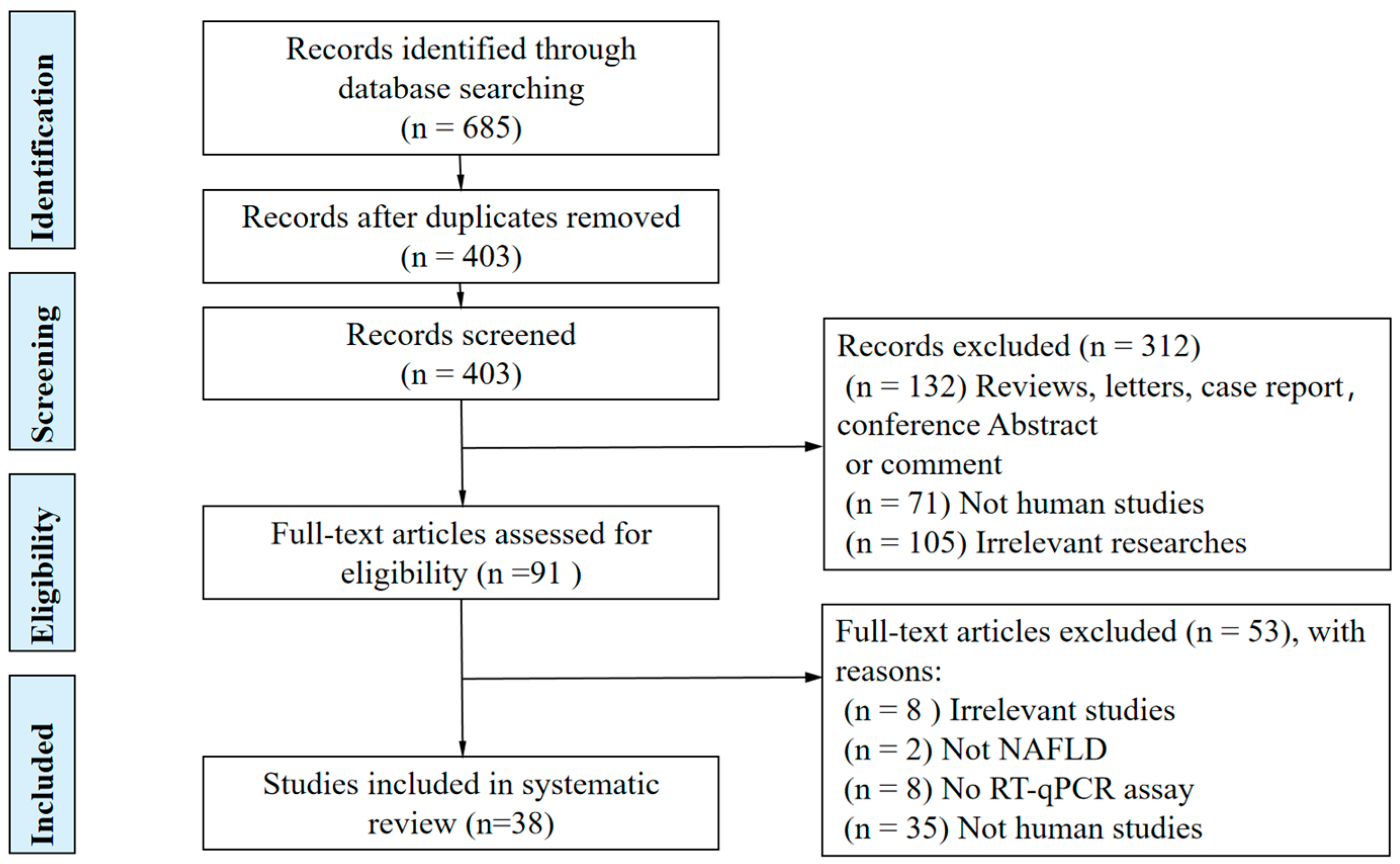
2. Methods
2.1. Literature Search Protocol and Search Strategy
2.2. Study Selection and Eligibility Criteria
2.3. Data Extraction
3. Results
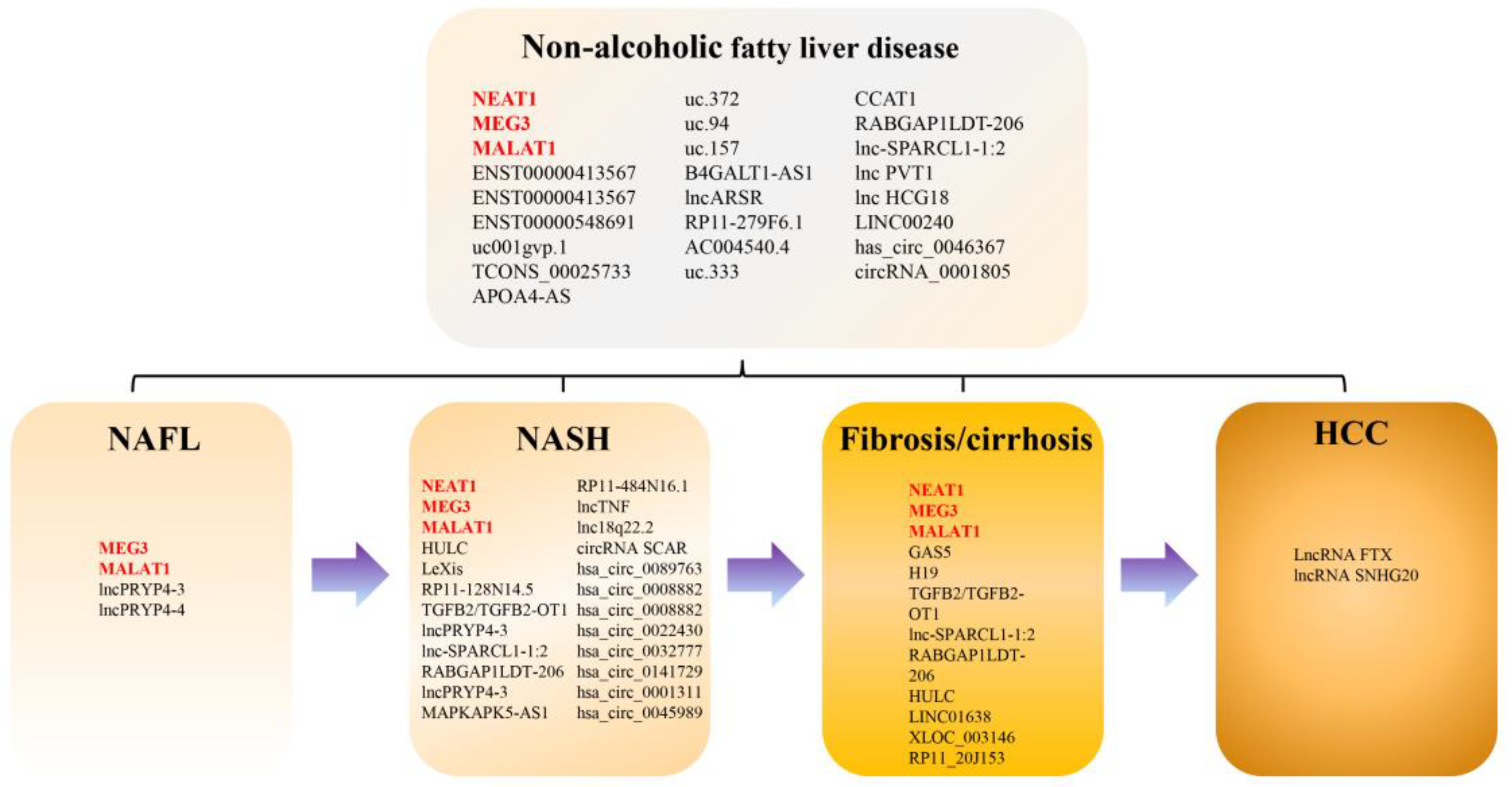
3.1. Summary of Studies on the lncRNA and circRNA Expression Profile in NAFLD
3.2. Differentially Expressed lncRNA in Patients with NAFLD
3.3. Differentially Expressed lncRNAs in Patients with NASH
3.4. Differentially Expressed lncRNAs in Patients with Advanced Fibrosis or Cirrhosis
3.5. Differentially Expressed lncRNAs in Patients with NAFLD-Related HCC
3.6. Differentially Expressed circRNAs in Patients with NAFLD and NASH
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cotter, T.G.; Rinella, M. Nonalcoholic Fatty Liver Disease 2020: The State of the Disease. Gastroenterology 2020, 158, 1851–1864. [Google Scholar] [CrossRef]
- Paik, J.M.; Kabbara, K.; Eberly, K.E.; Younossi, Y.; Henry, L.; Younossi, Z.M. Global burden of NAFLD and chronic liver disease among adolescents and young adults. Hepatology 2022, 75, 1204–1217. [Google Scholar] [CrossRef]
- Le, M.H.; Yeo, Y.H.; Li, X.; Li, J.; Zou, B.; Wu, Y.; Ye, Q.; Huang, D.Q.; Zhao, C.; Zhang, J.; et al. 2019 Global NAFLD Prevalence: A Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2021, 20, 2809–2817. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, J.V.; Mark, H.E.; Anstee, Q.M.; Arab, J.P.; Batterham, R.L.; Castera, L.; Cortez-Pinto, H.; Crespo, J.; Cusi, K.; Dirac, M.A.; et al. Advancing the global public health agenda for NAFLD: A consensus statement. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 60–78. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Targher, G.; Tilg, H.; Byrne, C.D. Non-alcoholic fatty liver disease: A multisystem disease requiring a multidisciplinary and holistic approach. Lancet Gastroenterol. Hepatol. 2021, 6, 578–588. [Google Scholar] [CrossRef]
- Dufour, J.F.; Anstee, Q.M.; Bugianesi, E.; Harrison, S.; Loomba, R.; Paradis, V.; Tilg, H.; Wong, V.W.; Zelber-Sagi, S. Current therapies and new developments in NASH. Gut 2022, 71, 2123–2134. [Google Scholar] [CrossRef] [PubMed]
- Pais, R.; Barritt, A.S.t.; Calmus, Y.; Scatton, O.; Runge, T.; Lebray, P.; Poynard, T.; Ratziu, V.; Conti, F. NAFLD and liver transplantation: Current burden and expected challenges. J. Hepatol. 2016, 65, 1245–1257. [Google Scholar] [CrossRef]
- Ekstedt, M.; Hagstrom, H.; Nasr, P.; Fredrikson, M.; Stal, P.; Kechagias, S.; Hultcrantz, R. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology 2015, 61, 1547–1554. [Google Scholar] [CrossRef]
- Lonardo, A.; Mantovani, A.; Petta, S.; Carraro, A.; Byrne, C.D.; Targher, G. Metabolic mechanisms for and treatment of NAFLD or NASH occurring after liver transplantation. Nat. Rev. Endocrinol. 2022, 18, 638–650. [Google Scholar] [CrossRef]
- Liu, C.H.; Ampuero, J.; Gil-Gomez, A.; Montero-Vallejo, R.; Rojas, A.; Munoz-Hernandez, R.; Gallego-Duran, R.; Romero-Gomez, M. miRNAs in patients with non-alcoholic fatty liver disease: A systematic review and meta-analysis. J. Hepatol. 2018, 69, 1335–1348. [Google Scholar] [CrossRef] [PubMed]
- Gjorgjieva, M.; Sobolewski, C.; Dolicka, D.; Correia de Sousa, M.; Foti, M. miRNAs and NAFLD: From pathophysiology to therapy. Gut 2019, 68, 2065–2079. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Qian, G.; Morral, N. Role of non-coding RNAs on liver metabolism and NAFLD pathogenesis. Hum. Mol. Genet. 2022, 31, R4–R21. [Google Scholar] [CrossRef]
- Han, S.; Zhang, T.; Kusumanchi, P.; Huda, N.; Jiang, Y.; Yang, Z.; Liangpunsakul, S. Long non-coding RNAs in liver diseases: Focusing on nonalcoholic fatty liver disease, alcohol-related liver disease, and cholestatic liver disease. Clin. Mol. Hepatol. 2020, 26, 705–714. [Google Scholar] [CrossRef]
- Huang, R.; Duan, X.; Fan, J.; Li, G.; Wang, B. Role of noncoding RNA in development of nonalcoholic fatty liver disease. BioMed Res. Int. 2019, 2019, 8690592. [Google Scholar] [CrossRef]
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target. Ther. 2021, 6, 185. [Google Scholar] [CrossRef]
- Guo, J.; Fang, W.; Sun, L.; Lu, Y.; Dou, L.; Huang, X.; Tang, W.; Yu, L.; Li, J. Ultraconserved element uc.372 drives hepatic lipid accumulation by suppressing miR-195/miR4668 maturation. Nat. Commun. 2018, 9, 612. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, J.; Yao, H.; Lin, Y.; Wei, J.; Hu, G.; Guo, J.; Li, J. Ultraconserved element uc.333 increases insulin sensitivity by binding to miR-223. Aging 2020, 12, 6667–6679. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Liu, H.; Lei, Z.; Li, Z.; Zhang, T.; Yang, M.; Zhou, K.; Sun, C. Long noncoding RNA CCAT1 inhibits miR-613 to promote nonalcoholic fatty liver disease via increasing LXRα transcription. J. Cell. Physiol. 2020, 235, 9819–9833. [Google Scholar] [CrossRef]
- Albadawy, R.; Agwa, S.H.A.; Khairy, E.; Saad, M.; El Touchy, N.; Othman, M.; Matboli, M. Clinical Significance of HSPD1/MMP14/ITGB1/miR-6881-5P/Lnc-SPARCL1-1:2 RNA Panel in NAFLD/NASH Diagnosis: Egyptian Pilot Study. Biomedicines 2021, 9, 1248. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.J.; Long, M.; Dai, R.J. Acetylation of H3K27 activated lncRNA NEAT1 and promoted hepatic lipid accumulation in non-alcoholic fatty liver disease via regulating miR-212-5p/GRIA3. Mol. Cell. Biochem. 2022, 477, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Niu, Q.; Liang, K.; Li, X.; Jiang, J.; Bian, C. Effect of LncPVT1/miR-20a-5p on Lipid Metabolism and Insulin Resistance in NAFLD. Diabetes Metab. Syndr. Obes. 2021, 14, 4599–4608. [Google Scholar] [CrossRef]
- Zhang, M.; Chi, X.; Qu, N.; Wang, C. Long noncoding RNA lncARSR promotes hepatic lipogenesis via Akt/SREBP-1c pathway and contributes to the pathogenesis of nonalcoholic steatohepatitis. Biochem. Biophys. Res. Commun. 2018, 499, 66–70. [Google Scholar] [CrossRef]
- Zhou, W.; Qiu, K. The correlation between lncRNA NEAT1 and serum hepcidin in the peripheral blood of non-alcoholic fatty liver disease patients. Am. J. Transl. Res. 2022, 14, 2593–2599. [Google Scholar]
- Cheng, X.; Shihabudeen Haider Ali, M.S.; Moran, M.; Viana, M.P.; Schlichte, S.L.; Zimmerman, M.C.; Khalimonchuk, O.; Feinberg, M.W.; Sun, X. Long non-coding RNA Meg3 deficiency impairs glucose homeostasis and insulin signaling by inducing cellular senescence of hepatic endothelium in obesity. Redox Biol. 2021, 40, 101863. [Google Scholar] [CrossRef]
- Xiang, J.; Deng, Y.Y.; Liu, H.X.; Pu, Y. LncRNA MALAT1 Promotes PPARα/CD36-Mediated Hepatic Lipogenesis in Nonalcoholic Fatty Liver Disease by Modulating miR-206/ARNT Axis. Front. Bioeng. Biotechnol. 2022, 10, 858558. [Google Scholar] [CrossRef]
- Sun, C.; Liu, X.; Yi, Z.; Xiao, X.; Yang, M.; Hu, G.; Liu, H.; Liao, L.; Huang, F. Genome-wide analysis of long noncoding RNA expression profiles in patients with non-alcoholic fatty liver disease. IUBMB Life 2015, 67, 847–852. [Google Scholar] [CrossRef]
- Qin, W.; Li, X.; Xie, L.; Li, S.; Liu, J.; Jia, L.; Dong, X.; Ren, X.; Xiao, J.; Yang, C.; et al. A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic. Acids Res. 2016, 44, 6423–6433. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, W.; Chen, Z.; Chen, J.; Meng, Y.; Feng, B.; Sun, L.; Dou, L.; Li, J.; Cui, Q.; et al. Long Noncoding RNA lncSHGL Recruits hnRNPA1 to Suppress Hepatic Gluconeogenesis and Lipogenesis. Diabetes 2018, 67, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Song, X.; Ling, Y.; Zhou, J.; Tao, Z.; Shen, Y. Comprehensive bioinformatics analysis of critical lncRNAs, mRNAs and miRNAs in non-alcoholic fatty liver disease. Mol. Med. Rep. 2019, 19, 2649–2659. [Google Scholar] [CrossRef] [PubMed]
- Albadawy, R.; Agwa, S.H.A.; Khairy, E.; Saad, M.; El Touchy, N.; Othman, M.; El Kassas, M.; Matboli, M. Circulatory Endothelin 1-Regulating RNAs Panel: Promising Biomarkers for Non-Invasive NAFLD/NASH Diagnosis and Stratification: Clinical and Molecular Pilot Study. Genes 2021, 12, 1813. [Google Scholar] [CrossRef]
- Xia, Y.; Zhang, Y.; Wang, H. Upregulated lncRNA HCG18 in Patients with Non-Alcoholic Fatty Liver Disease and Its Regulatory Effect on Insulin Resistance. Diabetes Metab. Syndr. Obes. 2021, 14, 4747–4756. [Google Scholar] [CrossRef]
- Xue, W.; Zhang, J.; Zhu, Y.; Huang, W. Identify Functional lncRNAs in Nonalcoholic Fatty Liver Disease by Constructing a ceRNA Network. ACS Omega 2022, 7, 22522–22530. [Google Scholar] [CrossRef]
- Guo, X.-Y.; Chen, J.-N.; Sun, F.; Wang, Y.-Q.; Pan, Q.; Fan, J.-G. circRNA_0046367 Prevents Hepatoxicity of Lipid Peroxidation: An Inhibitory Role against Hepatic Steatosis. Oxidative Med. Cell. Longev. 2017, 2017, 3960197. [Google Scholar] [CrossRef]
- Li, J.; Qi, J.; Tang, Y.; Liu, H.; Zhou, K.; Dai, Z.; Yuan, L.; Sun, C. A nanodrug system overexpressed circRNA_0001805 alleviates nonalcoholic fatty liver disease via miR-106a-5p/miR-320a and ABCA1/CPT1 axis. J. Nanobiotechnol. 2021, 19, 363. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, Z.; Trottier, J.; Barbier, O.; Wang, L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology 2017, 65, 604–615. [Google Scholar] [CrossRef]
- Zou, D.; Liu, L.; Zeng, Y.; Wang, H.; Dai, D.; Xu, M. LncRNA MEG3 up-regulates SIRT6 by ubiquitinating EZH2 and alleviates nonalcoholic fatty liver disease. Cell Death Discov. 2022, 8, 103. [Google Scholar] [CrossRef]
- Sookoian, S.; Flichman, D.; Garaycoechea, M.E.; San Martino, J.; Castaño, G.O.; Pirola, C.J. Metastasis-associated lung adenocarcinoma transcript 1 as a common molecular driver in the pathogenesis of nonalcoholic steatohepatitis and chronic immune-mediated liver damage. Hepatol. Commun. 2018, 2, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, Q.; Zhang, L.; Zhu, M.; Niu, J.; Xue, F.; Yang, L.; Qu, Q.; Lao, Y.; Ding, Z.; et al. LncPRYP4-3 serves as a novel diagnostic biomarker for dissecting subtypes of metabolic associated fatty liver disease by targeting RPS4Y2. Clin. Exp. Med. 2020, 20, 587–600. [Google Scholar] [CrossRef]
- Di Mauro, S.; Scamporrino, A.; Petta, S.; Urbano, F.; Filippello, A.; Ragusa, M.; Di Martino, M.T.; Scionti, F.; Grimaudo, S.; Pipitone, R.M.; et al. Serum coding and non-coding RNAs as biomarkers of NAFLD and fibrosis severity. Liver Int. 2019, 39, 1742–1754. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Liu, J.; Deng, H.; Ma, R.; Liao, J.-Y.; Liang, H.; Hu, J.; Li, J.; Guo, Z.; Cai, J.; et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell 2020, 183, 76–93.e22. [Google Scholar] [CrossRef] [PubMed]
- Park, J.G.; Kim, G.; Jang, S.Y.; Lee, Y.R.; Lee, E.; Lee, H.W.; Han, M.H.; Chun, J.M.; Han, Y.S.; Yoon, J.S.; et al. Plasma Long Noncoding RNA LeXis is a Potential Diagnostic Marker for Non-Alcoholic Steatohepatitis. Life 2020, 10, 230. [Google Scholar] [CrossRef]
- Atanasovska, B.; Rensen, S.S.; van der Sijde, M.R.; Marsman, G.; Kumar, V.; Jonkers, I.; Withoff, S.; Shiri-Sverdlov, R.; Greve, J.W.M.; Faber, K.N.; et al. A liver-specific long noncoding RNA with a role in cell viability is elevated in human nonalcoholic steatohepatitis. Hepatology 2017, 66, 794–808. [Google Scholar] [CrossRef]
- Leti, F.; Legendre, C.; Still, C.D.; Chu, X.; Petrick, A.; Gerhard, G.S.; DiStefano, J.K. Altered expression of MALAT1 lncRNA in nonalcoholic steatohepatitis fibrosis regulates CXCL5 in hepatic stellate cells. Transl. Res. 2017, 190, 25–39.e21. [Google Scholar] [CrossRef]
- Atanasovska, B.; Rensen, S.S.; Marsman, G.; Shiri-Sverdlov, R.; Withoff, S.; Kuipers, F.; Wijmenga, C.; van de Sluis, B.; Fu, J. Long Non-Coding RNAs Involved in Progression of Non-Alcoholic Fatty Liver Disease to Steatohepatitis. Cells 2021, 10, 1883. [Google Scholar] [CrossRef]
- Han, M.H.; Lee, J.H.; Kim, G.; Lee, E.; Lee, Y.R.; Jang, S.Y.; Lee, H.W.; Chun, J.M.; Han, Y.S.; Yoon, J.S.; et al. Expression of the Long Noncoding RNA GAS5 Correlates with Liver Fibrosis in Patients with Nonalcoholic Fatty Liver Disease. Genes 2020, 11, 545. [Google Scholar] [CrossRef]
- Yu, F.; Zheng, J.; Mao, Y.; Dong, P.; Lu, Z.; Li, G.; Guo, C.; Liu, Z.; Fan, X. Long Non-coding RNA Growth Arrest-specific Transcript 5 (GAS5) Inhibits Liver Fibrogenesis through a Mechanism of Competing Endogenous RNA. J. Biol. Chem. 2015, 290, 28286–28298. [Google Scholar] [CrossRef]
- Gerhard, G.S.; Davis, B.; Wu, X.; Hanson, A.; Wilhelmsen, D.; Piras, I.S.; Still, C.D.; Chu, X.; Petrick, A.T.; DiStefano, J.K. Differentially expressed mRNAs and lncRNAs shared between activated human hepatic stellate cells and nash fibrosis. Biochem. Biophys. Rep. 2020, 22, 100753. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, Z.; Wu, J.; Zhang, L.; Lee, S.; Shin, D.J.; Tran, M.; Wang, L. Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology 2018, 67, 1768–1783. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, G.N. Epidemiology and risk-stratification of NAFLD-associated HCC. J. Hepatol. 2021, 75, 1476–1484. [Google Scholar] [CrossRef]
- Wu, H.; Zhong, Z.; Wang, A.; Yuan, C.; Ning, K.; Hu, H.; Wang, C.; Yin, X. LncRNA FTX represses the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma via regulating the M1/M2 polarization of Kupffer cells. Cancer Cell Int. 2020, 20, 266. [Google Scholar] [CrossRef]
- Wang, B.; Li, X.; Hu, W.; Zhou, Y.; Din, Y. Silencing of lncRNA SNHG20 delays the progression of nonalcoholic fatty liver disease to hepatocellular carcinoma via regulating liver Kupffer cells polarization. IUBMB Life 2019, 71, 1952–1961. [Google Scholar] [CrossRef]
- Kim, S.S.; Baek, G.O.; Son, J.A.; Ahn, H.R.; Yoon, M.K.; Cho, H.J.; Yoon, J.H.; Nam, S.W.; Cheong, J.Y.; Eun, J.W. Early detection of hepatocellular carcinoma via liquid biopsy: Panel of small extracellular vesicle-derived long noncoding RNAs identified as markers. Mol. Oncol. 2021, 15, 2715–2731. [Google Scholar] [CrossRef]
- Chen, T. Circulating Non-Coding RNAs as Potential Diagnostic Biomarkers in Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2022, 9, 1029–1040. [Google Scholar] [CrossRef]
- Chen, X.; Tan, X.R.; Li, S.J.; Zhang, X.X. LncRNA NEAT1 promotes hepatic lipid accumulation via regulating miR-146a-5p/ROCK1 in nonalcoholic fatty liver disease. Life Sci. 2019, 235, 116829. [Google Scholar] [CrossRef]
- Jin, S.S.; Lin, C.J.; Lin, X.F.; Zheng, J.Z.; Guan, H.Q. Silencing lncRNA NEAT1 reduces nonalcoholic fatty liver fat deposition by regulating the miR-139-5p/c-Jun/SREBP-1c pathway. Ann. Hepatol. 2022, 27, 100584. [Google Scholar] [CrossRef]
- Bu, F.T.; Wang, A.; Zhu, Y.; You, H.M.; Zhang, Y.F.; Meng, X.M.; Huang, C.; Li, J. LncRNA NEAT1: Shedding light on mechanisms and opportunities in liver diseases. Liver Int. 2020, 40, 2612–2626. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Huang, F.Z.; Liu, H.Z.; Zhang, T.Y.; Yang, M.S.; Sun, C.Z. LncRNA MEG3 functions as a ceRNA in regulating hepatic lipogenesis by competitively binding to miR-21 with LRP6. Metabolism 2019, 94, 1–8. [Google Scholar] [CrossRef] [PubMed]
- McAllister-Lucas, L.M.; Ruland, J.; Siu, K.; Jin, X.; Gu, S.; Kim, D.S.; Kuffa, P.; Kohrt, D.; Mak, T.W.; Nunez, G.; et al. CARMA3/Bcl10/MALT1-dependent NF-κB activation mediates angiotensin II-responsive inflammatory signaling in nonimmune cells. Proc. Natl. Acad. Sci. USA 2007, 104, 139–144. [Google Scholar] [CrossRef]
- Lu, X.; Jiang, M.; Tian, J.; Liu, W.; Wu, F.; Yu, L.; Feng, G.; Zhong, S.; Xiang, Y.; Wen, H. Growth Arrest-Specific Transcript 5 (GAS5) Exerts Important Roles on the Treatment of BM45 Cells of Liver Cirrhosis. Mol. Ther. Nucleic Acids 2020, 22, 1154–1163. [Google Scholar] [CrossRef] [PubMed]
- Baranova, A.; Maltseva, D.; Tonevitsky, A. Adipose may actively delay progression of NAFLD by releasing tumor-suppressing, anti-fibrotic miR-122 into circulation. Obes. Rev. 2019, 20, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Munro, T.; Mattick, J.S. The potential of long noncoding RNA therapies. Trends Pharm. Sci. 2022, 43, 269–280. [Google Scholar] [CrossRef]
- DiStefano, J.K.; Gerhard, G.S. Long Noncoding RNAs and Human Liver Disease. Annu. Rev. Pathol. 2021, 17, 1–21. [Google Scholar] [CrossRef]

| NAFLD vs. Healthy Control | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| lncRNA | Species | Expression Direction | Sample Size (NAFLD/Healthy Control) | Fold-Change | p Value | Possible Mechanism | Sequencing | qRT-PCR | Study |
| NEAT1 | peripheral blood mononuclear cells | ⤤ | 119/100 | 2.4 | <0.05 | - | - | √ | Zhou, 2022 [27] |
| NEAT1 | serum | ⤤ | 10/10 | 5 | <0.001 | lncNEAT1/miR-212-5p/GRIA3 | - | √ | Hu, 2022 [24] |
| MEG3 | liver | ⤤ | 5/7 | 1.6 | <0.05 | managing obesity-associated hepatic endothelial senescence and insulin resistance | RNA sequencing | √ | Cheng, 2021 [28] |
| MALAT1 | liver | ⤤ | 20/10 | 1.73 | <0.05 | MALAT1/miR-206/ARNT | - | √ | Xiang, 2022 [29] |
| ENST00000413567 | liver | ⤦ | 5/5 | −4.22 (0.05) | <0.05 | - | Microarray | √ | Sun, 2015 [30] |
| NR_026836 | liver | ⟷ | 5/5 | >0.05 | - | Microarray | √ | Sun, 2015 [30] | |
| ENST00000548691 | liver | ⤤ | 5/5 | 4.74 (26) | <0.05 | - | Microarray | √ | Sun, 2015 [30] |
| ENST00000571619 | liver | ⟷ | 5/5 | >0.05 | - | Microarray | √ | Sun, 2015 [30] | |
| uc001gvp.1 | liver | ⤦ | 5/5 | −3.53 (0.09) | <0.05 | - | Microarray | √ | Sun, 2015 [30] |
| TCONS_00025733 | liver | ⤦ | 5/5 | −1.07 (0.48) | <0.05 | - | Microarray | √ | Sun, 2015 [30] |
| ENST00000491676 | liver | ⟷ | 5/5 | >0.05 | - | Microarray | √ | Sun, 2015 [30] | |
| APOA4-AS | liver | ⤤ | 8/5 | 8.3 | <0.05 | HuR–APOA4-AS complex stabilizes APOA4 mRNA | RNA sequencing | √ | Qin, 2016 [31] |
| uc.372 | liver | ⤤ | 11/11 | 2.06 | <0.001 | suppressing miR-195/miR4668 maturation | Microarray | √ | Guo, 2018 [20] |
| uc.94 | liver | ⤦ | 5/5 | 0.42 | <0.05 | - | Microarray | √ | Guo, 2018 [20] |
| uc.157 | liver | ⤤ | 5/5 | 1.32 | <0.05 | - | Microarray | √ | Guo, 2018 [20] |
| lncRNA B4GALT1-AS1(the human homologous sequence of mouse lncSHGL) | liver | ⤦ | 6/6 | 0.74 | <0.05 | lncSHGL/hnRNPA1/CaM/Akt | Microarray | √ | Wang, 2018 [32] |
| lncARSR | liver | ⤤ | 12/22 | 3.76 | <0.01 | regulating hepatic lipogenesis via Akt/SREBP-1c pathway | - | √ | Zhang, 2018 [26] |
| serum | ⤤ | 12/22 | 5.75 | <0.01 | - | - | √ | Zhang, 2018 [26] | |
| RP11-279F6.1 | liver | ⤤ | 2/2 | 2.27 | <0.01 | - | Microarray | √ | Wu, 2019 [33] |
| AC004540.4 | liver | ⤤ | 2/2 | 2 | <0.01 | - | Microarray | √ | Wu, 2019 [33] |
| uc.333 | liver | ⤦ | 8/8 | 0.13 | <0.01 | improving IR by binding to miR-223(FOXO1/AKT/GSK) | Microarray and lncRNA probe mapping | √ | Zhang, 2020 [21] |
| CCAT1 | liver | ⤤ | 17/10 | 2.58 | <0.05 | CCAT1/miR-613/LXRα | Microarray | √ | Huang, 2020 [22] |
| lncRNA RABGAP1LDT-206 | serum | ⤤ | 100/100 | <0.001 | - | Microarray | √ | Albadawy, 2021 [34] | |
| lnc-SPARCL1-1:2 | serum | ⤤ | 25/80 | 1.8 | <0.05 | lnc-SPARCL1-1:2/miR-6881-5P/HSPD1/MMP14/ITGB1 | - | √ | Albadawy, 2021 [23] |
| lnc PVT1 | serum | ⤤ | 81/78 | 1.46 | <0.001 | lncPVT1/miR-20a-5p | - | √ | Zhang, 2021 [25] |
| lnc HCG18 | serum | ⤤ | 116/101 | 1.89 | <0.001 | HCG18-miR-197-3p | - | √ | Xia, 2021 [35] |
| LINC00240 | liver | ⤤ | 3/3 | 5.25 | <0.01 | increasing the ROS content | GSE107231 dataset | √ | Xue, 2022 [36] |
| has_circ_0046367 | liver | ⤦ | 5/3 | 0.4 | <0.01 | circRNA_0046367/miR-34a/PPARα | - | √ | Guo, 2017 [37] |
| circRNA_0001805 | liver | ⤦ | 25/9 | 0.63 | <0.05 | circRNA_0001805 -miR-106a-5p/miR-320a-ABCA1/CPT1 | High-throughput sequencing | √ | Li, 2021 [38] |
| NAFL vs. Healthy Control | |||||||||
| lncRNA | Species | Expression Direction | Sample Size (NAFL/Healthy Control) | Fold-Change | p Value | Possible Mechanism | Sequencing | √ | Study |
| MEG3 | liver | ⟷ | 8/6 | - | >0.05 | - | - | √ | Zhang, 2017 [39] |
| MEG3 | liver | ⤦ | 15/10 | 0.6 | <0.05 | MEG3 up-regulates SIRT6 by ubiquitinating EZH2 | - | √ | Zou, 2022 [40] |
| MALAT1 | liver | ⟷ | 32/13 | - | 0.9 | - | Systems biology multiscale modeling | √ | Sookoian, 2018 [41] |
| LncPRYP4-3 | serum | ⤤ | 30/30 | 10 | <0.0001 | targeting RPS4Y2 | Microarray and lncRNA probe mapping | √ | Yang, 2020 [42] |
| LncPRYP4-4 | serum | ⤤ | 30/30 | 9.6 | <0.05 | - | Microarray and lncRNA probe mapping | √ | Yang, 2020 [42] |
| NASH vs. Healthy Control | |||||||||
| lncRNA | Species | Expression Direction | Sample Size (NASH/Healthy control) | Fold-Change | p Value | Possible Mechanism | Sequencing | √ | Study |
| MEG3 | liver | ⤤ | 7/7 | 1.9 | <0.05 | managing obesity-associated hepatic endothelial senescence and insulin resistance | RNA sequencing | √ | Cheng, 2021 [28] |
| RP11-128N14.5 | serum | ⤤ | 45/25 | 2.33 | ≤0.05 | - | Microarray and lncRNA probe mapping | √ | Di Mauro, 2019 [43] |
| LncPRYP4-3 | serum | ⤤ | 30/30 | 4.7 | <0.05 | targeting RPS4Y2 | Microarray and lncRNA probe mapping | √ | Yang, 2020 [42] |
| LncPRYP4-4 | serum | ⟷ | 30/30 | - | >0.05 | - | Microarray and lncRNA probe mapping | √ | Yang, 2020 [42] |
| lnc-SPARCL1-1:2 | serum | ⤦ | 55/80 | - | <0.01 | lnc-SPARCL1-1:2/miR-6881-5P/HSPD1/MMP14/ITGB1 | - | √ | Albadawy, 2021 [23] |
| lncRNA RABGAP1LDT-206 | serum | ⤤ | 60/100 | - | <0.001 | - | Microarray | √ | Albadawy, 2021 [34] |
| circRNA SCAR | liver mitochondrial | ⤦ | 18/20 | 0.16 | <0.001 | binding to ATP5B to inhibit mitochondrial ROS | Microarray | √ | Zhao, 2020 [44] |
| hsa_circ_0089763 | liver mitochondrial | ⤦ | 18/20 | 0.28 | <0.001 | - | Microarray | √ | Zhao, 2020 [44] |
| hsa_circ_0008882 | liver mitochondrial | ⤦ | 18/20 | 0.39 | <0.001 | - | Microarray | √ | Zhao, 2020 [44] |
| hsa_circ_0073378 | liver mitochondrial | ⤦ | 18/20 | 0.68 | <0.05 | - | Microarray | √ | Zhao, 2020 [44] |
| hsa_circ_0022430 | liver mitochondrial | ⤦ | 18/20 | 0.65 | <0.01 | - | Microarray | √ | Zhao, 2020 [44] |
| hsa_circ_0032777 | liver mitochondrial | ⤦ | 18/20 | 0.53 | <0.01 | - | Microarray | √ | Zhao, 2020 [44] |
| hsa_circ_0141729 | liver mitochondrial | ⤦ | 18/20 | 0.67 | <0.05 | - | Microarray | √ | Zhao, 2020 [44] |
| hsa_circ_0001311 | liver mitochondrial | ⤦ | 18/20 | 0.74 | <0.01 | - | Microarray | √ | Zhao, 2020 [44] |
| hsa_circ_0045989 | liver mitochondrial | ⤦ | 18/20 | 0.56 | <0.05 | - | Microarray | √ | Zhao, 2020 [44] |
| NASH vs. NAFL | |||||||||
| lncRNA | Species | Expression Direction | Sample Size (NASH/NAFL) | Fold-Change | p Value | Possible Mechanism | Sequencing | √ | Study |
| MEG3 | liver | ⤦ | 6/9 | 0.48 | <0.05 | MEG3 up-regulates SIRT6 by ubiquitinating EZH2 | - | √ | Zou, 2022 [40] |
| MALAT1 | liver | ⤤ | 15/32 | 1.75 | 0.029 | - | Systems biology multiscale modeling | √ | Sookoian, 2018 [41] |
| LncPRYP4-3 | serum | ⤦ | 30/30 | 0.47 | <0.05 | targeting RPS4Y2 | Microarray and lncRNA probe mapping | √ | Yang, 2020 [42] |
| LncPRYP4-4 | serum | ⟷ | 30/30 | - | >0.05 | - | Microarray and lncRNA probe mapping | √ | Yang, 2020 [42] |
| LeXis | plasma | ⤤ | 35/9 | 1.75 | 0.025 | - | - | √ | Park, 2020 [45] |
| liver | ⟷ | 35/9 | 0.539 | - | - | √ | Park, 2020 [45] | ||
| lnc-SPARCL1-1:2 | serum | ⤤ | 55/11 | 1.13 | <0.01 | lnc-SPARCL1-1:2/miR-6881-5P/HSPD1/MMP14/ITGB1 | - | √ | Albadawy, 2021 [23] |
| Steatosis | ||||||
|---|---|---|---|---|---|---|
| lncRNA | Species | Sample Size | Expression | Fold-Change | p Value | Study |
| GAS5 | liver | 30/21 | ⟷ | - | 0.602 | Han, 2020 [49] |
| plasma | 30/21 | ⟷ | - | 0.274 | Han, 2020 [49] | |
| LeXis | liver | 35/9 | ⤦ | 0.49 | 0.017 | Park, 2020 [45] |
| LeXis | plasma | 35/9 | ⟷ | - | 0.399 | Park, 2020 [45] |
| Lobular Inflammation | ||||||
| lncRNA | Species | Sample Size (with/without) | Expression | Fold-Change | p Value | Study |
| MALAT1 | liver | 34/15 | ⤤ | 1.91 | 0.0025 | Sookoian, 2018 [41] |
| MALAT1 | liver | 53/24 | ⟷ | - | >0.05 | Leti, 2017 [47] |
| NEAT1 | liver | 53/24 | ⤤ | 1.33 | 1.0 × 10−4 | Leti, 2017 [47] |
| HULC | liver | 53/24 | ⤤ | 2.3 | 2.53 × 10−5 | Leti, 2017 [47] |
| lncTNF | liver | 35/9 | ⤤ | R = 0.58 | 9.7 × 10−7 | Atanasovska, 2021 [48] |
| LeXis | liver | 35/9 | ⟷ | - | 0.914 | Park, 2020 [45] |
| LeXis | plasma | 35/9 | ⟷ | - | 0.404 | Park, 2020 [45] |
| lnc18q22.2 | liver | 17/8 | ⤤ | R = 0.62 | 1.38 × 10−4 | Atanasovska, 2017 [46] |
| GAS5 | liver | 29/32 | ⟷ | - | 0.602 | Han, 2020 [49] |
| plasma | 29/32 | ⟷ | - | 0.274 | Han, 2020 [49] | |
| Ballooning Degeneration | ||||||
| lncRNA | Species | Sample Size (with/without) | Expression | Fold-Change | p Value | Study |
| MALAT1 | liver | 34/15 | ⤤ | 3.01 | 0.0001 | Sookoian, 2018 [41] |
| NAS Score | ||||||
| lncRNA | Species | Sample Size | Expression | Fold-Change | p Value | Study |
| NAS > 5 vs. HC | ||||||
| RP11-128N14.5 | serum | 25/25 | ⤤ | 2.69 | <0.02 | Di Mauro, 2019 [43] |
| TGFB2/TGFB2-OT1 | serum | 25/25 | ⤤ | 26.1 | <0.05 | Di Mauro, 2019 [43] |
| NAS>5 vs. NAS<=4 | ||||||
| RP11-128N14.5(internal) | serum | 25/38 | ⤤ | 1.25 | <0.05 | Di Mauro, 2019 [43] |
| TGFB2/TGFB2-OT1(internal) | serum | 25/38 | ⤤ | 1.58 | <0.05 | Di Mauro, 2019 [43] |
| RP11-128N14.5(external) | serum | 27/23 | ⤤ | 4.3 | 0.04 | Di Mauro, 2019 [43] |
| TGFB2/TGFB2-OT1(external) | serum | 27/23 | ⤤ | 6.2 | 0.03 | Di Mauro, 2019 [43] |
| LeXis | liver | 35/9 | ⟷ | - | 0.872 | Park, 2020 [45] |
| plasma | 35/9 | ⟷ | - | 0.363 | Park, 2020 [45] | |
| lnc18q22.2 | liver | 17/8 | ⤤ | R = 0.58 | 8.64 × 10−4 | Atanasovska, 2017 [46] |
| GAS5 | liver | 24/27 | ⟷ | - | 0.674 | Han, 2020 [49] |
| plasma | 24/27 | ⟷ | - | 0.448 | Han, 2020 [49] | |
| NASH Grade | ||||||
| lncRNA | Species | Sample Size (with/without) | Expression | Fold-Change | p Value | Study |
| lnc18q22.2 | liver | 17/8 | ⤤ | 0.65 | 4.55 × 10−4 | Atanasovska, 2017 [46] |
| Fibrosis | lncRNA | Species | Sample Size | Expression | Fold-Change | p Value | Study |
|---|---|---|---|---|---|---|---|
| F0 vs. F1–4 | MALAT1 | liver | 13 HC/47 NAFLD | ⤤ | 5 | 1 × 10−7 | Sookoian, 2018 [41] |
| MEG3 | liver | 6 HC/6 fibrosis | ⤤ | 2.2 | <0.01 | Zhang, 2017 [39] | |
| MEG3 | liver | 10HC/15 fibrosis | ⤦ | 0.3 | <0.01 | Zou, 2022 [40] | |
| F0–2 vs. F3 | GAS5 | liver | 39 F0-2/6 F3 | ⟷ | - | 0.131 | Han, 2020 [49] |
| GAS5 | plasma | 39 F0-2/6 F3 | ⤤ | 2.02 | <0.001 | Han, 2020 [49] | |
| F0–2 vs. F3–4 | TGFB2/TGFB2-OT1 | serum | 37 F0-2/26 F3-4 | ⤤ | 1.82 | ≤0.001 | Di Mauro, 2019 [43] |
| RP11-128N14.5 | serum | 37 F0-2/26 F3-4 | ⟷ | - | >0.05 | Di Mauro, 2019 [43] | |
| F0–1 vs. F4 | lnc-SPARCL1-1:2 | serum | 25 F0-1/10 F4 | ⤤ | - | <0.05 | Albadawy, 2021 [23] |
| lncRNA RABGAP1LDT-206 | serum | 34 F3/11 F4 | ⤤ | - | <0.05 | Albadawy, 2021 [34] | |
| F2 vs. F4 | lnc-SPARCL1-1:2 | serum | 20 F2/10 F4 | ⤤ | - | <0.05 | Albadawy, 2021 [23] |
| lncRNA RABGAP1LDT-206 | serum | 26 F2/11 F4 | ⤤ | - | <0.05 | Albadawy, 2021 [34] | |
| F3 vs. F4 | GAS5 | liver | 6 F3/6 F4 | ⟷ | - | 0.818 | Han, 2020 [49] |
| GAS5 | plasma | 6 F3/6 F4 | ⤦ | 0.54 | 0.026 | Han, 2020 [49] | |
| lnc-SPARCL1-1:2 | serum | 24 F3/10 F4 | ⤤ | - | <0.05 | Albadawy, 2021 [23] | |
| lncRNA RABGAP1LDT-206 | serum | 29 F3/11 F4 | ⤤ | - | <0.05 | Albadawy, 2021 [34] | |
| (advanced fibrosis) vs. (not advanced fibrosis) | LeXis | serum | 33/11 | ⟷ | - | 0.328 | Park, 2020 [45] |
| liver | 33/11 | ⟷ | - | 0.14 | Park, 2020 [45] | ||
| NEAT1 | liver | 24/53 | ⤤ | 1.29 | 3 × 10−4 | Leti, 2017 [47] | |
| HULC | liver | 24/53 | ⤤ | 3.6 | 1.08 × 10−8 | Leti, 2017 [47] | |
| MALAT1 | liver | 24/53 | ⤤ | 3.6 | 8.02 × 10−6 | Leti, 2017 [47] | |
| LINC01638 | liver | 10/10 | ⤤ | 2.81 | ≤0.001 | Gerhard, 2020 [51] | |
| LINC01605 | liver | 10/10 | ⟷ | - | >0.05 | Gerhard, 2020 [51] | |
| XLOC_003146 | liver | 10/10 | ⤤ | 1.65 | ≤0.0001 | Gerhard, 2020 [51] | |
| RP11_20J153 | liver | 10/10 | ⤤ | 1.34 | ≤0.0001 | Gerhard, 2020 [51] | |
| HC vs. cirrhosis | GAS5 | liver | 15/20 | ⤦ | 0.43 | <0.01 | Yu, 2015 [50] |
| MEG3 | liver | 6/8 | ⤤ | 2.2 | <0.01 | Zhang, 2017 [39] | |
| H19 | liver | 6/17 | ⤤ | 3.67 | <0.05 | Liu, 2018 [52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Q.; Liu, C.-H.; Wu, D.; Jiang, W.; Zhang, N.; Tang, H. LncRNA and circRNA in Patients with Non-Alcoholic Fatty Liver Disease: A Systematic Review. Biomolecules 2023, 13, 560. https://doi.org/10.3390/biom13030560
Zeng Q, Liu C-H, Wu D, Jiang W, Zhang N, Tang H. LncRNA and circRNA in Patients with Non-Alcoholic Fatty Liver Disease: A Systematic Review. Biomolecules. 2023; 13(3):560. https://doi.org/10.3390/biom13030560
Chicago/Turabian StyleZeng, Qingmin, Chang-Hai Liu, Dongbo Wu, Wei Jiang, Nannan Zhang, and Hong Tang. 2023. "LncRNA and circRNA in Patients with Non-Alcoholic Fatty Liver Disease: A Systematic Review" Biomolecules 13, no. 3: 560. https://doi.org/10.3390/biom13030560
APA StyleZeng, Q., Liu, C.-H., Wu, D., Jiang, W., Zhang, N., & Tang, H. (2023). LncRNA and circRNA in Patients with Non-Alcoholic Fatty Liver Disease: A Systematic Review. Biomolecules, 13(3), 560. https://doi.org/10.3390/biom13030560





