New Anti-Hypoxic Metabolites from Co-Culture of Marine-Derived Fungi Aspergillus carneus KMM 4638 and Amphichorda sp. KMM 4639
Abstract
1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Fungal Strains
2.3. DNA Extraction and Amplification
2.4. Phylogenetic Analysis
2.5. Cultivation of Fungus
2.6. Extraction and Isolation
2.7. Spectral Data
2.8. Stereo Configuration Analysis of Amino Acids in Compounds 1, 2, 5 and 6
2.9. Cell Lines and Culture Conditions
2.10. In Vitro MTT-Based Cytotoxicity Assay
2.11. CoCl2-Mimic Hypoxia Modeling
2.12. Reactive Oxygen Species (ROS) Level Assay
2.13. Superoxide Dismutase Activity Detection
2.14. Statistical Data Evaluation
3. Results
3.1. Molecular Identification of the KMM 4639 Fungal Strain
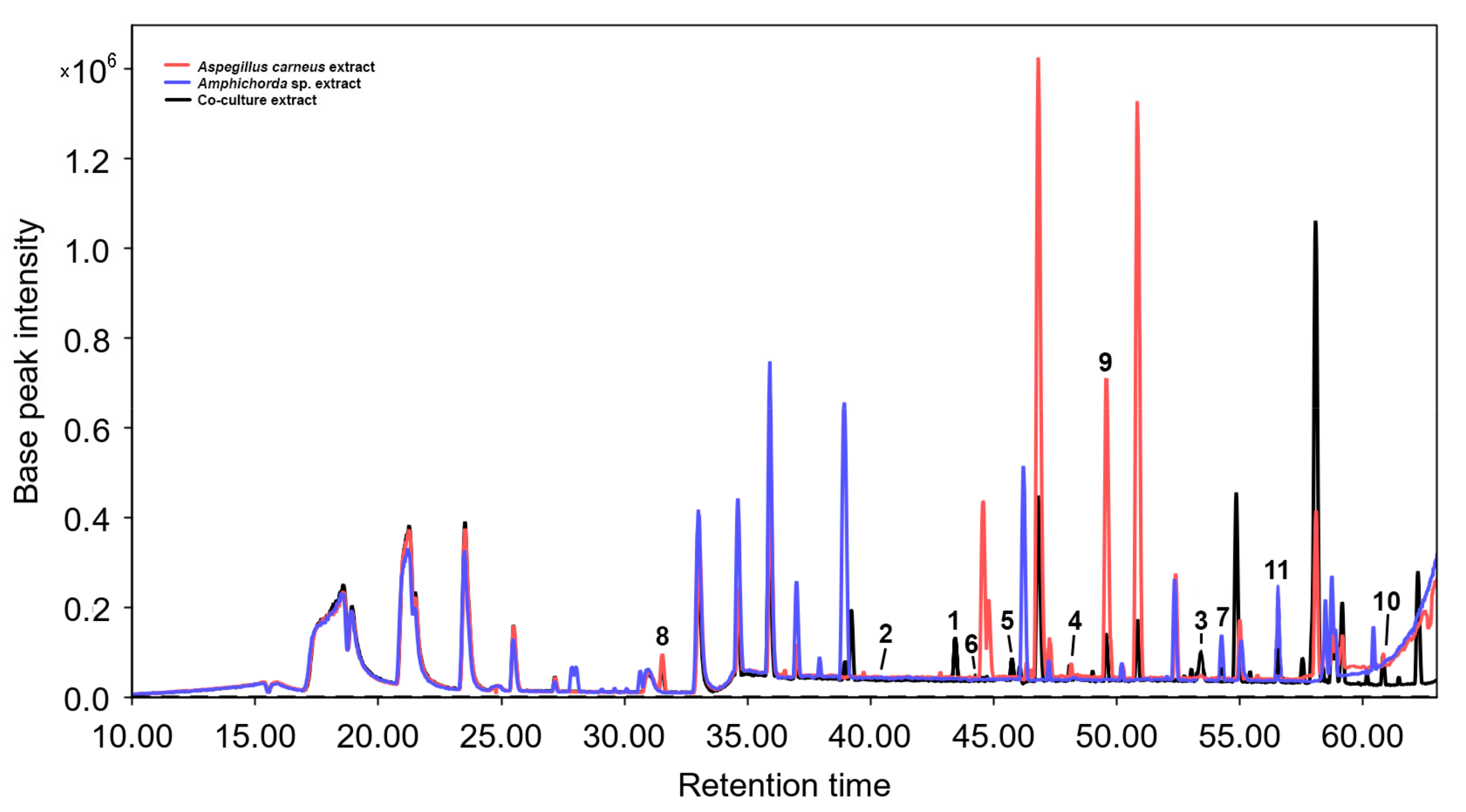
3.2. Isolated Compouns from Co-Culture
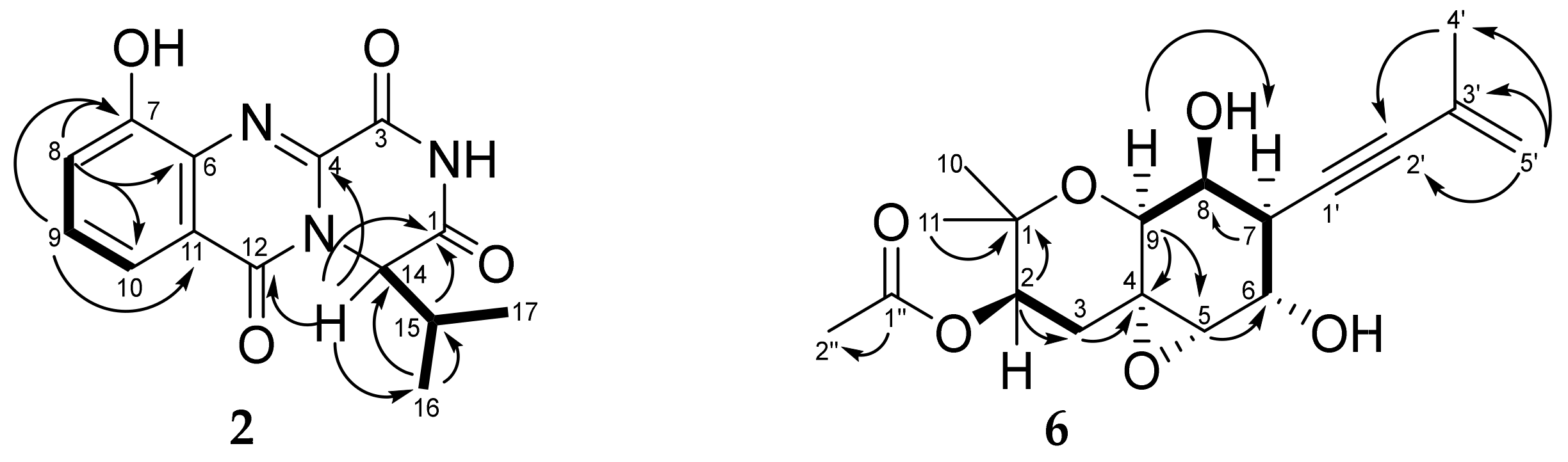
3.3. Structure Characterization of New Compounds
3.4. Cytotoxic Activity of Isolated Compounds
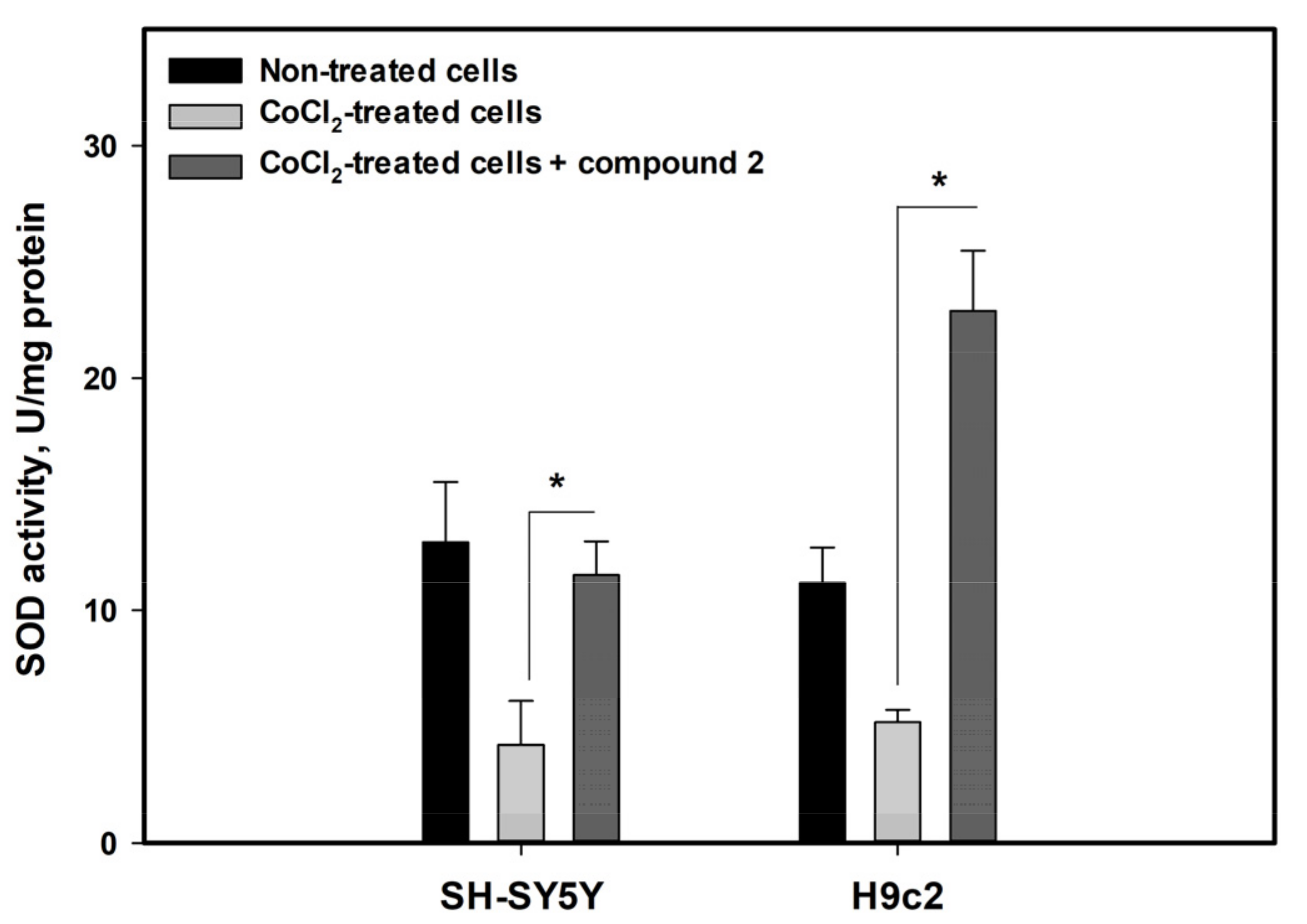
3.5. Effects of Isolated Compounds in CoCl2-Mimic Hypoxia
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carlsen, L.; Bruggemann, R. The 17 United Nations’ sustainable development goals: A status by 2020. Int. J. Sustain. Dev. World Ecol. 2022, 29, 219–229. [Google Scholar] [CrossRef]
- Knowles, S.L.; Raja, H.A.; Roberts, C.D.; Oberlies, N.H. Fungal-fungal co-culture: A primer for generating chemical diversity. Nat. Prod. Rep. 2022, 1557–1573. [Google Scholar] [CrossRef]
- Xu, K.; Yuan, X.L.; Li, C.; Li, X.D. Recent discovery of heterocyclic alkaloids from marine-derived Aspergillus species. Mar. Drugs 2020, 18, 54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wei, Q.; Yuan, X.; Xu, K. Newly reported alkaloids produced by marine-derived Penicillium species (covering 2014–2018). Bioorg. Chem. 2020, 99, 103840. [Google Scholar] [CrossRef] [PubMed]
- Zhuravleva, O.I.; Afiyatullov, S.S.; Denisenko, V.A.; Ermakova, S.P.; Slinkina, N.N.; Dmitrenok, P.S.; Kim, N.Y. Secondary metabolites from a marine-derived fungus Aspergillus carneus Blochwitz. Phytochemistry 2012, 80, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.H.; Xu, M.Y.; Li, H.J.; Li, J.Q.; Chen, Y.X.; Ma, W.Z.; Li, Y.P.; Xu, J.; Yang, D.P.; Lan, W.J. Amino Acid-Directed Strategy for Inducing the Marine-Derived Fungus Scedosporium apiospermum F41-1 to Maximize Alkaloid Diversity. Org. Lett. 2017, 19, 4888–4891. [Google Scholar] [CrossRef]
- Tuan, C.D.; Hung, N.V.; Minh, L.T.H.; Lien, H.T.H.; Chae, J.W.; Yun, H.Y.; Kim, Y.H.; Cuong, P.V.; Huong, D.T.M. A New Indole Glucoside and Other Constituents from the Sea Cucumber-Derived Aspergillus fumigatus M580 and Their Biological Activities. Rec. Nat. Prod. 2022, 16, 633–638. [Google Scholar] [CrossRef]
- Fan, Y.Q.; Li, P.H.; Chao, Y.X.; Chen, H.; Du, N.; He, Q.X.; Liu, K.C. Alkaloids with cardiovascular effects from the marine-derived fungus Penicillium expansum Y32. Mar. Drugs 2015, 13, 6489–6504. [Google Scholar] [CrossRef]
- Guo, X.C.; Zhang, Y.H.; Gao, W.B.; Pan, L.; Zhu, H.J.; Cao, F. Absolute configurations and chitinase inhibitions of quinazoline-containing diketopiperazines from the marine-derived fungus penicillium polonicum. Mar. Drugs 2020, 18, 479. [Google Scholar] [CrossRef]
- Limbadri, S.; Luo, X.; Lin, X.; Liao, S.; Wang, J.; Zhou, X.; Yang, B.; Liu, Y. Bioactive novel indole alkaloids and steroids from deep sea-derived fungus aspergillus fumigatus SCSIO 41012. Molecules 2018, 23, 2379. [Google Scholar] [CrossRef]
- Zhuravleva, O.I.; Belousova, E.B.; Oleinikova, G.K.; Antonov, A.S.; Khudyakova, Y.V.; Rasin, A.B.; Popov, R.S.; Menchinskaya, E.S.; Trinh, P.T.H.; Yurchenko, A.N.; et al. Cytotoxic Drimane-Type Sesquiterpenes from Co-Culture of the Marine-Derived Fungi Aspergillus carneus KMM 4638 and Beauveria felina (=Isaria felina) KMM 4639. Mar. Drugs 2022, 20, 584. [Google Scholar] [CrossRef]
- Smetanina, O.F.; Yurchenko, A.N.; Afiyatullov, S.S.; Kalinovsky, A.I.; Pushilin, M.A.; Khudyakova, Y.V.; Slinkina, N.N.; Ermakova, S.P.; Yurchenko, E.A. Oxirapentyns B–D produced by a marine sediment-derived fungus Isaria felina (DC.) Fr. Phytochem. Lett. 2012, 5, 165–169. [Google Scholar] [CrossRef]
- Cutler, H.G.; Arrendale, R.F.; Cole, P.D.; Springer, J.P.; Arison, B.H.; Roberts, R.G. Cinereain: A novel metabolite with plant growth regulating properties from Botrytis cinerea. Agric. Biol. Chem. 1988, 52, 1725–1733. [Google Scholar] [CrossRef]
- Capon, R.J.; Skene, C.; Stewart, M.; Ford, J.; O’Hair, R.A.J.; Williams, L.; Lacey, E.; Gill, J.H.; Heiland, K.; Friedel, T. Aspergillicins A–E: Five novel depsipeptides from the marine-derived fungus Aspergillus carneus. OBC Org. Biomol. Chem. 2003, 1, 1856–1862. [Google Scholar] [CrossRef]
- Sabareesh, V.; Ranganayaki, R.S.; Raghothama, S.; Bopanna, M.P.; Balaram, H.; Srinivasan, M.C.; Balaram, P. Identification and characterization of a library of microheterogeneous cyclohexadepsipeptides from the fungus Isaria. J. Nat. Prod. 2007, 70, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Scholin, C.A.; Herzog, M.; Sogin, M.; Anderson, D.M. Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rRNA gene. J. Phycol. 1994, 30, 999–1011. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Fujii, K.; Shimoya, T.; Ikai, Y.; Oka, H.; Harada, K.-I. Further application of advanced Marfey's method for determination of absolute configuration of primary amino compound. Tetrahedron Lett. 1998, 39, 2579–2582. [Google Scholar] [CrossRef]
- Zhuravleva, O.I.; Afiyatullov, S.S.; Yurchenko, E.A.; Denisenko, V.A.; Kirichuk, N.N.; Dmitrenok, P.S. New Metabolites from the Algal Associated Marine-derived Fungus Aspergillus carneus. Nat. Prod. Commun. 2013, 8, 1071–1074. [Google Scholar] [CrossRef]
- Yurchenko, A.N.; Smetanina, O.F.; Kalinovsky, A.I.; Pushilin, M.A.; Glazunov, V.P.; Khudyakova, Y.V.; Kirichuk, N.N.; Ermakova, S.P.; Dyshlovoy, S.A.; Yurchenko, E.A.; et al. Oxirapentyns F-K from the marine-sediment-derived fungus Isaria felina KMM 4639. J. Nat. Prod. 2014, 77, 1321–1328. [Google Scholar] [CrossRef]
- Friedman, M. Chemistry, Nutrition, and Microbiology of D-Amino Acids. J. Agric. Food Chem. 1999, 47, 3457–3479. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.; Biggins, J.B.; Bowman, B.R.; Verdine, G.L.; Gloer, J.B.; Alspaugh, J.A.; Bills, G.F. Identification of cyclosporin C from Amphichorda felina using a Cryptococcus neoformans differential temperature sensitivity assay. Appl. Microbiol. Biotechnol. 2018, 102, 2337–2350. [Google Scholar] [CrossRef] [PubMed]
- Kallow, W.; Neuhof, T.; Arezi, B.; Jungblut, P.; von Döhren, H. Penicillin biosynthesis: Intermediates of biosynthesis of δ-l-α-aminoadipyl-l-cysteinyl-d-valine formed by ACV synthetase from Acremonium chrysogenum. FEBS Lett. 1997, 414, 74–78. [Google Scholar] [CrossRef]
- Liang, M.; Lyu, H.N.; Ma, Z.Y.; Li, E.W.; Cai, L.; Yin, W.B. Genomics-driven discovery of a new cyclodepsipeptide from the guanophilic fungus Amphichorda guana. Org. Biomol. Chem. 2021, 19, 1960–1964. [Google Scholar] [CrossRef]
- Jiang, M.; Wu, Z.; Wu, Q.; Yin, H.; Guo, H.; Yuan, S.; Liu, Z.; Chen, S.; Liu, L. Amphichoterpenoids A–C, unprecedented picoline-derived meroterpenoids from the ascidian-derived fungus Amphichorda felina SYSU-MS7908. Chin. Chem. Lett. 2021, 32, 1893–1896. [Google Scholar] [CrossRef]
- Du, F.Y.; Zhang, P.; Li, X.M.; Li, C.S.; Cui, C.M.; Wang, B.G. Cyclohexadepsipeptides of the isaridin class from the marine-derived fungus Beauveria felina EN-135. J. Nat. Prod. 2014, 77, 1164–1169. [Google Scholar] [CrossRef] [PubMed]
- Raffa, N.; Keller, N.P. A call to arms: Mustering secondary metabolites for success and survival of an opportunistic pathogen. PLoS Pathog. 2019, 15, e1007606. [Google Scholar] [CrossRef] [PubMed]
- Vitale, G.A.; Coppola, D.; Palma Esposito, F.; Buonocore, C.; Ausuri, J.; Tortorella, E.; de Pascale, D. Antioxidant Molecules from Marine Fungi: Methodologies and Perspectives. Antioxidants 2020, 9, 1183. [Google Scholar] [CrossRef] [PubMed]
- Smetanina, O.F.; Yurchenko, A.N.; Girich, E.V.; Trinh, P.T.H.; Antonov, A.S.; Dyshlovoy, S.A.; von Amsberg, G.; Kim, N.Y.; Chingizova, E.A.; Pislyagin, E.A.; et al. Biologically Active Echinulin-Related Indolediketopiperazines from the Marine Sediment-Derived Fungus Aspergillus niveoglaucus. Molecules 2020, 25, 61. [Google Scholar] [CrossRef] [PubMed]
- Zhuravleva, O.I.; Antonov, A.S.; Trang, V.T.D.; Pivkin, M.V.; Khudyakova, Y.V.; Denisenko, V.A.; Popov, R.S.; Kim, N.Y.; Yurchenko, E.A.; Gerasimenko, A.V.; et al. New Deoxyisoaustamide Derivatives from the Coral-Derived Fungus Penicillium dimorphosporum KMM 4689. Mar. Drugs 2021, 19, 553. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.K.; Subramaniyan, S.A.; Hwang, I. Molecular and Cellular Response of Co-cultured Cells toward Cobalt Chloride (CoCl2)-Induced Hypoxia. ACS Omega 2019, 4, 20882–20893. [Google Scholar] [CrossRef]
- Muñoz-Sánchez, J.; Chánez-Cárdenas, M.E. The use of cobalt chloride as a chemical hypoxia model. J. Appl. Toxicol. 2019, 39, 556–570. [Google Scholar] [CrossRef]
- Bao, M.; Huang, W.; Zhao, Y.; Fang, X.; Zhang, Y.; Gao, F.; Huang, D.; Wang, B.; Shi, G. Verapamil Alleviates Myocardial Ischemia/Reperfusion Injury by Attenuating Oxidative Stress via Activation of SIRT1. Front. Pharmacol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.J.; Li, C.G. Tanshinone IIA and cryptotanshinone prevent mitochondrial dysfunction in hypoxia-induced H9c2 Cells: Association to mitochondrial ROS, intracellular nitric oxide, and calcium levels. Evid.-Based Complement. Altern. Med. 2013, 2013, 610694. [Google Scholar] [CrossRef]
- Thomas, L.W.; Ashcroft, M. Exploring the molecular interface between hypoxia-inducible factor signalling and mitochondria. Cell. Mol. Life Sci. 2019, 76, 1759–1777. [Google Scholar] [CrossRef] [PubMed]
- Osuru, H.P.; Lavallee, M.; Thiele, R.H. Molecular and Cellular Response of the Myocardium (H9C2 Cells) Towards Hypoxia and HIF-1α Inhibition. Front. Cardiovasc. Med. 2022, 9, 711421. [Google Scholar] [CrossRef]
- Baskaran, R.; Kalaiselvi, P.; Huang, C.-Y.; Padma, V.V. Neferine, a bisbenzylisoquinoline alkaloid, offers protection against cobalt chloride-mediated hypoxia-induced oxidative stress in muscle cells. Integr. Med. Res. 2015, 4, 231–241. [Google Scholar] [CrossRef]
- Baskaran, R.; Poornima, P.; Priya, L.B.; Huang, C.-Y.; Padma, V.V. Neferine prevents autophagy induced by hypoxia through activation of Akt/mTOR pathway and Nrf2 in muscle cells. Biomed. Pharmacother. 2016, 83, 1407–1413. [Google Scholar] [CrossRef]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Rosa, A.C.; Corsi, D.; Cavi, N.; Bruni, N.; Dosio, F. Superoxide Dismutase Administration: A Review of Proposed Human Uses. Molecules 2021, 26, 1844. [Google Scholar] [CrossRef] [PubMed]

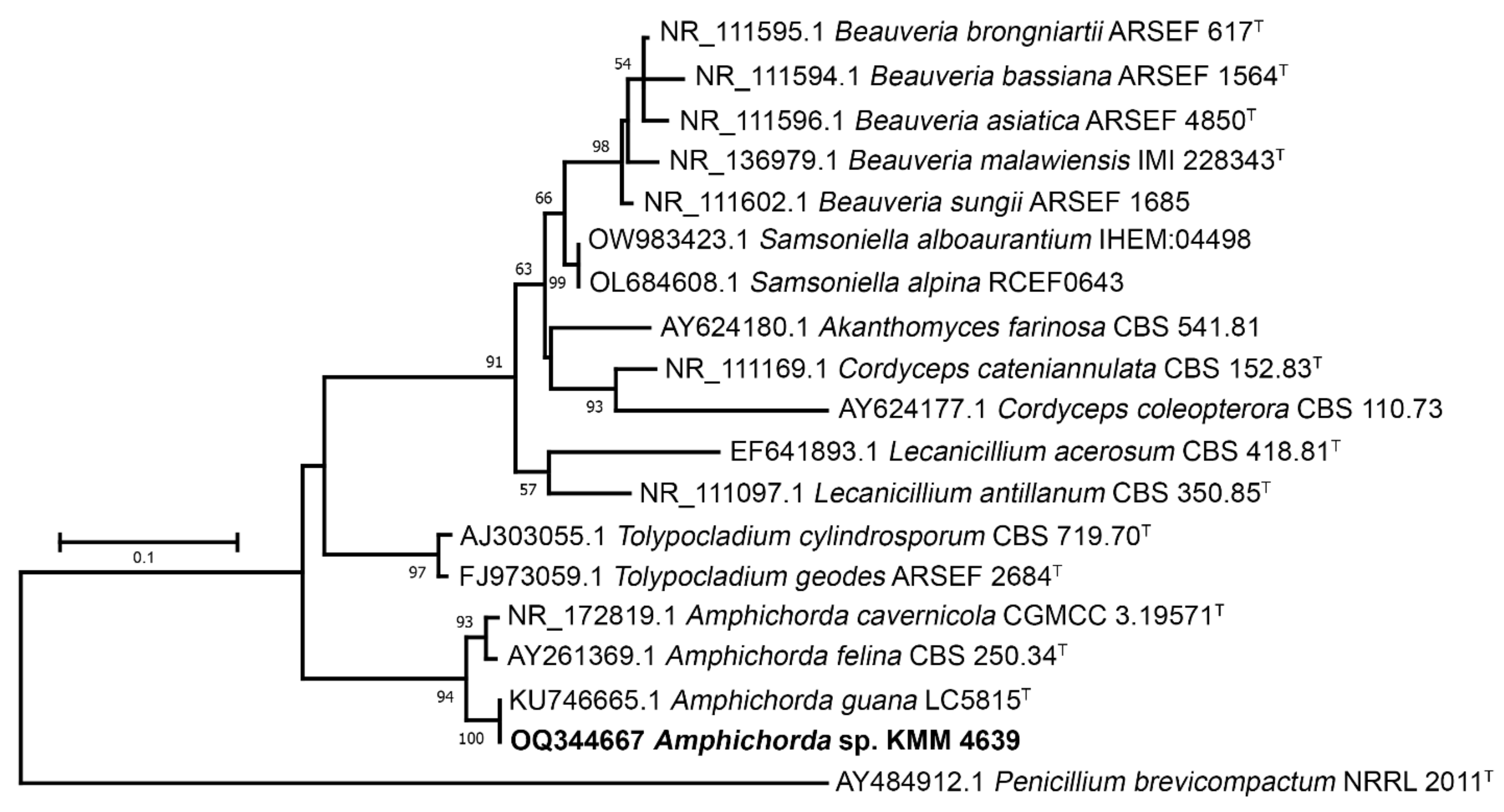




| Taxon | Collection Number | GenBank Accession Numbers | |
|---|---|---|---|
| ITS | β-Tubulin | ||
| Akanthomyces farinosa | CBS 541.81 | AY624180 | AY624218 |
| Amphichorda cavernicola | CGMCC 3.19571T | NR 172819 | ‒ |
| Amphichorda guana | CGMCC 3.17908T | KU746665 | KU746757 |
| Amphichorda felina | CBS 250.34T | AY261369 | ‒ |
| Amphichorda sp. | KMM 4639 | OQ344667 | OQ418107 |
| Tolypocladium cylindrosporum | CBS 719.70T | AJ303055 | ‒ |
| Tolypocladium geodes | ARSEF 2684T | FJ973059 | ‒ |
| Beauveria malawiensis | IMI 228343T | NR_136979 | ‒ |
| Beauveria sungii | ARSEF 1685T | NR_111602 | ‒ |
| Beauveria asiatica | ARSEF 4850T | NR_111596 | ‒ |
| Beauveria brongniartii | ARSEF 617T | NR_111595 | ‒ |
| Beauveria bassiana | ARSEF 1564T | NR_111594 | ‒ |
| Cordyceps cateniannulata | CBS 152.83T | NR_111169 | KY574462 |
| Cordyceps coleopterora | CBS 110.73 | AY624177 | AY624216 |
| Lecanicillium acerosum | CBS 418.81T | EF641893 | ‒ |
| Lecanicillium antillanum | CBS 350.85T | NR_111097 | MG993038 |
| Penicillium brevicompactum | NRRL 2011T | AY484912 | DQ645784 |
| Samsoniella alboaurantium | IHEM:04498 | OW983423 | ‒ |
| Samsoniella alpina | RCEF0643 | OL684608 | ‒ |
| Position | 1 a | 2 b | ||
|---|---|---|---|---|
| δC, Type | δH, Mult, J in Hz | δC, Type | δH, Mult, J in Hz | |
| 1 | 165.2, C | 165.1, C | ||
| 2 | 8.48, brs | 8.54, s | ||
| 3 | 156.5, C | 156.0, C | ||
| 4 | 139.2, C | 138.0, C | ||
| 5 | ||||
| 6 | 146.0, C | 134.3, C | ||
| 7 | 129.8, CH | 8.06, d (8.2) | 153.6, C | |
| 8 | 135.5, CH | 7.91, td (8.1, 1.6) | 119.1, CH | 7.40, d (8.0) |
| 9 | 130.1, CH | 7.69, td (7.9, 1.1) | 131.5, CH | 7.58, t (8.0) |
| 10 | 127.2, CH | 8.36, dd (8.0, 1.2) | 117.7, CH | 7.81, d (8.0) |
| 11 | 121.9, C | 121.9, C | ||
| 12 | 159.7, C | 159.3, C | ||
| 13 | ||||
| 14 | 61.7, CH | 5.57, dd (4.2, 0.8) | 61.8, CH | 5.55, d (4.3) |
| 15 | 33.7, CH | 2.27, m | 33.6, CH | 2.46, m |
| 16 | 16.9, CH3 | 0.92, d (6.9) | 16.8, CH3 | 0.93, d (6.9) |
| 17 | 19.0, CH3 | 1.23, d (6.9) | 19.0, CH3 | 1.23, d (6.9) |
| Position | 6 a | |
|---|---|---|
| δC, Type | δH, Mult, J in Hz | |
| 1 | 74.3, C | |
| 2 | 73.8, CH | 4.90 t (3.2) |
| 3 | 32.3, CH2 | α: 2.55, dd (14.3, 2.8) β: 1.57, dd (14.3, 3.4) |
| 4 | 57.9, C | |
| 5 | 64.3, CH | 3.08, s |
| 6 | 67.8, CH | 4.17, dd (9.9, 3.2) |
| 7 | 37.5, CH | 2.90, dd (10.2, 2.0) |
| 8 | 72.4, CH | 3.84, dt (10.4, 2.5) |
| 9 | 68.2, CH | 4.09, d (2.9) |
| 10 | 25.4, CH3 | 1.19, s |
| 11 | 22.0, CH3 | 1.40, s |
| 1′ | 87.1, C | |
| 2′ | 84.9, C | |
| 3′ | 126.5, C | |
| 4′ | 122.1, CH2 | 5.30, brs 5.22, m |
| 5′ | 23.7, CH3 | 1.90, brs |
| 1″ | 170.3, C | |
| 2″ | 20.8, CH3 | 2.11, s |
| 6-OH | OH | 2.21, d (3.6) |
| 8-OH | OH | 2.31, d (10.4) |
| Compound | Cell Lines | |||
|---|---|---|---|---|
| PC-3 | MCF-7 | SH-SY5Y | H9c2 | |
| IC50, µM | ||||
| 1 | >100 | >100 | >100 | >100 |
| 2 | >100 | >100 | >100 | >100 |
| 3 | >100 | 92.5 ± 3.1 | >100 | >100 |
| 4 | >100 | 68.7 ± 1.6 | 72.9 ± 2.8 | >100 |
| 5 | 83.8 ± 5.5 | 86.3 ± 2.3 | >100 | >100 |
| 7 | >100 | >100 | >100 | >100 |
| 8 | >100 | >100 | 93.8 ± 3.8 | >100 |
| 9 | >100 | >100 | >100 | >100 |
| 10 | >100 | >100 | >100 | >100 |
| 11 | >100 | >100 | >100 | >100 |
| Compound | Source | ||
|---|---|---|---|
| Amphichorda sp. KMM 4639 | Aspergillus carneus KMM 4638 | Co-Culture | |
| 1 | – | – | + |
| 2 | – | – | + |
| 3 | – | – | + |
| 4 | – | – | + |
| 5 | – | – | + |
| 6 | – | – | + |
| 7 | + [12] | – | + |
| 8 | – | – | + |
| 9 | – | + [5] | + |
| 10 | – | + [14] | + |
| 11 | + [15] | – | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belousova, E.B.; Zhuravleva, O.I.; Yurchenko, E.A.; Oleynikova, G.K.; Antonov, A.S.; Kirichuk, N.N.; Chausova, V.E.; Khudyakova, Y.V.; Menshov, A.S.; Popov, R.S.; et al. New Anti-Hypoxic Metabolites from Co-Culture of Marine-Derived Fungi Aspergillus carneus KMM 4638 and Amphichorda sp. KMM 4639. Biomolecules 2023, 13, 741. https://doi.org/10.3390/biom13050741
Belousova EB, Zhuravleva OI, Yurchenko EA, Oleynikova GK, Antonov AS, Kirichuk NN, Chausova VE, Khudyakova YV, Menshov AS, Popov RS, et al. New Anti-Hypoxic Metabolites from Co-Culture of Marine-Derived Fungi Aspergillus carneus KMM 4638 and Amphichorda sp. KMM 4639. Biomolecules. 2023; 13(5):741. https://doi.org/10.3390/biom13050741
Chicago/Turabian StyleBelousova, Elena B., Olesya I. Zhuravleva, Ekaterina A. Yurchenko, Galina K. Oleynikova, Alexandr S. Antonov, Natalya N. Kirichuk, Viktoria E. Chausova, Yuliya V. Khudyakova, Alexander S. Menshov, Roman S. Popov, and et al. 2023. "New Anti-Hypoxic Metabolites from Co-Culture of Marine-Derived Fungi Aspergillus carneus KMM 4638 and Amphichorda sp. KMM 4639" Biomolecules 13, no. 5: 741. https://doi.org/10.3390/biom13050741
APA StyleBelousova, E. B., Zhuravleva, O. I., Yurchenko, E. A., Oleynikova, G. K., Antonov, A. S., Kirichuk, N. N., Chausova, V. E., Khudyakova, Y. V., Menshov, A. S., Popov, R. S., Menchinskaya, E. S., Pislyagin, E. A., Mikhailov, V. V., & Yurchenko, A. N. (2023). New Anti-Hypoxic Metabolites from Co-Culture of Marine-Derived Fungi Aspergillus carneus KMM 4638 and Amphichorda sp. KMM 4639. Biomolecules, 13(5), 741. https://doi.org/10.3390/biom13050741








