Pairwise Engineering of Tandemly Aligned Self-Splicing Group I Introns for Analysis and Control of Their Alternative Splicing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. β-Galactosidase Assay
2.3. Reverse Transcription-PCR (RT-PCR)
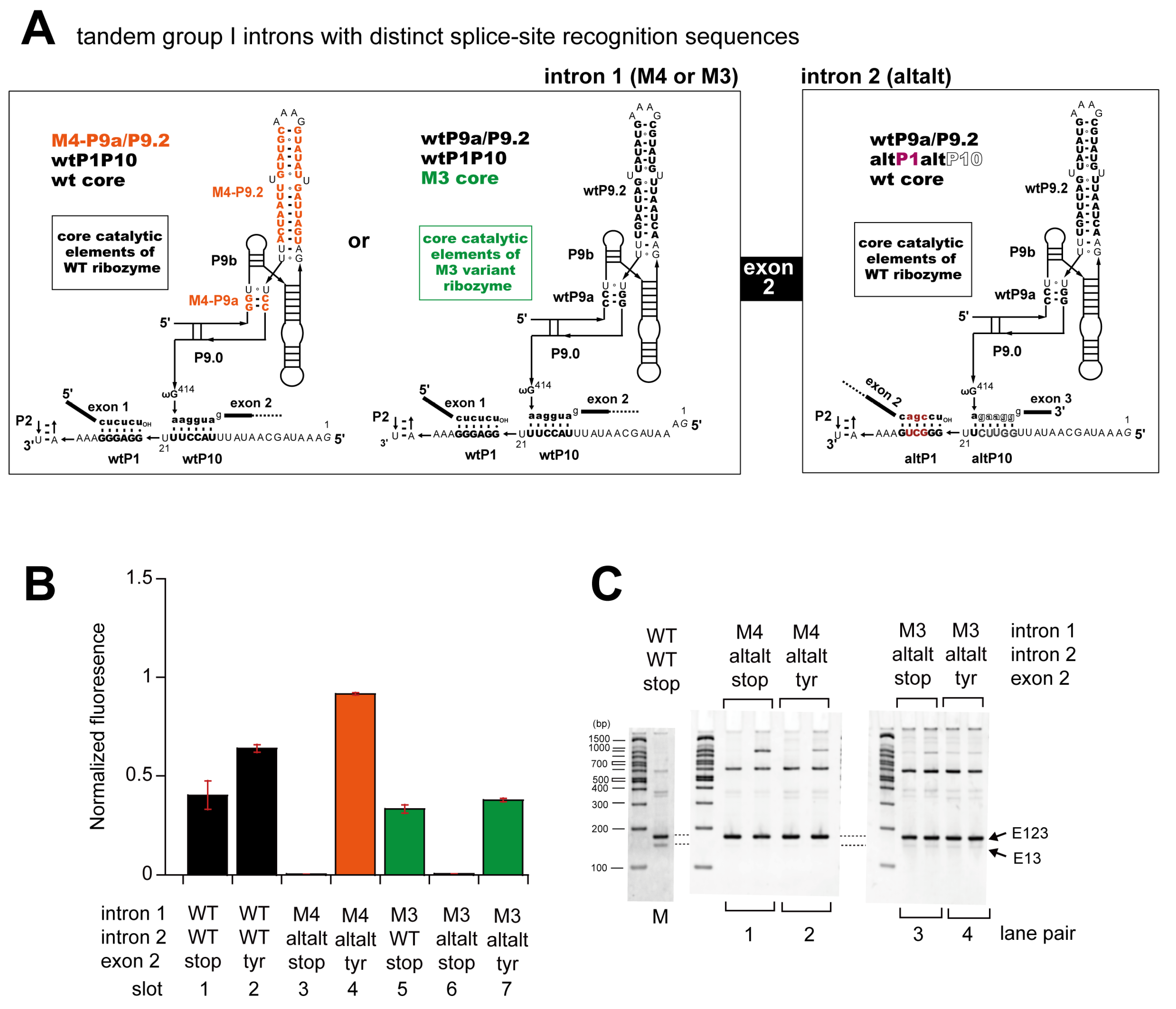
3. Results
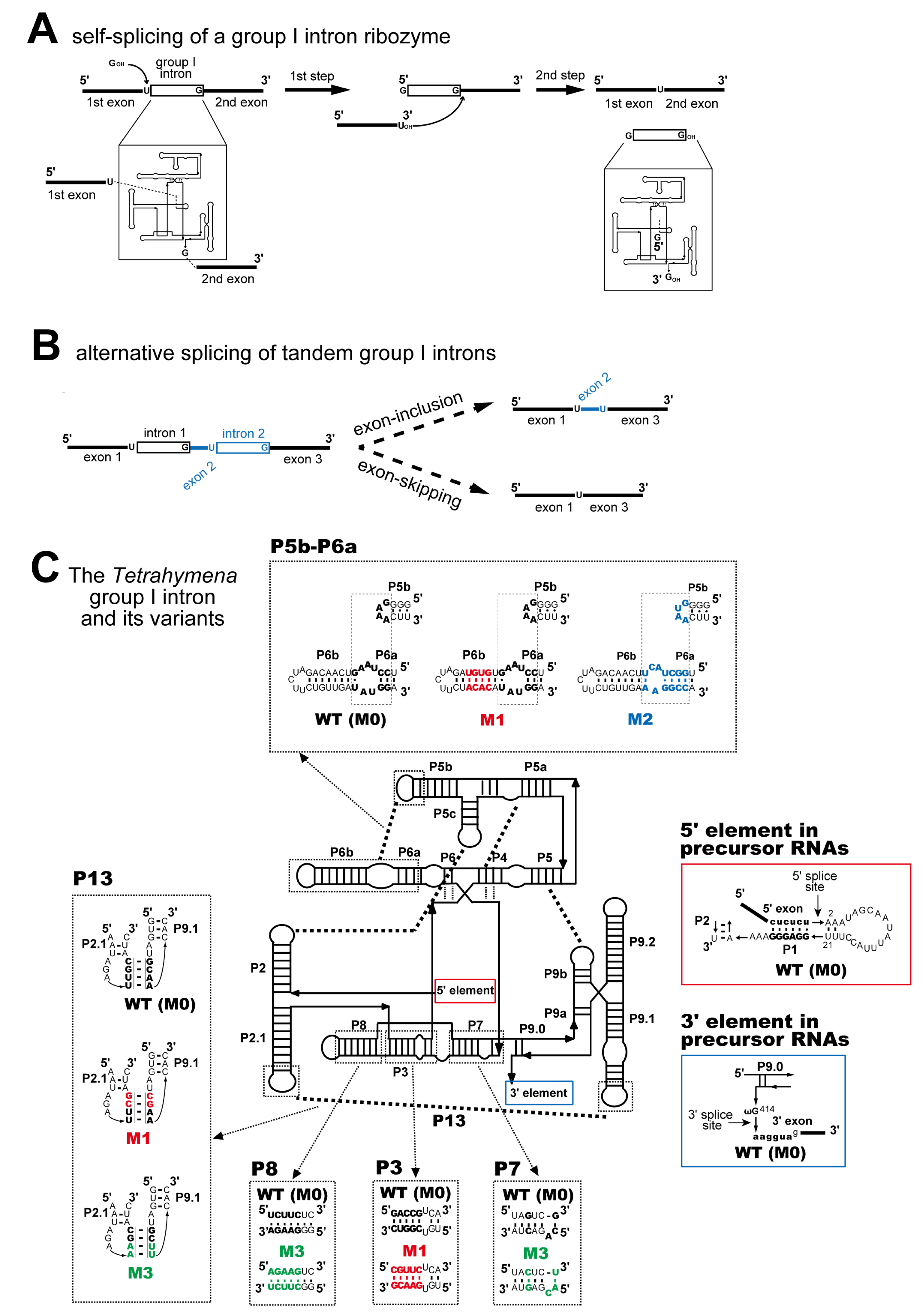
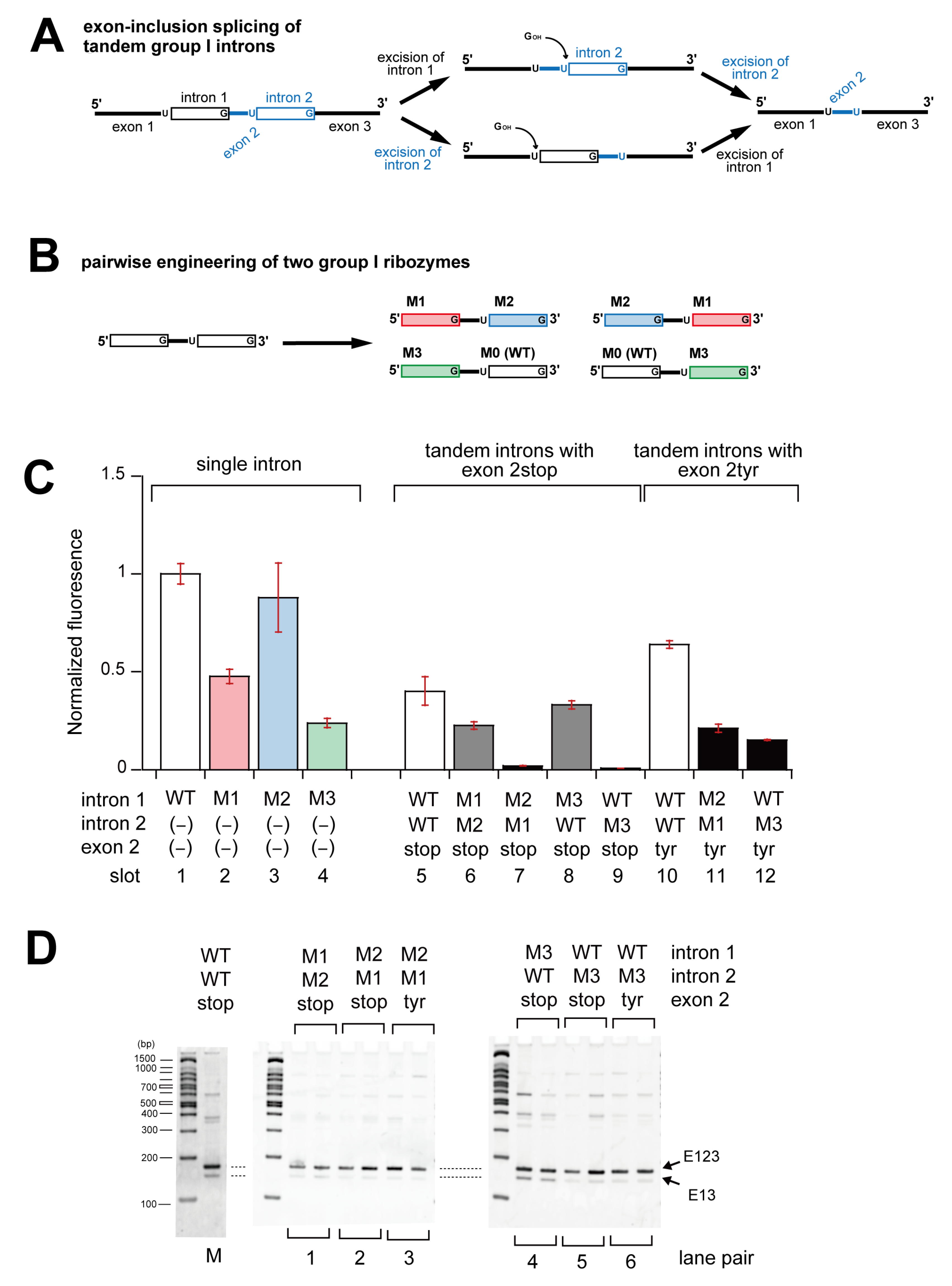
3.1. In Vivo Splicing of Precursor RNAs Containing Tetrahymena Introns in Tandem
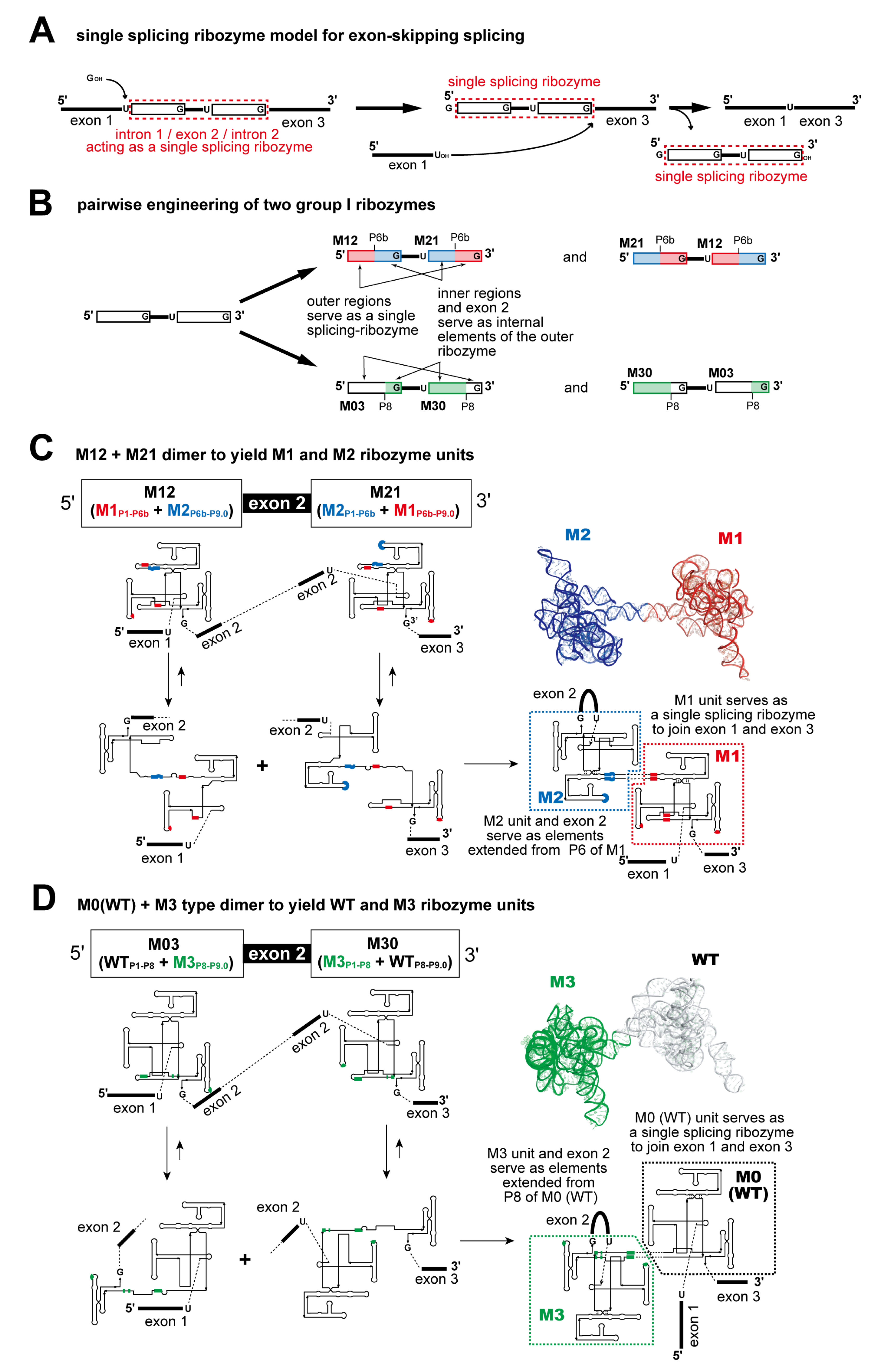
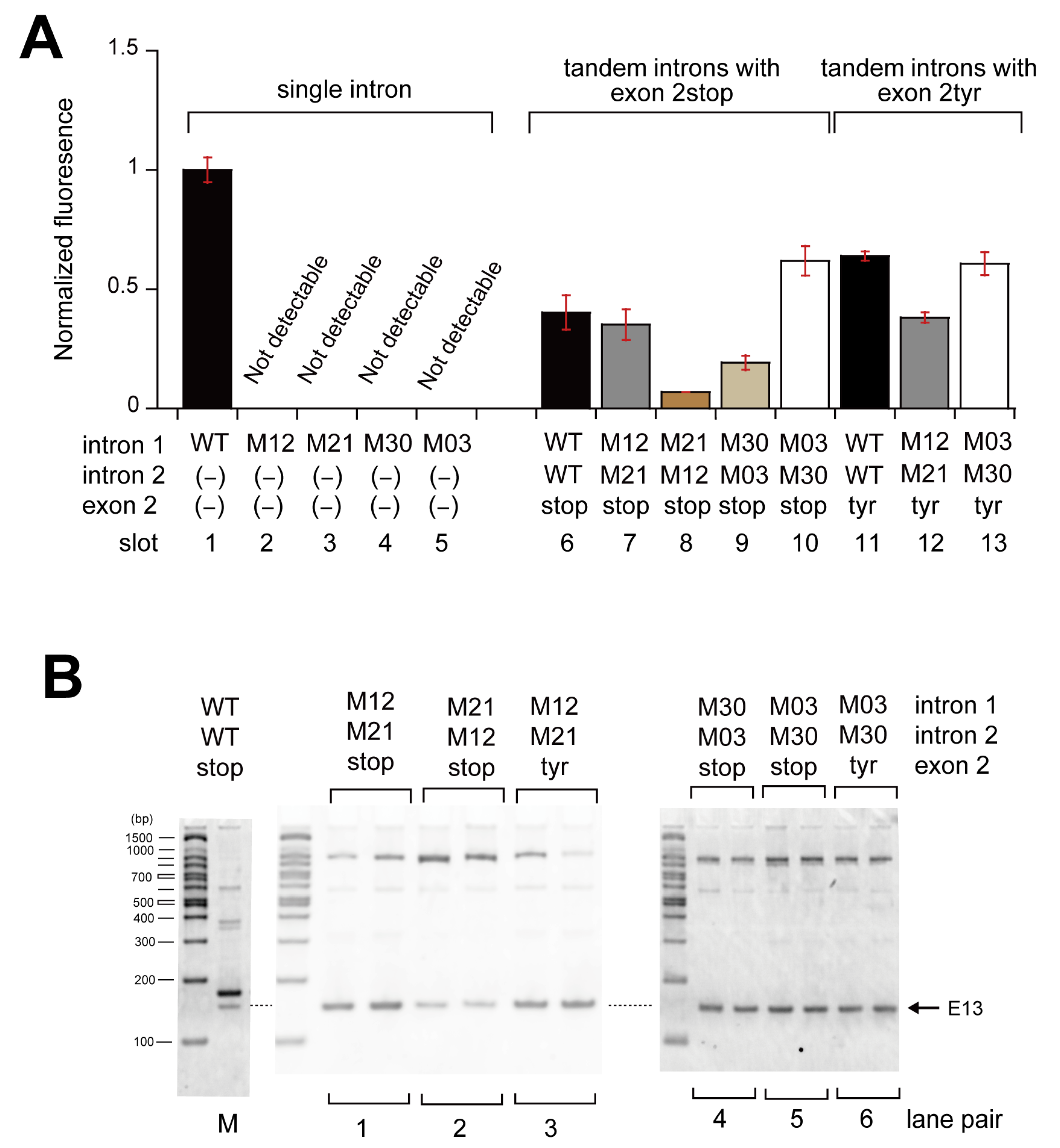
3.2. Pairwise Engineering of Tandem Introns to Suppress Exon-Inclusion Splicing
3.3. Pairwise Engineering of a Tandem Intron Ribozyme to Suppress Exon-Skipping Splicing
3.4. Engineering of Splice Site Recognition Elements as an Alternative Strategy to Suppress Exon-Skipping Splicing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elliott, D.; Ladomery, M. Pre-mRNA splicing by the spliceosome. In Molecular Biology of RNA, 2nd ed.; Elliott, D., Ladomery, M., Eds.; Oxford University Press: Oxford, UK, 2015; pp. 84–110. [Google Scholar]
- Wan, R.; Bai, R.; Zhan, X.; Shi, Y. How is precursor messenger RNA spliced by the spliceosome? Annu. Rev. Biochem. 2020, 89, 333–358. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.E.; Charenton, C.; Nagai, K. RNA splicing by the spliceosome. Annu. Rev. Biochem. 2020, 89, 359–388. [Google Scholar] [CrossRef]
- Saldanha, R.; Mohr, G.; Belfort, M.; Lambowitz, A.M. Group I and group II introns. FASEB J. 1993, 7, 15–24. [Google Scholar] [CrossRef]
- Woodson, S.A. Structure and assembly of group I introns. Curr. Opin. Struct. Biol. 2005, 15, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Vicens, Q.; Cech, T.R. Atomic level architecture of group I introns revealed. Trends Biochem. Sci. 2006, 31, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Pyle, A.M. Group II intron self-splicing. Annu. Rev. Biophys. 2016, 45, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Galej, W.P.; Toor, N.; Newman, A.J.; Nagai, K. Molecular mechanism and evolution of nuclear pre-mRNA and group II Intron splicing: Insights from cryo-electron microscopy structures. Chem. Rev. 2018, 118, 4156–4176. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Cech, T.R. Self-splicing and enzymatic activity of an intervening sequence RNA from Tetrahymena (Nobel Lecture). Angew. Chem. Int. Ed. 1990, 29, 759–768. [Google Scholar] [CrossRef]
- Das, R.; Russell, R. How to kinetically dissect an RNA machine. Biochemistry 2021, 60, 3485–3490. [Google Scholar] [CrossRef]
- Su, Z.; Zhang, K.; Kappel, K.; Li, S.; Palo, M.Z.; Pintilie, G.D.; Rangan, R.; Luo, B.; Wei, Y.; Das, R.; et al. Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 Å resolution. Nature 2021, 596, 603–607. [Google Scholar] [CrossRef]
- Breitbart, R.E.; Andreadis, A.; Nadal-Ginard, B. Alternative splicing: A ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu. Rev. Biochem. 1987, 56, 467–495. [Google Scholar] [CrossRef] [PubMed]
- Elliott, D.; Ladomery, M. Regulated alternative splicing. In Molecular Biology of RNA, 2nd ed.; Elliott, D., Ladomery, M., Eds.; Oxford University Press: Oxford, UK, 2015; pp. 111–137. [Google Scholar]
- Robart, A.R.; Montgomery, N.K.; Smith, K.L.; Zimmerly, S. Principles of 3’ splice site selection and alternative splicing for an unusual group II intron from Bacillus anthracis. RNA 2004, 10, 854–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toor, N.; Robart, A.R.; Christianson, J.; Zimmerly, S. Self-splicing of a group IIC intron: 5′ exon recognition and alternative 5′ splicing events implicate the stem-loop motif of a transcriptional terminator. Nucleic Acids Res. 2006, 34, 6461–6471. [Google Scholar] [CrossRef] [PubMed]
- Chee, G.J.; Takami, H. Alternative splicing by participation of the group II intron ORF in extremely halotolerant and alkaliphilic Oceanobacillus iheyensis. Microbes Environ. 2011, 26, 54–60. [Google Scholar] [CrossRef] [Green Version]
- McNeil, B.A.; Simon, D.M.; Zimmerly, S. Alternative splicing of a group II intron in a surface layer protein gene in Clostridium tetani. Nucleic Acids Res. 2014, 42, 1959–1969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cummings, D.J.; Michel, F.; Domenico, J.M.; McNally, K.L. Mitochondrial DNA sequence analysis of the cytochrome oxidase subunit II gene from Podospora anserina. A group IA intron with a putative alternative splice site. J. Mol. Biol. 1990, 212, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Ruoff, B.; Johansen, S.; Vogt, V.M. Characterization of the self-splicing products of a mobile intron from the nuclear rDNA of Physarum polycephalum. Nucleic Acids Res. 1992, 20, 5899–5906. [Google Scholar] [CrossRef] [Green Version]
- Michel, F.; Jaeger, L.; Westhof, E.; Kuras, R.; Tihy, F.; Xu, M.Q.; Shub, D.A. Activation of the catalytic core of a group I intron by a remote 3′ splice junction. Genes Dev. 1992, 6, 1373–1385. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, L.; Westhof, E.; Michel, F. Function of a pseudoknot in the suppression of an alternative splicing event in a group I intron. Biochimie 1996, 78, 466–473. [Google Scholar] [CrossRef]
- Wallweber, G.J.; Mohr, S.; Rennard, R.; Caprara, M.G.; Lambowitz, A.M. Characterization of Neurospora mitochondrial group I introns reveals different CYT-18 dependent and independent splicing strategies and an alternative 3′ splice site for an intron ORF. RNA 1997, 3, 114–131. [Google Scholar] [PubMed]
- Pichler, A.; Schroeder, R. Folding problems of the 5’ splice site containing the P1 stem of the group I thymidylate synthase intron: Substrate binding inhibition in vitro and mis-splicing in vivo. J. Biol. Chem. 2002, 277, 17987–17993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landthaler, M.; Shub, D.A. Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: Introns in multiple genes, a single gene with three introns, and exon-skipping by group I ribozymes. Proc. Natl. Acad. Sci. USA 1999, 96, 7005–7010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hicks, L.D.; Warrier, I.; Raghavan, R.; Minnick, M.F. Ribozyme stability, exon-skipping, and a potential role for RNA helicase in group I intron splicing by Coxiella burnetii. J. Bacteriol. 2011, 193, 5292–5299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Matsumura, S.; Furuta, H.; Ikawa, Y. Tecto-GIRz: Engineered group I ribozyme the catalytic ability of which can be controlled by self-dimerization. ChemBioChem 2016, 17, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Hirata, Y.; Tominaga, Y.; Furuta, H.; Matsumura, S.; Ikawa, Y. Heterodimerization of group I ribozymes enabling exon recombination through pairs of cooperative trans-splicing reactions. ChemBioChem 2017, 18, 1659–1667. [Google Scholar] [CrossRef]
- Ikawa, Y.; Matsumura, S. Engineered group I ribozymes as RNA-based modular tools to control gene expression. In Applied RNA Bioscience; Masuda, S., Izawa, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 203–220. [Google Scholar]
- Williamson, C.L.; Desai, N.M.; Burke, J.M. Compensatory mutations demonstrate that P8 and P6 are RNA secondary structure elements important for processing of a group I intron. Nucleic Acids Res. 1989, 17, 675–689. [Google Scholar] [CrossRef] [Green Version]
- Urano, Y.; Kamiya, M.; Kanda, K.; Ueno, T.; Hirose, K.; Nagano, T. Evolution of fluorescein as a platform for finely tunable fluorescence probes. J. Am. Chem. Soc. 2005, 127, 4888–4894. [Google Scholar] [CrossRef]
- Inuzuka, S.; Kakizawa, H.; Nishimura, K.; Naito, T.; Miyazaki, K.; Furuta, H.; Matsumura, S.; Ikawa, Y. Recognition of cyclic-di-GMP by a riboswitch conducts translational repression through masking the ribosome-binding site distant from the aptamer domain. Genes Cells 2018, 23, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Joyce, G.F.; van der Horst, G.; Inoue, T. Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic Acids Res. 1989, 17, 7879–7889. [Google Scholar] [CrossRef] [Green Version]
- Inoue, K.; Ohno, M.; Shimura, Y. Aspects of splice site selection in constitutive and alternative pre-mRNA splicing. Gene Expr. J. Liver Res. 1995, 4, 177–182. [Google Scholar]
- Dvinge, H. Regulation of alternative mRNA splicing: Old players and new perspectives. FEBS Lett. 2018, 592, 2987–3006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.R.; Baker, J.L.; Weinberg, Z.; Sudarsan, N.; Breaker, R.R. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 2010, 329, 845–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, K.M.; Syrett, H.A.; Knudsen, S.M.; Ellington, A.D. Group I aptazymes as genetic regulatory switches. BMC Biotechnol. 2002, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, S.; Ikawa, Y.; Shiraishi, H.; Inoue, T. Design and development of a catalytic ribonucleoprotein. EMBO J. 2001, 20, 5453–5460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikawa, Y.; Tsuda, K.; Matsumura, S.; Atsumi, S.; Inoue, T. Putative intermediary stages for the molecular evolution from a ribozyme to a catalytic RNP. Nucleic Acids Res. 2003, 31, 1488–1496. [Google Scholar] [CrossRef] [Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ueda, T.; Nishimura, K.-i.; Nishiyama, Y.; Tominaga, Y.; Miyazaki, K.; Furuta, H.; Matsumura, S.; Ikawa, Y. Pairwise Engineering of Tandemly Aligned Self-Splicing Group I Introns for Analysis and Control of Their Alternative Splicing. Biomolecules 2023, 13, 654. https://doi.org/10.3390/biom13040654
Ueda T, Nishimura K-i, Nishiyama Y, Tominaga Y, Miyazaki K, Furuta H, Matsumura S, Ikawa Y. Pairwise Engineering of Tandemly Aligned Self-Splicing Group I Introns for Analysis and Control of Their Alternative Splicing. Biomolecules. 2023; 13(4):654. https://doi.org/10.3390/biom13040654
Chicago/Turabian StyleUeda, Tomoki, Kei-ichiro Nishimura, Yuka Nishiyama, Yuto Tominaga, Katsushi Miyazaki, Hiroyuki Furuta, Shigeyoshi Matsumura, and Yoshiya Ikawa. 2023. "Pairwise Engineering of Tandemly Aligned Self-Splicing Group I Introns for Analysis and Control of Their Alternative Splicing" Biomolecules 13, no. 4: 654. https://doi.org/10.3390/biom13040654
APA StyleUeda, T., Nishimura, K.-i., Nishiyama, Y., Tominaga, Y., Miyazaki, K., Furuta, H., Matsumura, S., & Ikawa, Y. (2023). Pairwise Engineering of Tandemly Aligned Self-Splicing Group I Introns for Analysis and Control of Their Alternative Splicing. Biomolecules, 13(4), 654. https://doi.org/10.3390/biom13040654





