Enhanced Biosafety of the Sleeping Beauty Transposon System by Using mRNA as Source of Transposase to Efficiently and Stably Transfect Retinal Pigment Epithelial Cells
Abstract
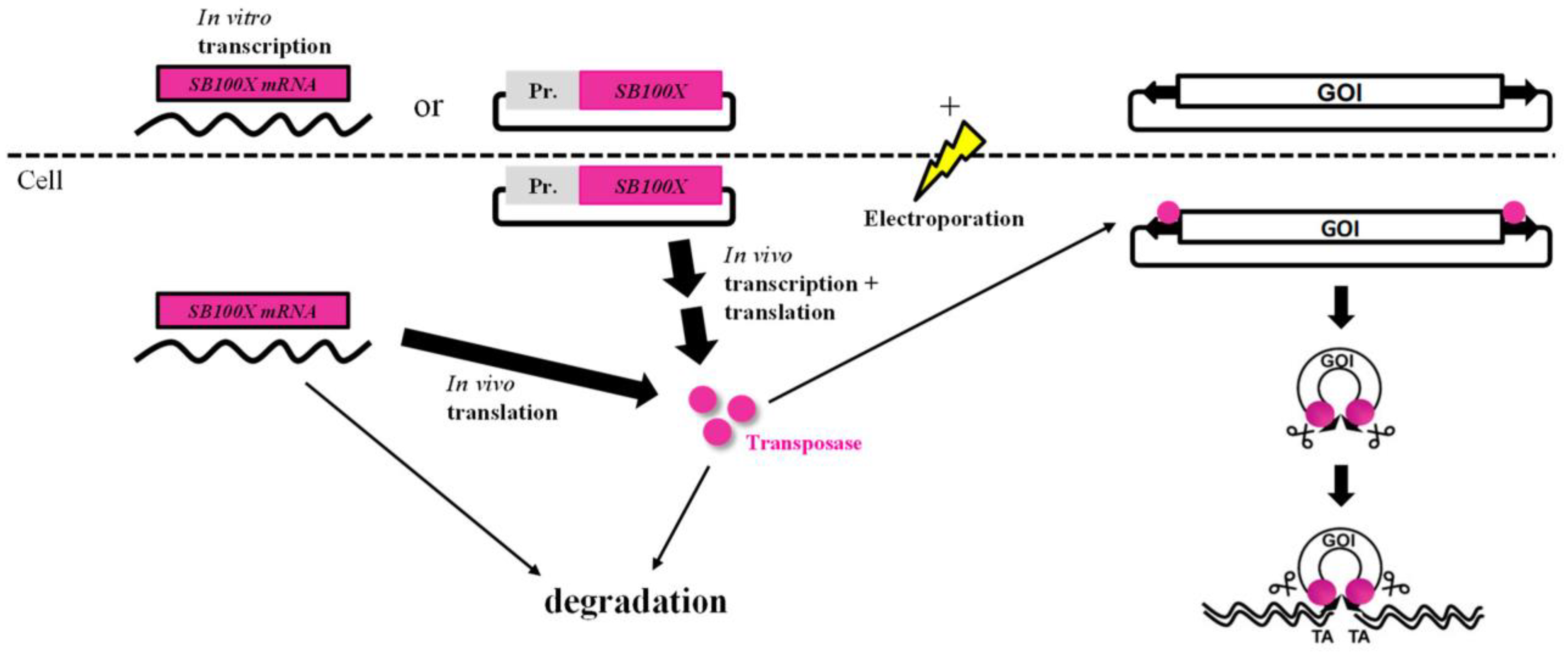
:1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. In Vitro mRNA Transcription
2.3. Cultivation of ARPE-19 Cells
2.4. Isolation and Cultivation of Primary Human RPE Cells
2.5. Electroporation and Cultivation of RPE Cells
2.6. Fluorescence Imaging
2.7. Flow Cytometric Analysis
2.8. Protein Purification, SDS-PAGE, and Western Blot Analysis
2.9. Polymerase Chain Reaction (PCR) and Real-Time Quantitative PCR
2.10. Statistical Analysis
3. Results
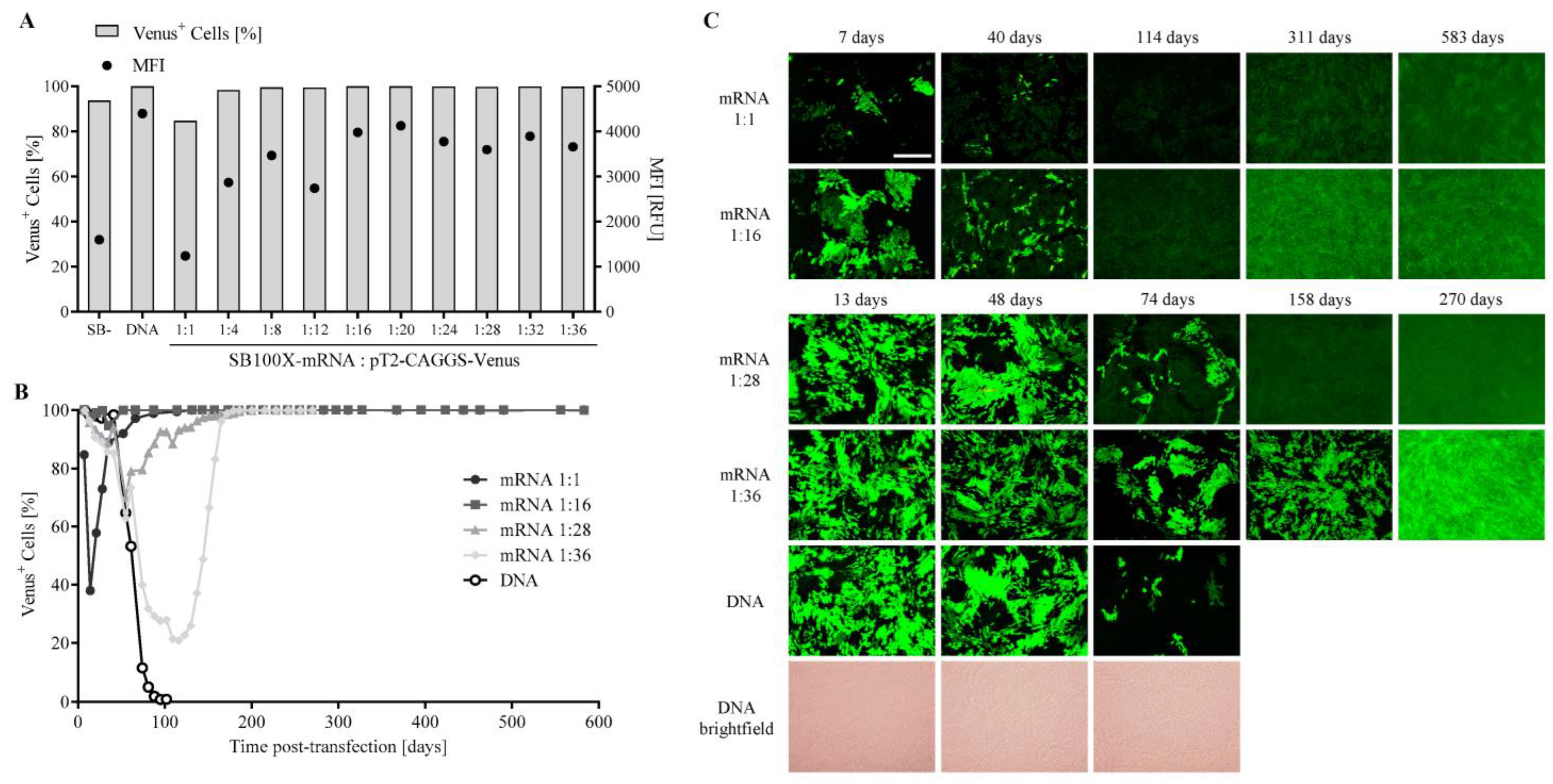
3.1. Transfection Efficiency and Long-Term Venus Expression in ARPE-19 Cells Using the SB100X Transposase as mRNA
3.2. Comparison of SB100X-DNA and -mRNA for the Transfection of ARPE-19 Cells and Analysis of SB100X Expression
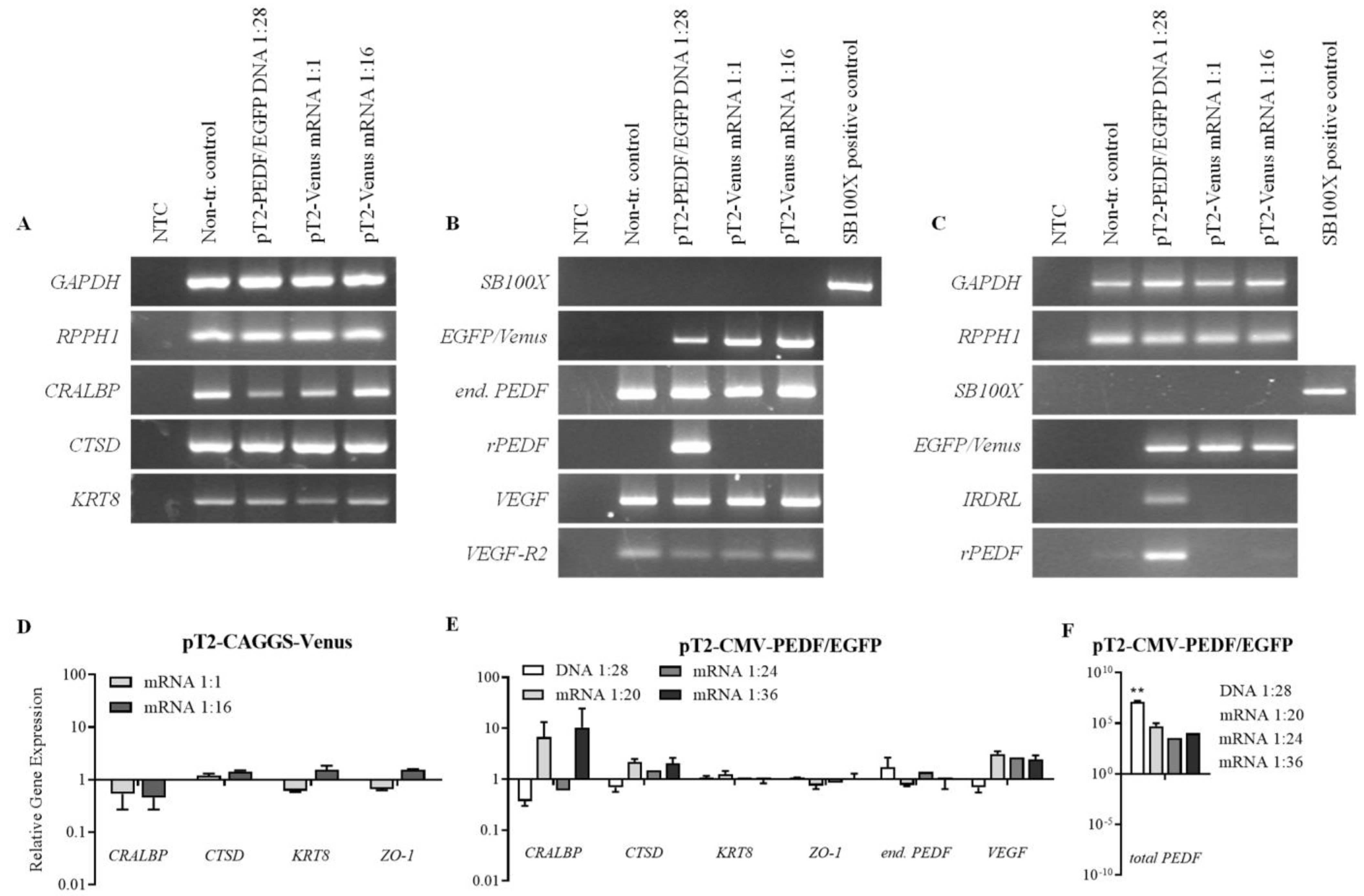
3.3. Transfection Efficiency and Long-Term PEDF Secretion in ARPE-19 Cells Using the SB100X Transposase as mRNA
3.4. Gene Expression Analysis of Transfected ARPE-19 Cells
3.5. Transfection of Primary Human RPE Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cai, J.; Jiang, W.G.; Grant, M.B.; Boulton, M. Pigment epithelium-derived factor inhibits angiogenesis via regulated intracellular proteolysis of vascular endothelial growth factor receptor 1. J. Biol. Chem. 2006, 281, 3604–3613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duh, E.J.; Yang, H.S.; Suzuma, I.; Miyagi, M.; Youngman, E.; Mori, K.; Katai, M.; Yan, L.; Suzuma, K.; West, K.; et al. Pigment epithelium-derived factor suppresses ischemia-induced retinal neovascularization and VEGF-induced migration and growth. Investig. Ophthalmol. Vis. Sci. 2002, 43, 821–829. [Google Scholar] [PubMed]
- Falk, T.; Gonzalez, R.; Sherman, S.J. The yin and yang of VEGF and PEDF: Multifaceted neurotrophic factors and their potential in the treatment of Parkinson’s disease. Int. J. Mol. Sci. 2010, 11, 2875–2900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno-Matsui, K.; Morita, I.; Tombran-Tink, J.; Mrazek, D.; Onodera, M.; Uetama, T.; Hayano, M.; Murota, S.I.; Mochizuki, M. Novel mechanism for age-related macular degeneration: An equilibrium shift between the angiogenesis factors VEGF and PEDF. J. Cell. Physiol. 2001, 189, 323–333. [Google Scholar] [CrossRef]
- Koponen, S.; Kokki, E.; Kinnunen, K.; Ylä-Herttuala, S. Viral-Vector-Delivered Anti-Angiogenic Therapies to the Eye. Pharmaceutics 2021, 13, 219. [Google Scholar] [CrossRef]
- Holekamp, N.M.; Bouck, N.; Volpert, O. Pigment epithelium-derived factor is deficient in the vitreous of patients with choroidal neovascularization due to age-related macular degeneration. Am. J. Ophthalmol. 2002, 134, 220–227. [Google Scholar] [CrossRef]
- Hubschman, J.P.; Reddy, S.; Schwartz, S.D. Age-related macular degeneration: Current treatments. Clin. Ophthalmol. 2009, 3, 155–166. [Google Scholar] [CrossRef] [Green Version]
- Andreoli, C.M.; Miller, J.W. Anti-vascular endothelial growth factor therapy for ocular neovascular disease. Curr. Opin. Ophthalmol. 2007, 18, 502–508. [Google Scholar] [CrossRef]
- Lalwani, G.A.; Rosenfeld, P.L.; Fung, A.E.; Dubovy, S.R.; Michels, S.; Feuer, W.; Davis, J.L.; Flynn, H.W.; Esquiabro, M. A variable-dosing regimen with intravitreal ranibizumab for neovascular age-related macular degeneration: Year 2 of the PrONTO study. Am. J. Ophthalmol. 2009, 148, 43–58. [Google Scholar] [CrossRef]
- Fernández-Robredo, P.; Sancho, A.; Johnen, S.; Recalde, S.; Gama, N.; Thumann, G.; Groll, J.; García-Layana, A. Current treatment limitations in age-related macular degeneration and future approaches based cell therapy and tissue engineering. J. Ophthalmol. 2014, 2014, 510285. [Google Scholar] [CrossRef] [Green Version]
- Bashshur, Z.F.; Haddad, Z.A.; Schakal, A.; Jaafar, R.F.; Saab, M.; Noureddin, B.N. Intravitreal bevacizumab for treatment of neovascular age-related macular degeneration: A one-year prospective study. Am. J. Ophthalmol. 2008, 145, 249–256. [Google Scholar] [CrossRef]
- Dugel, P.U.; Koh, A.; Ogura, Y.; Jaffe, G.J.; Schmidt-Erfurth, U.; Brown, D.M.; Gomes, A.V.; Warburton, J.; Weichselberger, A.; Holz, F.G. HAWK and HARRIER: Phase 3, Multicenter, Randomized, Double-Masked Trials of Brolucizumab for Neovascular Age-Related Macular Degeneration. Ophthalmology 2020, 127, 72–84. [Google Scholar] [CrossRef]
- ElSheikh, R.H.; Chauhan, M.Z.; Sallam, A.B. Current and Novel Therapeutic Approaches for Treatment of Neovascular Age-Related Macular Degeneration. Biomolecules 2022, 12, 1629. [Google Scholar] [CrossRef]
- Edington, M.; Connolly, J.; Chong, N.V. Pharmacokinetics of intravitreal anti-VEGF drugs in vitrectomized versus non-vitrectomized eyes. Expert Opin. Drug Metab. Toxicol. 2017, 13, 1217–1224. [Google Scholar] [CrossRef]
- Jakubiak, P.; Alvarez-Sánchez, R.; Fueth, M.; Broders, O.; Kettenberger, H.; Stubenrauch, K.; Caruso, A. Ocular Pharmacokinetics of Intravitreally Injected Protein Therapeutics: Comparison among Standard-of-Care Formats. Mol. Pharm. 2021, 18, 2208–2217. [Google Scholar] [CrossRef]
- Cox, J.T.; Eliott, D.; Sobrin, L. Inflammatory Complications of Intravitreal Anti-VEGF Injections. J. Clin. Med. 2021, 10, 981. [Google Scholar] [CrossRef]
- Pauleikhoff, D.; Scheider, A.; Wiedmann, P.; Gelisken, F.; Scholl, H.P.; Roider, I.; Mohr, A.; Zlateva, G.; Xu, X. Neovascular age-related macular degeneration in Germany. Encroachment on the quality of life and the financial implications. Ophthalmol. Z. Dtsch. Ophthalmol. Ges. 2009, 106, 242–251. [Google Scholar] [CrossRef]
- Coleman, H.R.; Chan, C.C.; Ferris, F.L., 3rd; Chew, E.Y. Age-related macular degeneration. Lancet 2008, 372, 1835–1845. [Google Scholar] [CrossRef]
- Krishnan, R.; Goverdhan, S.; Lochhead, J. Submacular haemorrhage after intravitreal bevacizumab compared with intravitreal ranibizumab in large occult choroidal neovascularization. Clin. Exp. Ophthalmol. 2009, 37, 384–388. [Google Scholar] [CrossRef]
- Carneiro, A.M.; Barthelmes, D.; Falcão, M.S.; Mendonça, L.S.; Fonseca, S.L.; Gonçalves, R.M.; Faria-Correia, F.; Falcão-Reis, F.M. Arterial thromboembolic events in patients with exudative age-related macular degeneration treated with intravitreal bevacizumab or ranibizumab. Ophthalmologica 2011, 225, 211–221. [Google Scholar] [CrossRef]
- Johnen, S.; Djalali-Talab, Y.; Kazanskaya, O.; Möller, T.; Harmening, N.; Kropp, M.; Izsvàk, Z.; Walter, P.; Thumann, G. Antiangiogenic and neurogenic activities of Sleeping Beauty-mediated PEDF-transfected RPE cells in vitro and in vivo. BioMed Res. Int. 2015, 2015, 863845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuerten, D.; Johnen, S.; Harmening, N.; Souteyrand, G.; Walter, P.; Thumann, G. Transplantation of PEDF-transfected pigment epithelial cells inhibits corneal neovascularization in a rabbit model. Graefe Arch. Clin. Exp. Ophthalmol. 2015, 253, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, L.; Recalde, S.; Hernandez, M.; Bezunartea, J.; Rodriguez-Madoz, J.R.; Johnen, S.; Diarra, S.; Marie, C.; Izsvák, Z.; Ivics, Z.; et al. Long-Term PEDF Release in Rat Iris and Retinal Epithelial Cells after Sleeping Beauty Transposon-Mediated Gene Delivery. Mol. Ther. Nucleic Acids 2017, 15, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, M.; Recalde, S.; Garcia-Garcia, L.; Bezunartea, J.; Miskey, C.; Johnen, S.; Diarra, S.; Sebe, A.; Rodriguez-Madoz, J.R.; Pouillot, S.; et al. Preclinical Evaluation of a Cell-Based Gene Therapy Using the Sleeping Beauty Transposon System in Choroidal Neovascularization. Mol. Ther. Methods Clin. Dev. 2019, 15, 403–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kropp, M.; Harmening, N.; Bascuas, T.; Johnen, S.; De Clerck, E.; Fernández, V.; Ronchetti, M.; Cadossi, R.; Zanini, C.; Scherman, D.; et al. GMP-Grade Manufacturing and Quality Control of a Non-Virally Engineered Advanced Therapy Medicinal Product for Personalized Treatment of Age-Related Macular Degeneration. Biomedicines 2022, 10, 2777. [Google Scholar] [CrossRef]
- Robbins, P.D.; Ghivizzani, S.C. Viral vectors for gene therapy. Pharmacol. Ther. 1998, 80, 35–47. [Google Scholar] [CrossRef]
- Liu, M.M.; Tuo, J.; Chan, C.-C. Gene therapy for ocular diseases. Br. J. Ophthalmol. 2011, 95, 604–612. [Google Scholar] [CrossRef] [Green Version]
- Giacca, M.; Zacchigna, S. Virus-mediated gene delivery for human gene therapy. J. Control. Release 2012, 161, 377–388. [Google Scholar] [CrossRef]
- Conley, S.M.; Cai, X.; Naash, M.I. Non-viral ocular gene therapy: Assessment and future directions. Curr. Opin. Mol. Ther. 2008, 10, 456–463. [Google Scholar]
- Kachi, S.; Oshima, Y.; Esumi, N.; Kachi, M.; Rogers, B.; Zack, D.J.; Campochiaro, P.A. Nonviral ocular gene transfer. Gene Ther. 2005, 12, 843–851. [Google Scholar] [CrossRef]
- Oliveira, A.V.; Rosa da Costa, A.M.; Silva, G.A. Non-viral strategies for ocular gene delivery. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 77, 1275–1289. [Google Scholar] [CrossRef] [PubMed]
- Bordet, T.; Behar-Cohen, F. Ocular gene therapies in clinical practice: Viral vectors and nonviral alternatives. Drug. Discov. Today 2019, 24, 1685–1693. [Google Scholar] [CrossRef]
- Weleber, R.G.; Pennesi, M.E.; Wilson, D.J.; Kaushal, S.; Erker, L.R.; Jensen, L.; McBride, M.T.; Flotte, T.R.; Humphries, M.; Calcedo, R.; et al. Results at 2 Years after Gene Therapy for RPE65-Deficient Leber Congenital Amaurosis and Severe Early-Childhood-Onset Retinal Dystrophy. Ophthalmology 2016, 123, 1606–1620. [Google Scholar] [CrossRef] [Green Version]
- FDA. FDA Approves Novel Gene Therapy to Treat Patients with a Rare Form of Inherited Vision Loss. 2017. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-novel-gene-therapy-treat-patients-rare-form-inherited-vision-loss (accessed on 17 November 2021).
- Deng, C.; Zhao, P.Y.; Branham, K.; Schlegel, D.; Fahim, A.T.; Jayasundera, T.K.; Khan, N.; Besirli, C.G. Real-world outcomes of voretigene neparvovec treatment in pediatric patients with RPE65-associated Leber congenital amaurosis. Graefe Arch. Clin. Exp. Ophthalmol. 2022, 260, 1543–1550. [Google Scholar] [CrossRef]
- Gerhardt, M.J.; Priglinger, C.S.; Rudolph, G.; Hufendiek, K.; Framme, C.; Jägle, H.; Salchow, D.J.; Anschütz, A.; Michalakis, S.; Priglinger, S.G. Gene Therapy with Voretigene Neparvovec Improves Vision and Partially Restores Electrophysiological Function in Pre-School Children with Leber Congenital Amaurosis. Biomedicines 2023, 11, 103. [Google Scholar] [CrossRef]
- Constable, I.J.; Pierce, C.M.; Lai, C.M.; Magno, A.L.; Degli-Esposti, M.A.; French, M.A.; McAllister, I.L.; Butler, S.; Barone, S.B.; Schwartz, S.D.; et al. Phase 2a Randomized Clinical Trial: Safety and Post Hoc Analysis of Subretinal rAAV.sFLT-1 for Wet Age-related Macular Degeneration. EBioMedicine 2016, 14, 168–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heier, J.S.; Kherani, S.; Desai, S.; Dugel, P.; Kaushal, S.; Cheng, S.H.; Delacono, C.; Purvis, A.; Richards, S.; Le-Halpere, A.; et al. Intravitreous injection of AAV2-sFLT01 in patients with advanced neovascular age-related macular degeneration: A phase 1, open-label trial. Lancet 2017, 390, 50–61. [Google Scholar] [CrossRef]
- Rakoczy, E.P.; Lai, C.M.; Magno, A.L.; Wikstrom, M.E.; French, M.A.; Pierce, C.M.; Schwartz, S.D.; Blumenkranz, M.S.; Chalberg, T.W.; Degli-Esposti, M.A.; et al. Gene therapy with recombinant adeno-associated vectors for neovascular age-related macular degeneration: 1 year follow-up of a phase 1 randomised clinical trial. Lancet 2015, 386, 2395–2403. [Google Scholar] [CrossRef] [Green Version]
- Campochiaro, P.A.; Lauer, A.K.; Sohn, E.H.; Mir, T.A.; Naylor, S.; Anderton, M.C.; Kelleher, M.; Harrop, R.; Ellis, S.; Mitrophanous, K.A. Lentiviral Vector Gene Transfer of Endostatin/Angiostatin for Macular Degeneration (GEM) Study. Hum. Gene Ther. 2017, 28, 99–111. [Google Scholar] [CrossRef]
- Guimaraes, T.A.C.; Georgiou, M.; Bainbridge, J.W.B.; Michaelides, M. Gene therapy for neovascular age-related macular degeneration: Rationale, clinical trials and future directions. Br. J. Ophthalmol. 2021, 105, 151–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiss, S. Ixo-Vec Intravitreal Gene Therapy for Neovascular AGe-Related Macular Degeneration. In Proceedings of the 46th Annual Macula Society Meeting, Miami, FL, USA, 15–18 February 2023. [Google Scholar]
- Hartman, Z.C.; Appledorn, D.M.; Amalfitano, A. Adenovirus vector induced innate immune responses: Impact upon efficacy and toxicity in gene therapy and vaccine applications. Virus Res. 2008, 132, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Song, S. Distinct immune responses to transgene products from rAAV1 and rAAV8 vectors. Proc. Natl. Acad. Sci. USA 2009, 106, 17158–17162. [Google Scholar] [CrossRef] [Green Version]
- Maestro, S.; Weber, N.D.; Zabaleta, N.; Aldabe, R.; Gonzalez-Aseguinolaza, G. Novel vectors and approaches for gene therapy in liver diseases. JHEP Rep. 2021, 3, 100300. [Google Scholar] [CrossRef]
- Deichmann, A.; Hacein-Bey-Abina, S.; Schmidt, M.; Garrigue, A.; Brugman, M.H.; Hu, J.; Glimm, H.; Gyapay, G.; Prum, B.; Fraser, C.C.; et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Investig. 2007, 117, 2225–2232. [Google Scholar] [CrossRef]
- De Ravin, S.S.; Su, L.; Theobald, N.; Choi, U.; Macpherson, J.L.; Poidinger, M.; Symonds, G.; Pond, S.M.; Ferris, A.L.; Hughes, S.H.; et al. Enhancers are major targets for murine leukemia virus vector integration. J. Virol. 2014, 88, 4504–4513. [Google Scholar] [CrossRef] [Green Version]
- Cavazza, A.; Moiani, A.; Mavilio, F. Mechanisms of retroviral integration and mutagenesis. Hum. Gene Ther. 2013, 24, 119–131. [Google Scholar] [CrossRef] [Green Version]
- Ginn, S.L.; Liao, S.H.; Dane, A.P.; Hu, M.; Hyman, J.; Finnie, J.W.; Zheng, M.; Cavazzana-Calvo, M.; Alexander, S.I.; Thrasher, A.J.; et al. Lymphomagenesis in SCID-X1 mice following lentivirus-mediated phenotype correction independent of insertional mutagenesis and gammac overexpression. Mol. Ther. 2010, 18, 965–976. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Garrigue, A.; Wang, G.P.; Soulier, J.; Lim, A.; Morillon, E.; Clappier, E.; Caccavelli, L.; Delabesse, E.; Beldjord, K.; et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Investig. 2008, 118, 3132–3142. [Google Scholar] [CrossRef]
- Mátés, L.; Chuah, M.K.L.; Belay, E.; Jerchow, B.; Manoj, N.; Acosta-Sanchez, A.; Grzela, D.P.S.A.; Becker, K.; Matrai, J.; Ma, L.; et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009, 41, 753–761. [Google Scholar] [CrossRef]
- Thumann, G.; Stöcker, M.; Maltusch, C.; Salz, A.K.; Barth, S.; Walter, P.; Johnen, S. High efficiency non-viral transfection of retinal and iris pigment epithelial cells with pigment epithelium-derived factor. Gene Ther. 2010, 17, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Johnen, S.; Izsvák, Z.; Stöcker, M.; Harmening, N.; Salz, A.K.; Walter, P.; Thumann, G. Sleeping Beauty transposon-mediated transfection of retinal and iris pigment epithelial cells. Investig. Ophthalmol. Vis. Sci. 2012, 53, 4787–4796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thumann, G.; Harmening, N.; Prat-Souteyrand, C.; Marie, M.; Pastor, M.; Sebe, A.; Miskey, C.; Hurst, L.D.; Diarra, S.; Kropp, M.; et al. Engineering of PEDF-Expressing Primary Pigment Epithelial Cells by the SB Transposon System Delivered by pFAR4 Plasmids. Mol. Ther. Nucleic Acids 2017, 17, 302–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastor, M.; Johnen, S.; Harmening, N.; Quiviger, M.; Pailloux, J.; Kropp, M.; Walter, P.; Ivics, Z.; Izsvák, Z.; Thumann, G.; et al. The Antibiotic-free pFAR4 Vector Paired with the Sleeping Beauty Transposon System Mediates Efficient Transgene Delivery in Human Cells. Mol. Ther. Nucleic Acids 2018, 11, 57–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudecek, M.; Izsvák, Z.; Johnen, S.; Renner, M.; Thumann, G.; Ivics, Z. Going non-viral. The Sleeping Beauty transposon system breaks on through to the clinical side. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 355–380. [Google Scholar] [CrossRef] [Green Version]
- Bascuas, T.; Zedira, H.; Kropp, M.; Harmening, N.; Asrih, M.; Prat-Souteyrand, C.; Tian, S.; Thumann, G. Human Retinal Pigment Epithelial Cells Overexpressing the Neuroprotective Proteins PEDF and GM-CSF to Treat Degeneration of the Neural Retina. Curr. Gene Ther. 2021, 22, 168–183. [Google Scholar] [CrossRef]
- Izsvàk, Z.; Ivics, Z.; Hackett, P.B. Characterization of a Tc1-like transposable element in zebrafish (Danio rerio). Mol. Gen. Genet. 1995, 247, 312–322. [Google Scholar] [CrossRef]
- Ivics, Z.; Izsvàk, Z.; Minter, A.; Hackett, P.B. Identification of functional domains and evolution of Tc1-like transposable elements. Proc. Natl. Acad. Sci. USA 1996, 93, 5008–5013. [Google Scholar] [CrossRef] [Green Version]
- Ivics, Z.; Hackett, P.B.; Plasterk, R.H.; Izsvàk, Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 1997, 91, 501–510. [Google Scholar] [CrossRef] [Green Version]
- Zayed, H.; Izsvàk, Z.; Walisko, O.; Ivics, Z. Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol. Ther. 2004, 9, 292–304. [Google Scholar] [CrossRef]
- Yant, S.R.; Wu, X.; Huang, Y.; Garrison, B.; Burgess, S.M.; Kay, M.A. High-Resolution Genome-Wide Mapping of Transposon Integration in Mammals. Mol. Cell. Biol. 2005, 25, 2085–2094. [Google Scholar] [CrossRef] [Green Version]
- Ammar, I.; Gogol-Döring, A.; Miskey, C.; Chen, W.; Cathomen, T.; Izsvák, Z.; Ivics, Z. Retargeting transposon insertions by theadeno-associated virus Rep protein. Nucleic Acids Res. 2012, 40, 6693–6712. [Google Scholar] [CrossRef] [Green Version]
- Voigt, K.; Gogol-Döring, A.; Miskey, C.; Chen, W.; Cathomen, T.; Izsvàk, Z.; Ivics, Z. Retargeting Sleeping Beauty transposon insertions by engineered zinc finger DNA-binding domains. Mol. Ther. 2012, 20, 1852–1862. [Google Scholar] [CrossRef] [Green Version]
- Gogol-Döring, A.; Ammar, I.; Gupta, S.; Bunse, M.; Miskey, C.; Chen, W.; Uckert, W.; Schulz, T.F.; Izsvàk, Z.; Ivics, Z. Genome-wide Profiling Reveals Remarkable Parallels Between Insertion Site Selection Properties of the MLV Retrovirus and the piggyBac Transposon in Primary Human CD4+ T Cells. Mol. Ther. 2016, 24, 592–606. [Google Scholar] [CrossRef] [Green Version]
- Izsvák, Z.; Hackett, P.B.; Cooper, L.J.; Ivics, Z. Translating Sleeping Beauty transposition into cellular therapies: Victories and challenges. Bioessays 2010, 32, 756–767. [Google Scholar] [CrossRef] [Green Version]
- Amberger, M.; Ivics, Z. Latest Advances for the Sleeping Beauty Transposon System: 23 Years of Insomnia but Prettier than Ever: Refinement and Recent Innovations of the Sleeping Beauty Transposon System Enabling Novel, Nonviral Genetic Engineering Applications. Bioessays 2020, 42, e2000136. [Google Scholar] [CrossRef]
- Irving, M.; Lanitis, E.; Migliorini, D.; Ivics, Z.; Guedan, S. Choosing the Right Tool for Genetic Engineering: Clinical Lessons from Chimeric Antigen Receptor-T Cells. Hum. Gene Ther. 2021, 32, 1044–1058. [Google Scholar] [CrossRef]
- Wilber, A.; Frandsen, J.L.; Geurts, J.L.; Largaespada, D.A.; Hackett, P.B.; McIvor, R.S. RNA as a source of transposase for Sleeping Beauty-mediated gene insertion and expression in somatic cells and tissues. Mol. Ther. 2006, 13, 625–630. [Google Scholar] [CrossRef]
- Wilber, A.; Wangensteen, K.J.; Chen, Y.; Zhuo, L.; Frandsen, J.L.; Bell, J.B.; Chen, Z.J.; Ekker, S.C.; McIvor, R.S.; Wang, X. Messenger RNA as a source of transposase for Sleeping Beauty transposon-mediated correction of hereditary tyrosinemia type I. Mol. Ther. 2007, 15, 1280–1287. [Google Scholar] [CrossRef]
- Huang, X.; Haley, K.; Wong, M.; Guo, H.; Lu, C.; Wilber, A.; Zhou, X. Unexpectedly high copy number of random integration but low frequency of persistent expression of the Sleeping Beauty transposase after trans delivery in primary human T cells. Hum. Gene Ther. 2010, 21, 1577–1590. [Google Scholar] [CrossRef] [Green Version]
- Tschorn, N.; van Heuvel, Y.; Stitz, J. Transgene Expression and Transposition Efficiency of Two-Component Sleeping Beauty Transposon Vector Systems Utilizing Plasmid or mRNA Encoding the Transposase. Mol. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Galla, M.; Schambach, A.; Falk, C.S.; Maetzig, T.; Kuehle, J.; Lange, K.; Zychlinski, D.; Heinz, N.; Brugman, M.H.; Göhring, G.; et al. Avoiding cytotoxicity of transposases by dose-controlled mRNA delivery. Nucleic Acids Res. 2011, 39, 7147–7160. [Google Scholar] [CrossRef] [PubMed]
- Wiehe, J.M.; Ponsaerts, P.; Rojewski, M.T.; Homann, J.M.; Greiner, J.; Kronawitter, D.; Schrezenmeier, H.; Hombach, V.; Wiesneth, M.; Zimmermann, O.; et al. mRNA-mediated gene delivery into human progenitor cells promotes highly efficient protein expression. J. Cell. Mol. Med. 2007, 11, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Hudecek, M.; Ivics, Z. Non-viral therapeutic cell engineering with the Sleeping Beauty transposon system. Curr. Opin. Genet. Dev. 2018, 52, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Dunn, K.C.; Aotaki-Keen, A.E.; Putkey, F.R.; Hjelmeland, L.M. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp. Eye Res. 1996, 62, 155–169. [Google Scholar] [CrossRef]
- Johnen, S.; Harmening, N.; Marie, C.; Scherman, D.; Izsvák, Z.; Ivics, Z.; Walter, P.; Thumann, G. Electroporation-Based Genetic Modification of Primary Human Pigment Epithelial Cells using the Sleeping Beauty Transposon System. J. Vis. Exp. 2021, 168. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Izsvàk, Z.; Chuah, M.K.L.; VandenDriessche, T.; Ivics, Z. Efficient stable gene transfer into human cells by the Sleeping Beauty transposon vectors. Methods 2009, 49, 287–297. [Google Scholar] [CrossRef]
- Kebriaei, P.; Izsvák, Z.; Narayanavari, S.A.; Singh, H.; Ivics, Z. Gene Therapy with the Sleeping Beauty Transposon System. Trends Genet. 2017, 33, 852–870. [Google Scholar] [CrossRef]
- Wileys, J. Gene Therapy Clinial Trials Worldwide: Journal of Gene Medicine. Available online: https://a873679.fmphost.com/fmi/webd/GTCT (accessed on 16 February 2021).
- ClinicalTrials.gov: National Library of Medicine. Available online: https://clinicaltrials.gov (accessed on 16 February 2021).
- Prommersberger, S.; Monjezi, R.; Botezatu, L.; Miskey, C.; Amberger, M.; Mestermann, K.; Hudecek, M.; Ivics, Z. Generation of CAR-T Cells with Sleeping Beauty Transposon Gene Transfer. Methods Mol. Biol. 2022, 2521, 41–66. [Google Scholar] [CrossRef]
- Wei, M.; Mi, C.-L.; Jing, C.-Q.; Wang, T.-Y. Progress of Transposon Vector System for Production of Recombinant Therapeutic Proteins in Mammalian Cells. Front. Bioeng. Biotechnol. 2022, 10, 879222. [Google Scholar] [CrossRef]
- Grabundzija, I.; Irgang, M.; Mátés, L.; Belay, E.; Matrai, J.; Gogol-Döring, A.; Kawakami, K.; Chen, W.; Ruiz, P.; Chuah, M.K.L.; et al. Comparative Analysis of Transposable Element Vector Systems in Human Cells. Mol. Ther. 2010, 18, 1200–1209. [Google Scholar] [CrossRef]
- Berg, K.; Schäfer, V.N.; Tschorn, N.; Stitz, J. Advanced Establishment of Stable Recombinant Human Suspension Cell Lines Using Genotype–Phenotype Coupling Transposon Vectors. In Genotype Phenotype Coupling: Methods and Protocols; Zielonka, S., Krah, S., Eds.; Springer: New York, NY, USA, 2020; pp. 351–361. [Google Scholar]
- Bloquel, C.; Fabre, E.; Bureau, M.F.; Scherman, D. Plasmid DNA electrotransfer for intracellular and secreted proteins expression: New methodological developments and applications. J. Gene Med. 2004, 6 (Suppl. S1), S11–S23. [Google Scholar] [CrossRef]
- Kreiss, P.; Cameron, B.; Rangara, R.; Mailhe, P.; Aguerre-Charriol, O.; Airiau, M.; Scherman, D.; Crouzet, J.; Pitard, B. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res. 1999, 27, 3792–3798. [Google Scholar] [CrossRef] [Green Version]
- Chabot, S.; Orio, J.; Schmeer, M.; Schleef, M.; Golzio, M.; Teissié, J. Minicircle DNA electrotransfer for efficient tissue-targeted gene delivery. Gene Ther. 2013, 20, 62–68. [Google Scholar] [CrossRef] [Green Version]
- Johnen, S.; Kazanskaya, O.; Armogan, N.; Stickelmann, C.; Stöcker, M.; Walter, P.; Thumann, G. Endogenic Regulation of Proliferation and Zinc Transporters by Pigment Epithelial Cells Nonvirally Transfected with PEDF. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5400–5407. [Google Scholar] [CrossRef] [Green Version]
- Bire, S.; Ley, D.; Casteret, S.; Mermod, N.; Bigot, Y.; Rouleux-Bonnin, F. Optimization of the piggyBac transposon using mRNA and insulators: Toward a more reliable gene delivery system. PLoS ONE 2013, 8, e82559. [Google Scholar] [CrossRef] [Green Version]




| Gene | Gene ID | Sequence (5′-3′) | Fragment Size | Annealing Temperature |
|---|---|---|---|---|
| GAPDH | 2597 | F: AAG GTC ATC CCT GAG CTG AAC | 353 nt | 56 °C |
| R: TTA CTC CTT GGA GGC CAT GTG | ||||
| KRT8 | 3856 | F: AAG GAT GCC AAC GCC AAG TTG | 360 nt | 58 °C |
| R: CGA TCT TCT TCA CAA CCA CGG | ||||
| VEGF | 7422 | F: AAG GAC CTA TGT CCT CAC ACC | 399 nt | 58 °C |
| R: TAG TGA CTG TCA CCG ATC AGG | ||||
| ZO-1 | 7082 | F: ACA CTG CTG AGT CCT TTG GTG | 398 nt | 58 °C |
| R: CTA GCC AAT ACC AAC AGT CCC | ||||
| VEGF-R2 | 3791 | F: ATG TGA AGC GGT CAA CAA AGT | 59 nt | 58 °C |
| R: CTG GTC ACG TGG AAG GAG AT | ||||
| IRDRL | Synthetic sequence | F: CTC GTT TTT CAA CTA CTC CAC AAA TTT CT | 85 nt | 58 °C |
| R: GTG TCA TGC ACA AAG TAG ATG TCC TA | ||||
| rPEDF | 5176 (cDNA sequence) | F: TAC TGA GGG ACA CAG ACA CA | 180 nt | 56 °C |
| R: AAG TCA TGC CCG CTT TTG AG | ||||
| End. PEDF | 5176 | F: GCT GGC TTT GAG TGG AAC GA | 244 nt | 58 °C |
| R: GTG TCC TGT GGA ATC TGC TG | ||||
| Total PEDF | 5176 | F: TAC TGA GGG ACA CAG ACA CA | 99 nt | 58 °C |
| R: CAA TGA TGA TGA TGA TGA TGG | ||||
| RPPH1 | 85495 | F: AGC TGA GTG CGT CCT GTC ACT | 63 nt | 58 °C |
| R: TCT GGC CCT AGT CTC AGA CCT T | ||||
| CTSD | 1509 | F: AGG CAA AGG CTA CAA GCT GTC | 459 nt | 58 °C |
| R: TGT GCT CTG GAT CAG CTC TAC | ||||
| CRALBP | 6017 | F: TCA CCA CGA CCT ACA ATG TGG | 243 nt | 58 °C |
| R: AAC TAC AGT TCA GCT GGC AGG | ||||
| SB100X | Synthetic sequence | F: CTT GCA AGC CGA AGA ACA CC | 400 nt | 58 °C |
| R: CAT TCC TCC TGA CAG AGC TG | ||||
| EGFP/Venus | Synthetic sequence | F: AGC TGA CCC TGA AGC TGA TCT | 328 nt | 58 °C |
| R: ACG TTG TGG CTG TTG TAG TTG T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harmening, N.; Johnen, S.; Izsvák, Z.; Ivics, Z.; Kropp, M.; Bascuas, T.; Walter, P.; Kreis, A.; Pajic, B.; Thumann, G. Enhanced Biosafety of the Sleeping Beauty Transposon System by Using mRNA as Source of Transposase to Efficiently and Stably Transfect Retinal Pigment Epithelial Cells. Biomolecules 2023, 13, 658. https://doi.org/10.3390/biom13040658
Harmening N, Johnen S, Izsvák Z, Ivics Z, Kropp M, Bascuas T, Walter P, Kreis A, Pajic B, Thumann G. Enhanced Biosafety of the Sleeping Beauty Transposon System by Using mRNA as Source of Transposase to Efficiently and Stably Transfect Retinal Pigment Epithelial Cells. Biomolecules. 2023; 13(4):658. https://doi.org/10.3390/biom13040658
Chicago/Turabian StyleHarmening, Nina, Sandra Johnen, Zsuzsanna Izsvák, Zoltan Ivics, Martina Kropp, Thais Bascuas, Peter Walter, Andreas Kreis, Bojan Pajic, and Gabriele Thumann. 2023. "Enhanced Biosafety of the Sleeping Beauty Transposon System by Using mRNA as Source of Transposase to Efficiently and Stably Transfect Retinal Pigment Epithelial Cells" Biomolecules 13, no. 4: 658. https://doi.org/10.3390/biom13040658
APA StyleHarmening, N., Johnen, S., Izsvák, Z., Ivics, Z., Kropp, M., Bascuas, T., Walter, P., Kreis, A., Pajic, B., & Thumann, G. (2023). Enhanced Biosafety of the Sleeping Beauty Transposon System by Using mRNA as Source of Transposase to Efficiently and Stably Transfect Retinal Pigment Epithelial Cells. Biomolecules, 13(4), 658. https://doi.org/10.3390/biom13040658











