On the Significance of the ADNT1 Carrier in Arabidopsis thaliana under Waterlogging Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Waterlogging Stress Conditions
2.3. Chlorophyll Fluorescence Measurements
2.4. Metabolite Measurements
2.5. Metabolite Profiling
2.6. Expression Analysis by qRT-PCR
2.7. Measurements of Adenylates Levels
2.8. Experimental Design and Statistical Analysis
3. Results
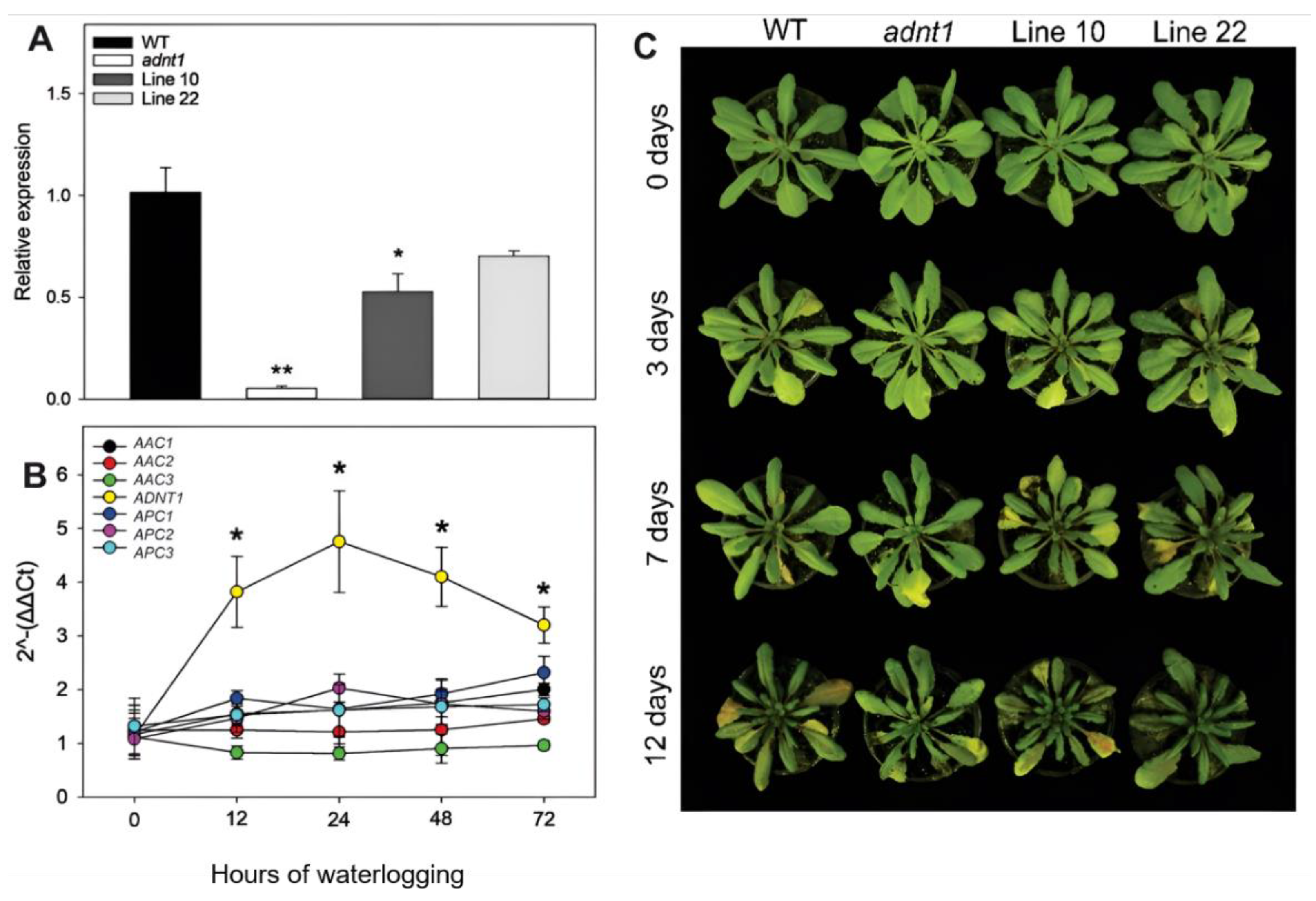
3.1. The Expression of Mitochondrial Adenylates Carriers during Waterlogging Stress
3.2. Phenotypic Characterization of ADNT1 Deficient Plants during Waterlogging
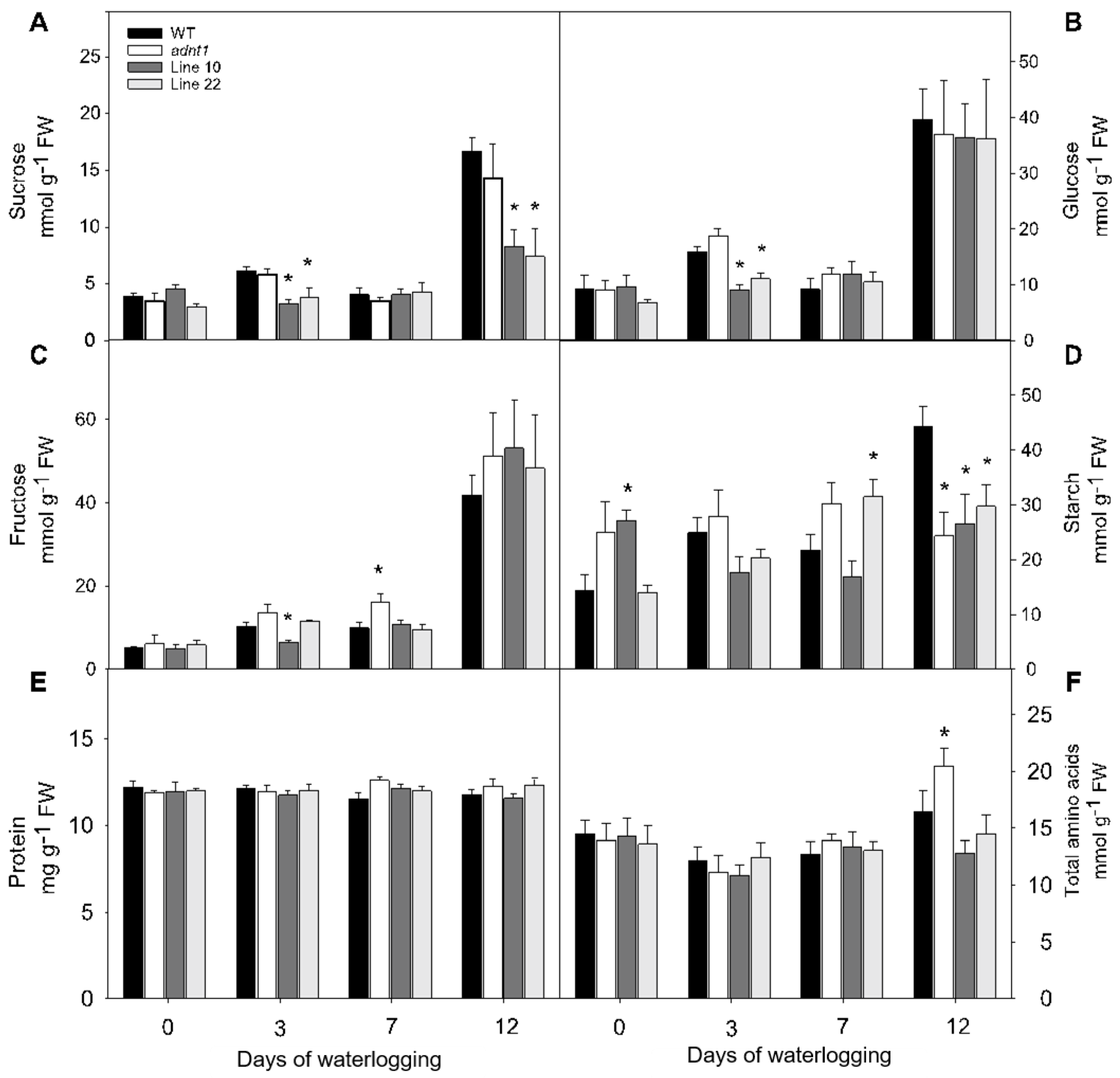
3.3. Changes in Primary Metabolites in Leaves of Plants Deficient in the Expression of ADNT1 under Root Waterlogging Stress
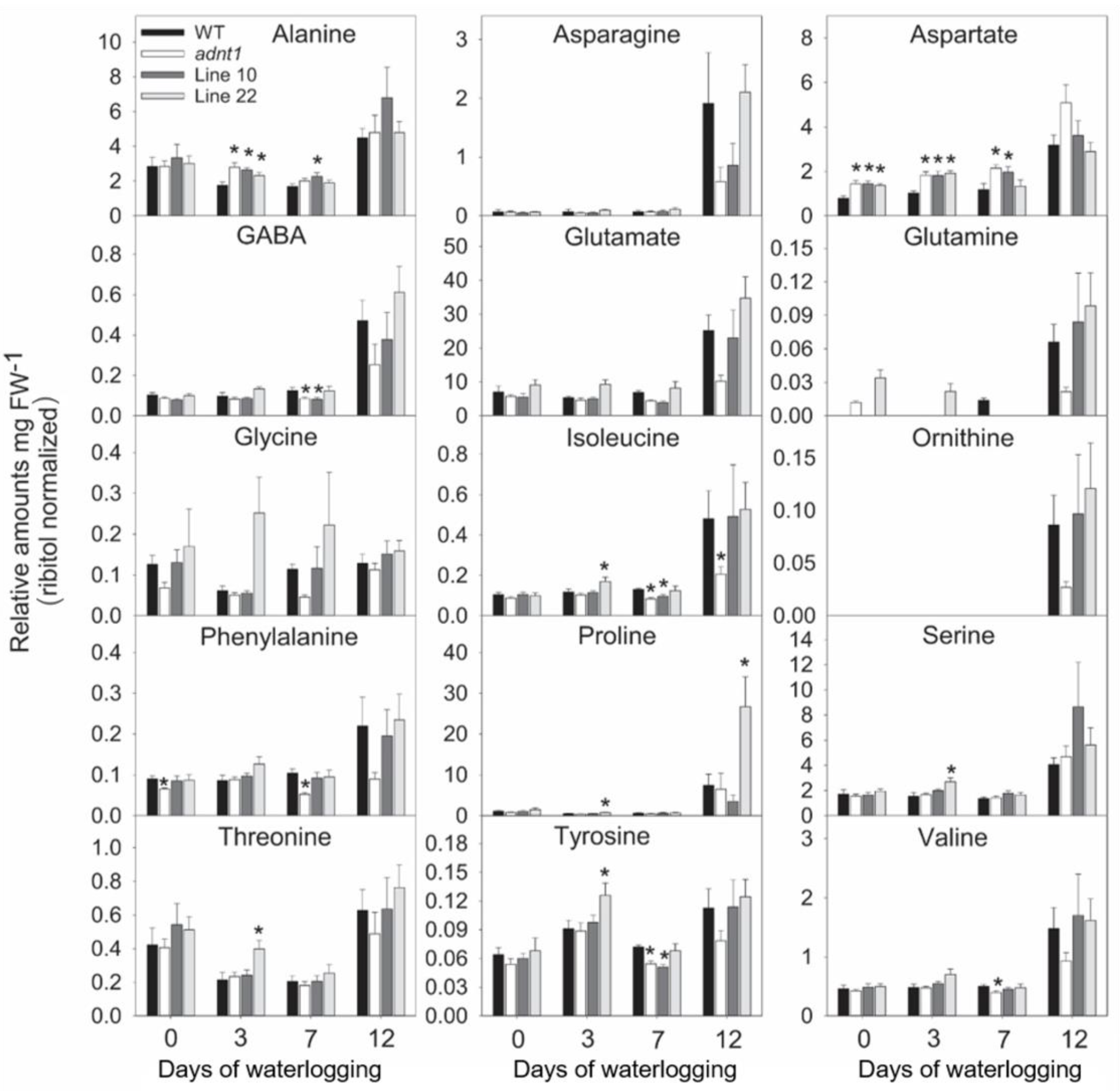
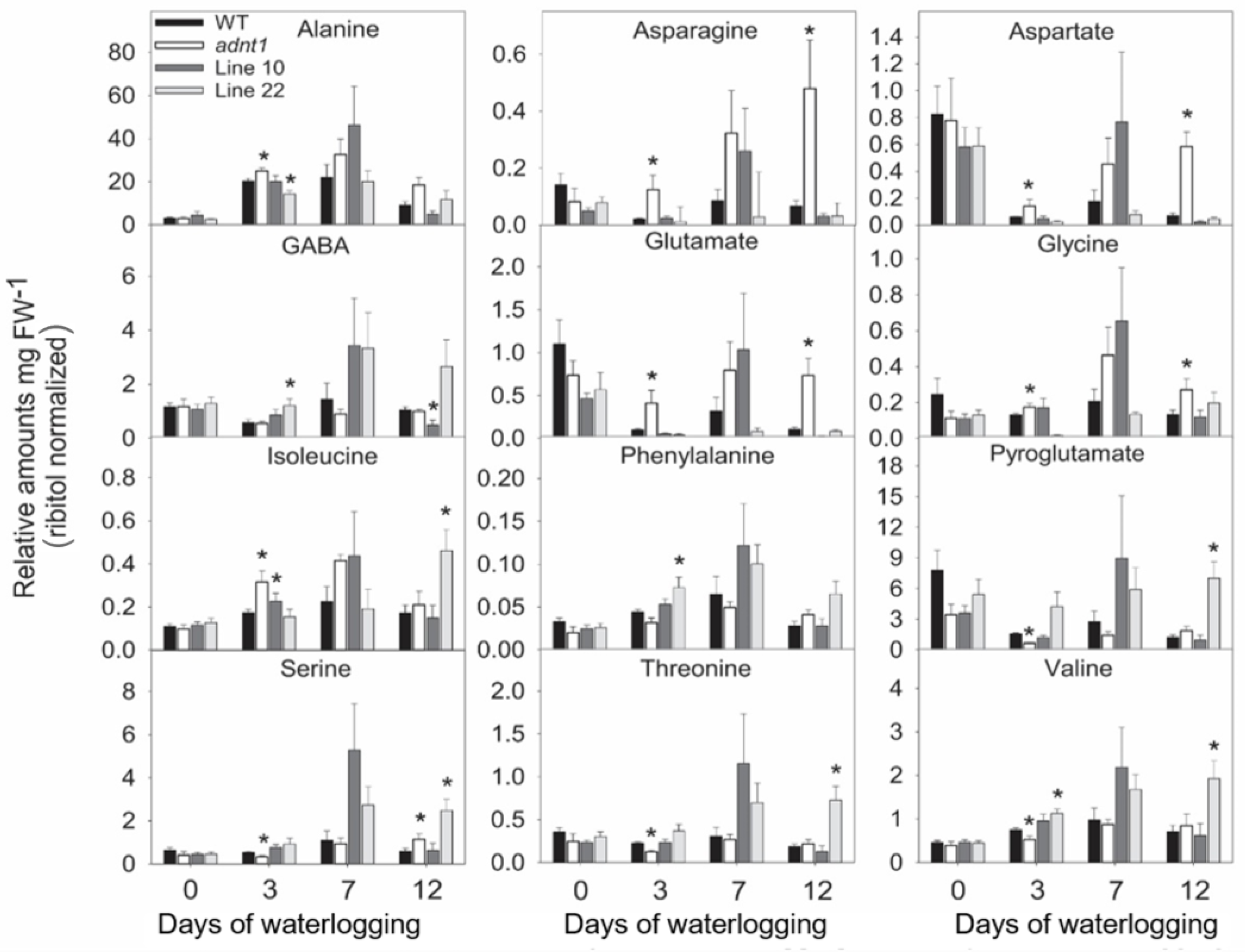
3.4. Deficiency of ADNT1 Leads to a Differential Metabolic Response Following Waterlogging Conditions
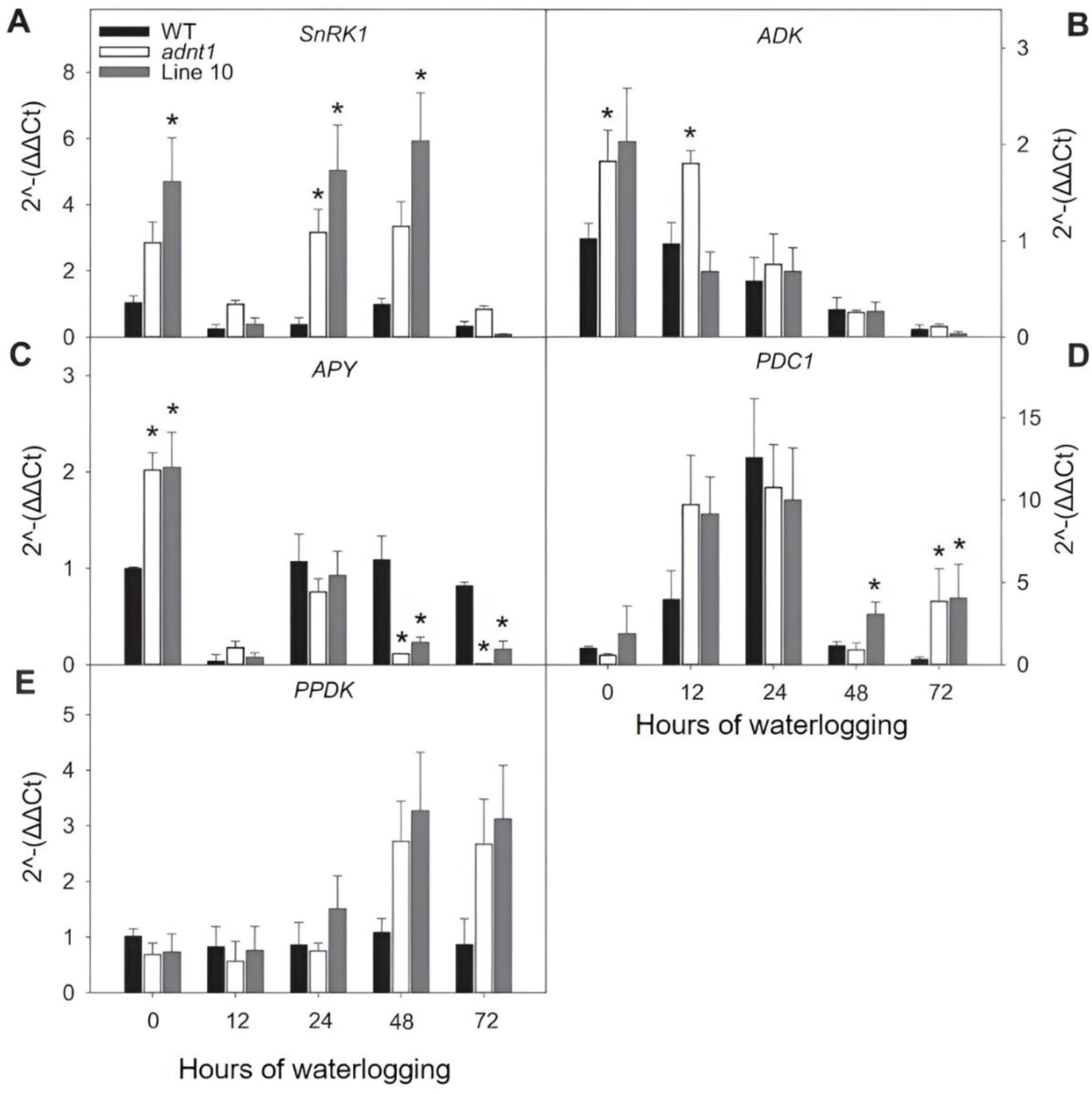
3.5. Expression of Genes Encoding Proteins Related to Energy Status in Roots of ADNT1 Deficient Plants under Waterlogging
3.6. Downregulation of ADNT1 Gene Impacts the Levels of AMP in Roots
4. Discussion
4.1. Deficiency of ADNT1 Expression Leads to an Early “Hypoxic Status” in A. thaliana Plants under Waterlogging
4.2. Reduced Expression of ADNT1 Affects the Expression of Genes Related to Waterlogging Stress Responses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haferkamp, I.; Fernie, A.R.; Neuhaus, H.E. Adenine nucleotide transport in plants: Much more than a mitochondrial issue. Trends Plant Sci. 2011, 16, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Gardeström, P.; Igamberdiev, A.U. The origin of cytosolic ATP in photosynthetic cells. Physiol. Plant. 2016, 157, 367–379. [Google Scholar] [CrossRef] [PubMed]
- De Col, V.; Fuchs, P.; Nietzel, T.; Elsä Sser, M.; Pao Voon, C.; Candeo, A.; Seeliger, I.; Fricker, M.D.; Grefen, C.; Møller, I.M.; et al. ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. eLife 2017, 6, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Santoro, A.; Carrari, F.; Blanco, E.; Nunes-Nesi, A.; Arrigoni, R.; Genchi, F.; Fernie, A.R.; Palmieri, F. Identification and characterization of ADNT1, a novel mitochondrial adenine nucleotide transporter from Arabidopsis. Plant Physiol. 2008, 148, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- da Fonseca-Pereira, P.; Neri-Silva, R.; de Monteiro-Batista, R.C.; Pérez-Díaz, J.L.; Palmieri, F.; Araújo, W.L.; Fernie, A.R.; Nunes-Nesi, A. The physiological role of mitochondrial ADNT1 carrier during senescence in Arabidopsis. Plant Stress. 2021, 2, 100019. [Google Scholar] [CrossRef]
- Geigenberger, P. Response of plant metabolism to too little oxygen. Curr. Opin. Plant Biol. 2003, 6, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Steinbeck, J.; Fuchs, P.; Lichtenauer, S.; Elsässer, M.; Schippers, J.H.M.; Nietzel, T.; Ruberti, C.; Van Aken, O.; Meyer, A.J.; et al. Multiparametric real-time sensing of cytosolic physiology links hypoxia responses to mitochondrial electron transport. New Phytol. 2019, 224, 1668–1684. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Lu, M.Y.J.; Shih, M.C. The SnRK1-eIFiso4G1 signaling relay regulates the translation of specific mRNAs in Arabidopsis under submergence. New Phytol. 2019, 222, 366–381. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Kleczkowski, L.A. Membrane potential, adenylate levels and Mg2+ are interconnected via adenylate kinase equilibrium in plant cells. Biochimica et Biophysica Acta 2003, 1607, 111–119. [Google Scholar] [CrossRef]
- Chen, L.; Su, Z.Z.; Huang, L.; Xia, F.N.; Qi, H.; Xie, L.J.; Xiao, S.; Chen, Q.F. The AMP-activated protein kinase kin10 is involved in the regulation of autophagy in Arabidopsis. Front. Plant Sci. 2017, 8, 1201. [Google Scholar] [CrossRef]
- Raymond, P.; Alani, A.; Pradet, A. ATP production by respiration and fermentation, and energy charge during aerobiosis and anaerobiosis in 12 fatty and starchy germinating seeds. Plant Physiol. 1985, 79, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Saglio, P.H.; Raymond, P.; Pradet, A. Metabolic activity and energy charge of excised maize root tips under anoxia control by soluble sugars. Plant Physiol. 1980, 66, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.; Aubert, S.; Gout, E.; Bligny, R.; Douce, R. Cooperation and competition between adenylate kinase, nucleoside diphosphokinase, electron transport, and ATP synthase in plant mitochondria studied by 31P-nuclear magnetic resonance. Plant Physiol. 1997, 113, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Loreti, E.; Shih, M.; Perata, P. Energy and sugar signaling during hypoxia. New Phytol. 2021, 229, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Oh, S.A.; Lee, S.Y.; Chung, I.K.; Lee, C.H.; Nam, H.G. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 1996, 30, 739–754. [Google Scholar] [CrossRef]
- Medeiros, D.B.; Barros, K.A.; Barros, J.A.S.; Omena-Garcia, R.P.; Arrivault, S.; Sanglard, L.M.V.P.; Detmann, K.C.; Silva, W.B.; Daloso, D.M.; DaMatta, F.M.; et al. Impaired malate and fumarate accumulation due to the mutation of the tonoplast dicarboxylate transporter has little effects on stomatal behavior. Plant Physiol. 2017, 175, 1068–1081. [Google Scholar] [CrossRef]
- Fernie, A.R.; Roessner, U.; Geigenberger, P. The sucrose analog palatinose leads to a stimulation of sucrose degradation and starch synthesis when supplied to discs of growing potato tubers. Plant Physiol. 2001, 125, 1967–1977. [Google Scholar] [CrossRef]
- Cross, J.M.; Von Korff, M.; Altmann, T.; Bartzetko, L.; Sulpice, R.; Gibon, Y.; Palacios, N.; Stitt, M. Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 2006, 142, 1574–1588. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Carrari, F.; Gibon, Y.; Sulpice, R.; Lytovchenko, A.; Fisahn, J.; Graham, J.; Ratcliffe, R.G.; Sweetlove, L.J.; Fernie, A.R. Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J. 2007, 50, 1093–1106. [Google Scholar] [CrossRef]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determinations of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Luedemann, A.; Strassburg, K.; Erban, A.; Kopka, J. Tag Finder for the quantitative analysis of gas chromatography—Mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 2008, 24, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmüller, E.; Dörmann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm metabolome database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef]
- Arvidsson, S.; Kwasniewski, M.; Riano-Pachon, D.M.; Mueller-Roeber, B. QuantPrime—A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008, 9, 465. [Google Scholar] [CrossRef]
- Jelitto, T.; Sonnewald, U.; Willmitzer, L.; Hajirezeai, M.; Stitt, M. Inorganic pyrophosphate content and metabolites in potato and tobacco plants expressing E. coli pyrophosphatase in their cytosol. Planta 1992, 188, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.C.J.; van Dongen, J.T. Making sense of low oxygen sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef]
- Chen, L.; Liao, B.; Qi, H.; Xie, L.J.; Huang, L.; Tan, W.J.; Zhai, N.; Yuan, L.B.; Zhou, Y.; Yu, L.J.; et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 2015, 11, 2233–2246. [Google Scholar] [CrossRef]
- da Fonseca-Pereira, P.; Neri-Silva, R.; Cavalcanti, J.H.F.; Brito, D.S.; Weber, A.P.M.; Araújo, W.L.; Nunes-Nesi, A. Data-Mining Bioinformatics: Connecting adenylate transport and metabolic responses to stress. Trends Plant Sci. 2018, 23, 961–974. [Google Scholar] [CrossRef]
- Rocha, M.; Licausi, F.; Araujo, W.L.; Nunes-Nesi, A.; Sodek, L.; Fernie, A.R.; van Dongen, J.T. Glycolysis and the Tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 2010, 152, 1501–1513. [Google Scholar] [CrossRef]
- Ricoult, C.; Cliquet, J.B.; Limami, A.M. Stimulation of alanine amino transferase (AlaAT) gene expression and alanine accumulation in embryo axis of the model legume Medicago truncatula contribute to anoxia stress tolerance. Physiol. Plant. 2005, 123, 30–39. [Google Scholar] [CrossRef]
- Miyashita, Y.; Good, A.G. Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Mustroph, A.; Barding, G.A.; Kaiser, K.A.; Larive, C.K.; Bailey-Serres, J. Characterization of distinct root and shoot responses to low-oxygen stress in Arabidopsis with a focus on primary C- and N-metabolism. Plant Cell Environ. 2014, 37, 2366–2380. [Google Scholar] [CrossRef] [PubMed]
- Klimova, T.; Chandel, N.S. Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death Differ. 2008, 15, 660–666. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Kleczkowski, L.A. Equilibration of adenylates in the mitochondrial intermembrane space maintains respiration and regulates cytosolic metabolism. J. Exp. Bot. 2006, 57, 2133–2141. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.N.; Lunn, J.E.; Urbanczyk-Wochniak, E.; Lytovchenko, A.; van Dongen, J.T.; Faix, B.; Schmälzlin, E.; Fernie, A.R.; Geigenberger, P. Decreased expression of cytosolic pyruvate kinase in potato tubers leads to a decline in pyruvate resulting in an in vivo repression of the alternative oxidase. Plant Physiol. 2008, 148, 1640–1654. [Google Scholar] [CrossRef]
- Jethva, J.; Schmidt, R.R.; Sauter, M.; Selinski, J. Try or die: Dynamics of plant respiration and how to survive low Oxygen conditions. Plants 2022, 11, 205. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Acj Voesenek, L.; Wang, Z.; Choi, G. Life in the balance: A signaling network controlling survival of flooding. Curr. Opin. Plant Biol. 2010, 13, 489–494. [Google Scholar] [CrossRef]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef]
- Baena-González, E.; Sheen, J. Convergent energy and stress signaling. Trends Plant Sci. 2008, 13, 474–482. [Google Scholar] [CrossRef]
- Geigenberger, P.; Riewe, D.; Fernie, A.R. The central regulation of plant physiology by adenylates. Trends Plant Sci. 2010, 15, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Im, J.H.; Cho, Y.H.; Kim, G.D.; Kang, G.H.; Hong, J.W.; Yoo, S.D. Inverse modulation of the energy sensor Snf1-related protein kinase 1 on hypoxia adaptation and salt stress tolerance in Arabidopsis thaliana. Plant Cell Environ. 2014, 37, 2303–2312. [Google Scholar] [CrossRef] [PubMed]
- Dzeja, P.; Terzic, A. Adenylate kinase and AMP signaling networks: Metabolic monitoring, signal communication and body energy sensing. Int. J. Mol. Sci. 2009, 10, 1729–1772. [Google Scholar] [CrossRef]
- Clark, G.; Roux, S.J. Role of Ca2+ in mediating plant responses to extracellular ATP and ADP. Int. J. Mol. Sci. 2018, 19, 3590. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Huang, S.N.; Wang, G.; Xuan, J.P.; Guo, Z.R. Overexpression of pyruvate decarboxylase 1 gene enhances waterlogging stress in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2016, 106, 244–252. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neri-Silva, R.; Monteiro-Batista, R.d.C.; Fonseca-Pereira, P.d.; Nunes, M.D.; Viana-Silva, A.L.; Palhares Ribeiro, T.; Pérez-Díaz, J.L.; Medeiros, D.B.; Araújo, W.L.; Fernie, A.R.; et al. On the Significance of the ADNT1 Carrier in Arabidopsis thaliana under Waterlogging Conditions. Biomolecules 2023, 13, 731. https://doi.org/10.3390/biom13050731
Neri-Silva R, Monteiro-Batista RdC, Fonseca-Pereira Pd, Nunes MD, Viana-Silva AL, Palhares Ribeiro T, Pérez-Díaz JL, Medeiros DB, Araújo WL, Fernie AR, et al. On the Significance of the ADNT1 Carrier in Arabidopsis thaliana under Waterlogging Conditions. Biomolecules. 2023; 13(5):731. https://doi.org/10.3390/biom13050731
Chicago/Turabian StyleNeri-Silva, Roberto, Rita de Cássia Monteiro-Batista, Paula da Fonseca-Pereira, Mateus Dias Nunes, Ana Luiza Viana-Silva, Tamara Palhares Ribeiro, Jorge L. Pérez-Díaz, David B. Medeiros, Wagner L. Araújo, Alisdair R. Fernie, and et al. 2023. "On the Significance of the ADNT1 Carrier in Arabidopsis thaliana under Waterlogging Conditions" Biomolecules 13, no. 5: 731. https://doi.org/10.3390/biom13050731
APA StyleNeri-Silva, R., Monteiro-Batista, R. d. C., Fonseca-Pereira, P. d., Nunes, M. D., Viana-Silva, A. L., Palhares Ribeiro, T., Pérez-Díaz, J. L., Medeiros, D. B., Araújo, W. L., Fernie, A. R., & Nunes-Nesi, A. (2023). On the Significance of the ADNT1 Carrier in Arabidopsis thaliana under Waterlogging Conditions. Biomolecules, 13(5), 731. https://doi.org/10.3390/biom13050731





