An Overview of Two Old Friends Associated with Platelet Redox Signaling, the Protein Disulfide Isomerase and NADPH Oxidase
Abstract
1. Introduction
2. Methods
2.1. Bioinformatic Analysis
2.1.1. Candidate Targets Collection
2.1.2. Protein–Protein Interactions (PPIs) Network Construction
2.1.3. Biological Function and Pathway Enrichment
3. Results
3.1. Screening of Candidate Targets of Protein Disulfide-Isomerase
3.1.1. Interactional Network Analysis of Targets
3.1.2. Analysis of Biological Processes Targets
3.1.3. Biological Function Enrichment
3.2. Screening of Candidate Targets for Protein NADPH Oxidases
3.2.1. Interactional Network Analysis of Targets
3.2.2. Biological Function of NOX Isoforms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| .NO | Nitric Oxide |
| -CXXC- | catalytic vicinal active site thiols |
| AAR2 | AAR2 Splicing Factor |
| BP | Biological Processes |
| CC | Cellular Component |
| CUL1 | Cullin 1 |
| CUL2 | Cullin 2 |
| CUL3 | Cullin 3 |
| DUOX1 | Dual Oxidase 1 |
| DUOX2 | Dual Oxidase 2 |
| ER | Endoplasmic Reticulum |
| ERP29 | Endoplasmic Reticulum Protein 29 |
| ERP44 | Endoplasmic Reticulum Protein 44 |
| GO | Gene Ontology |
| H2O2 | Hydrogen Peroxide |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KRAS | KRAS Proto-Oncogene, GTPase |
| MF | Molecular Functions |
| NOX | NADPH oxidase |
| NOX1 | NADPH Oxidase 1 |
| NOX2 | NADPH Oxidase 2 |
| NOX3 | NADPH Oxidase 3 |
| NOX4 | NADPH Oxidase 4 |
| NOX5 | NADPH Oxidase 5 |
| O2.− | Superoxide |
| P4HB | Prolyl 4-Hydroxylase Subunit Beta |
| PDI | Protein Disulphide Isomerase |
| PDIA3 | Protein Disulfide Isomerase Family A Member 3 |
| PDIA4 | Protein Disulfide Isomerase Family A Member 4 |
| PDIA6 | Protein Disulfide Isomerase Family A Member 6 |
| RAC1 | Rac Family Small GTPase 1 |
| RAC2 | Rac Family Small GTPase 2 |
| ROS | Reactive Oxygen Species |
| SOD1 | Superoxide Dismutase 1 |
| TMX3 | Thioredoxin Related Transmembrane Protein 3 |
| TNFRSF1A | Tumor Necrosis Factor Receptor 1 |
| UBE2M | Ubiquitin Conjugating Enzyme E2 M |
References
- Eitan, F.; Mutaz, D. Oxidative stress and platelet dysfunction. Thromb. Haemost. Res. 2018, 2, 1017. [Google Scholar]
- Freedman, J.E. Oxidative Stress and Platelets. Arterioscler. Thromb. Vasc. Biol. 2008, 28, s11–s16. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Arthur, J.F.; Gardiner, E.E.; Andrews, R.K.; Zeng, L.; Xu, K. Regulation of platelet activation and thrombus formation by reactive oxygen species. Redox Biol. 2018, 14, 126–130. [Google Scholar] [CrossRef]
- Arauna, D.; Garcia, F.; Rodriguez-Manas, L.; Marrugat, J.; Saez, C.; Alarcon, M.; Wehinger, S.; Espinosa-Parrilla, Y.; Palomo, I.; Fuentes, E. Older adults with frailty syndrome present an altered platelet function and an increased level of circulating oxidative stress and mitochondrial dysfunction biomarker GDF-15. Free Radic. Biol. Med. 2020, 149, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Masselli, E.; Pozzi, G.; Vaccarezza, M.; Mirandola, P.; Galli, D.; Vitale, M.; Carubbi, C.; Gobbi, G. ROS in platelet biology: Functional aspects and methodological insights. Int. J. Mol. Sci. 2020, 21, 4866. [Google Scholar] [CrossRef]
- Bae, Y.S.; Oh, H.; Rhee, S.G.; Yoo, Y.D. Regulation of reactive oxygen species generation in cell signaling. Mol. Cells 2011, 32, 491–509. [Google Scholar] [CrossRef]
- Trevelin, S.C.; Lopes, L.R. Protein disulfide isomerase and Nox: New partners in redox signaling. Curr. Pharm. Des. 2015, 21, 5951–5963. [Google Scholar] [CrossRef]
- Ali Khan, H.; Mutus, B. Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front. Chem. 2014, 2, 70. [Google Scholar] [CrossRef]
- Soares Moretti, A.I.; Martins Laurindo, F.R. Protein disulfide isomerases: Redox connections in and out of the endoplasmic reticulum. Arch. Biochem. Biophys. 2017, 617, 106–119. [Google Scholar] [CrossRef]
- Ferrari, D.M.; Söling, H.-D. The protein disulphide-isomerase family: Unravelling a string of folds. Biochem. J. 1999, 339, 1–10. [Google Scholar] [CrossRef]
- Gaspar, R.S.; Sage, T.; Little, G.; Kriek, N.; Pula, G.; Gibbins, J.M. Protein disulphide isomerase and NADPH oxidase 1 cooperate to control platelet function and are associated with cardiometabolic disease risk factors. Antioxidants 2021, 10, 497. [Google Scholar] [CrossRef] [PubMed]
- Przyborowski, K.; Kurpinska, A.; Wojkowska, D.; Kaczara, P.; Suraj-Prazmowska, J.; Karolczak, K.; Malinowska, A.; Pelesz, A.; Kij, A.; Kalvins, I.; et al. Protein disulfide isomerase-A1 regulates intraplatelet reactive oxygen species–thromboxane A2-dependent pathway in human platelets. J. Thromb. Haemost. 2022, 20, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.E.; Cifuentes, M.E.; Kiarash, A.; Quinn, M.T.; Pagano, P.J. Novel Competitive Inhibitor of NAD(P)H Oxidase Assembly Attenuates Vascular O2− and Systolic Blood Pressure in Mice. Circ. Res. 2001, 89, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, E.; Gibbins, J.M.; Holbrook, L.M.; Palomo, I. NADPH oxidase 2 (NOX2): A key target of oxidative stress-mediated platelet activation and thrombosis. Trends Cardiovasc. Med. 2018, 28, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Leto, T.L.; Morand, S.; Hurt, D.; Ueyama, T. Targeting and regulation of reactive oxygen species generation by Nox family NADPH oxidases. Antioxid. Redox Signal. 2009, 11, 2607–2619. [Google Scholar] [CrossRef]
- Xu, S.; Sankar, S.; Neamati, N. Protein disulfide isomerase: A promising target for cancer therapy. Drug Discov. Today 2014, 19, 222–240. [Google Scholar] [CrossRef]
- Flaumenhaft, R. Protein disulfide isomerase as an antithrombotic target. Trends Cardiovasc. Med. 2013, 23, 264–268. [Google Scholar] [CrossRef]
- Powell, L.E.; Foster, P.A. Protein disulphide isomerase inhibition as a potential cancer therapeutic strategy. Cancer Med. 2021, 10, 2812–2825. [Google Scholar] [CrossRef]
- Radermacher, K.A.; Wingler, K.; Kleikers, P.; Altenhöfer, S.; JR Hermans, J.; Kleinschnitz, C.; HHW Schmidt, H. The 1027th target candidate in stroke: Will NADPH oxidase hold up? Exp. Transl. Stroke Med. 2012, 4, 1–11. [Google Scholar] [CrossRef]
- Panday, A.; Sahoo, M.K.; Osorio, D.; Batra, S. NADPH oxidases: An overview from structure to innate immunity-associated pathologies. Cell. Mol. Immunol. 2015, 12, 5–23. [Google Scholar] [CrossRef]
- Joshi, S.; Khan, S.R. NADPH oxidase: A therapeutic target for hyperoxaluria-induced oxidative stress–an update. Future Med. Chem. 2019, 11, 2975–2978. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.-J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Kotlyar, M.; Pastrello, C.; Sheahan, N.; Jurisica, I. Integrated interactions database: Tissue-specific view of the human and model organism interactomes. Nucleic Acids Res. 2015, 44, D536–D541. [Google Scholar] [CrossRef] [PubMed]
- Alonso-López, D.; Campos-Laborie, F.J.; Gutiérrez, M.A.; Lambourne, L.; Calderwood, M.A.; Vidal, M.; De Las Rivas, J. APID database: Redefining protein–protein interaction experimental evidences and binary interactomes. Database 2019, 2019, baz005. [Google Scholar] [CrossRef]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2013, 42, D358–D363. [Google Scholar] [CrossRef]
- Boyanova, D.; Nilla, S.; Birschmann, I.; Dandekar, T.; Dittrich, M. PlateletWeb: A systems biologic analysis of signaling networks in human platelets. Blood 2012, 119, e22–e34. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Xin, Z.; Cai, Y.; Dang, L.T.; Burke, H.M.S.; Revote, J.; Charitakis, N.; Bienroth, D.; Nim, H.T.; Li, Y.-F.; Ramialison, M. MonaGO: A novel gene ontology enrichment analysis visualisation system. BMC Bioinform. 2022, 23, 69. [Google Scholar] [CrossRef]
- Li, M.; Ren, C.; Wu, C.; Li, X.; Li, X.; Mao, W. Bioinformatics Analysis Reveals Diagnostic Markers and Vital Pathways Involved in Acute Coronary Syndrome. Cardiol. Res. Pract. 2020, 2020, 3162581. [Google Scholar] [CrossRef]
- Di Lena, P.; Martelli, P.L.; Fariselli, P.; Casadio, R. NET-GE: A novel NETwork-based Gene Enrichment for detecting biological processes associated to Mendelian diseases. BMC Genom. 2015, 16 (Suppl. S8), S6. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, M.; Verissimo-Filho, S.; Wittig, I.; Schickling, B.M.; Hahner, F.; Schurmann, C.; Netto, L.E.S.; Rosa, J.C.; Brandes, R.P.; Sartoretto, S.; et al. Redox Activation of Nox1 (NADPH Oxidase 1) Involves an Intermolecular Disulfide Bond Between Protein Disulfide Isomerase and p47(phox) in Vascular Smooth Muscle Cells. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 224–236. [Google Scholar] [CrossRef] [PubMed]
- de A. Paes, A.M.; Veríssimo-Filho, S.; Guimaraes, L.L.; Silva, A.C.B.; Takiuti, J.T.; Santos, C.X.; Janiszewski, M.; Laurindo, F.R.; Lopes, L.R. Protein disulfide isomerase redox-dependent association with p47phox: Evidence for an organizer role in leukocyte NADPH oxidase activation. J. Leukoc. Biol. 2011, 90, 799–810. [Google Scholar]
- Gaspar, R.S.; Mansilla, S.; Vieira, V.A.; da Silva, L.B.; Gibbins, J.M.; Castro, L.; Trostchansky, A.; Paes, A.M.A. The protein disulphide isomerase inhibitor CxxCpep modulates oxidative burst and mitochondrial function in platelets. Free Radic. Biol. Med. 2021, 172, 668–674. [Google Scholar] [CrossRef]
- Laurindo, F.R.; Pescatore, L.A.; Fernandes Dde, C. Protein disulfide isomerase in redox cell signaling and homeostasis. Free Radic. Biol. Med. 2012, 52, 1954–1969. [Google Scholar] [CrossRef]
- Walsh, T.G.; Berndt, M.C.; Carrim, N.; Cowman, J.; Kenny, D.; Metharom, P. The role of Nox1 and Nox2 in GPVI-dependent platelet activation and thrombus formation. Redox Biol. 2014, 2, 178–186. [Google Scholar] [CrossRef]
- Piguet, P.F.; Vesin, C.; Da Kan, C. Activation of platelet caspases by TNF and its consequences for kinetics. Cytokine 2002, 18, 222–230. [Google Scholar] [CrossRef]
- Limb, G.; Webster, L.; Soomro, H.; Janikoun, S.; Shilling, J. Platelet expression of tumour necrosis factor-alpha (TNF-α), TNF receptors and intercellular adhesion molecule-1 (ICAM-1) in patients with proliferative diabetic retinopathy. Clin. Exp. Immunol. 1999, 118, 213–218. [Google Scholar] [CrossRef]
- Inaba, K. Disulfide bond formation system in Escherichia coli. J. Biochem. 2009, 146, 591–597. [Google Scholar] [CrossRef] [PubMed]
- LaMantia, M.; Miura, T.; Tachikawa, H.; Kaplan, H.A.; Lennarz, W.J.; Mizunaga, T. Glycosylation site binding protein and protein disulfide isomerase are identical and essential for cell viability in yeast. Proc. Natl. Acad. Sci. USA 1991, 88, 4453–4457. [Google Scholar] [CrossRef] [PubMed]
- Hatahet, F.; Ruddock, L.W. Protein disulfide isomerase: A critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 2009, 11, 2807–2850. [Google Scholar] [CrossRef]
- Irvine, A.G.; Wallis, A.K.; Sanghera, N.; Rowe, M.L.; Ruddock, L.W.; Howard, M.J.; Williamson, R.A.; Blindauer, C.A.; Freedman, R.B. Protein disulfide-isomerase interacts with a substrate protein at all stages along its folding pathway. PLoS ONE 2014, 9, e82511. [Google Scholar] [CrossRef] [PubMed]
- Moretti, A.I.S.; Pavanelli, J.C.; Nolasco, P.; Leisegang, M.S.; Tanaka, L.Y.; Fernandes, C.G.; Wosniak, J., Jr.; Kajihara, D.; Dias, M.H.; Fernandes, D.C.; et al. Conserved Gene Microsynteny Unveils Functional Interaction Between Protein Disulfide Isomerase and Rho Guanine-Dissociation Inhibitor Families. Sci. Rep. 2017, 7, 17262. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, W.; Ren, J.; Fang, J.; Ke, H.; Gong, W.; Feng, W.; Wang, C.C. Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase. Antioxid. Redox Signal. 2013, 19, 36–45. [Google Scholar] [CrossRef]
- Zhao, G.; Lu, H.; Li, C. Proapoptotic activities of protein disulfide isomerase (PDI) and PDIA3 protein, a role of the Bcl-2 protein Bak. J. Biol. Chem. 2015, 290, 8949–8963. [Google Scholar] [CrossRef]
- Zhang, Y.; Baig, E.; Williams, D.B. Functions of ERp57 in the folding and assembly of major histocompatibility complex class I molecules. J. Biol. Chem. 2006, 281, 14622–14631. [Google Scholar] [CrossRef] [PubMed]
- Young, H.S.; McGowan, L.M.; Jepson, K.A.; Adams, J.C. Impairment of cell adhesion and migration by inhibition of protein disulphide isomerases in three breast cancer cell lines. Biosci. Rep. 2020, 40, BSR20193271. [Google Scholar] [CrossRef]
- Wang, L.; Essex, D. A new antithrombotic strategy: Inhibition of the C-terminal active site of protein disulfide isomerase. J. Thromb. Haemost. JTH 2017, 15, 770. [Google Scholar] [CrossRef]
- Furlan-Freguia, C.; Marchese, P.; Gruber, A.; Ruggeri, Z.M.; Ruf, W. P2X7 receptor signaling contributes to tissue factor-dependent thrombosis in mice. J. Clin. Investig. 2011, 121, 2932–2944. [Google Scholar] [CrossRef]
- Valdivia, A.; Duran, C.; San Martin, A. The role of Nox-mediated oxidation in the regulation of cytoskeletal dynamics. Curr. Pharm. Des. 2015, 21, 6009–6022. [Google Scholar] [CrossRef]
- Sonkar, V.K.; Kumar, R.; Jensen, M.; Wagner, B.A.; Sharathkumar, A.A.; Miller, F.J., Jr.; Fasano, M.; Lentz, S.R.; Buettner, G.R.; Dayal, S. Nox2 NADPH oxidase is dispensable for platelet activation or arterial thrombosis in mice. Blood Adv. 2019, 3, 1272–1284. [Google Scholar] [CrossRef] [PubMed]
- Pescatore, L.A.; Bonatto, D.; Forti, F.L.; Sadok, A.; Kovacic, H.; Laurindo, F.R. Protein disulfide isomerase is required for platelet-derived growth factor-induced vascular smooth muscle cell migration, Nox1 NADPH oxidase expression, and RhoGTPase activation. J. Biol. Chem. 2012, 287, 29290–29300. [Google Scholar] [CrossRef] [PubMed]
- De Bessa, T.; Oliveira, P.; Wosniak, J.; Debbas, V.; Santos, C.; Shah, A.; Laurindo, F.R. Protein Disulfide Isomerase-A1 (PDIA1) Governs the Morphology of Endoplasmic Reticulum-Plasma Membrane Contact Sites. Free Radic. Biol. Med. 2022, 180, s29. [Google Scholar] [CrossRef]
- Sirker, A.; Zhang, M.; Shah, A.M. NADPH oxidases in cardiovascular disease: Insights from in vivo models and clinical studies. Basic Res. Cardiol. 2011, 106, 735–747. [Google Scholar] [CrossRef]
- Rada, B.; Leto, T.L. Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib. Microbiol. 2008, 15, 164–187. [Google Scholar] [CrossRef]
- Choo, H.J.; Saafir, T.B.; Mkumba, L.; Wagner, M.B.; Jobe, S.M. Mitochondrial calcium and reactive oxygen species regulate agonist-initiated platelet phosphatidylserine exposure. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2946–2955. [Google Scholar] [CrossRef]
- Gyulkhandanyan, A.V.; Mutlu, A.; Freedman, J.; Leytin, V. Markers of platelet apoptosis: Methodology and applications. J. Thromb. Thrombolysis 2012, 33, 397–411. [Google Scholar] [CrossRef]
- McArthur, K.; Chappaz, S.; Kile, B.T. Apoptosis in megakaryocytes and platelets: The life and death of a lineage. Blood 2018, 131, 605–610. [Google Scholar] [CrossRef]
- Gaspar, R.S.; da Silva, S.A.; Stapleton, J.; Fontelles, J.L.L.; Sousa, H.R.; Chagas, V.T.; Alsufyani, S.; Trostchansky, A.; Gibbins, J.M.; Paes, A.M.A. Myricetin, the Main Flavonoid in Syzygium cumini Leaf, Is a Novel Inhibitor of Platelet Thiol Isomerases PDI and ERp5. Front. Pharm. 2019, 10, 1678. [Google Scholar] [CrossRef]
- Santos, G.B.; Gonzalez-Perilli, L.; Mastrogiovanni, M.; Aicardo, A.; Cerdeira, C.D.; Trostchansky, A.; Brigagao, M. Nitroxide 4-hydroxy-2,2’,6,6’-tetramethylpiperidine 1-oxyl (Tempol) inhibits the reductase activity of protein disulfide isomerase via covalent binding to the Cys(400) residue on CXXC redox motif at the a’active site. Chem.-Biol. Interact. 2017, 272, 117–124. [Google Scholar] [CrossRef]
- Gonzalez-Perilli, L.; Mastrogiovanni, M.; de Castro Fernandes, D.; Rubbo, H.; Laurindo, F.; Trostchansky, A. Nitroarachidonic acid (NO(2)AA) inhibits protein disulfide isomerase (PDI) through reversible covalent adduct formation with critical cysteines. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
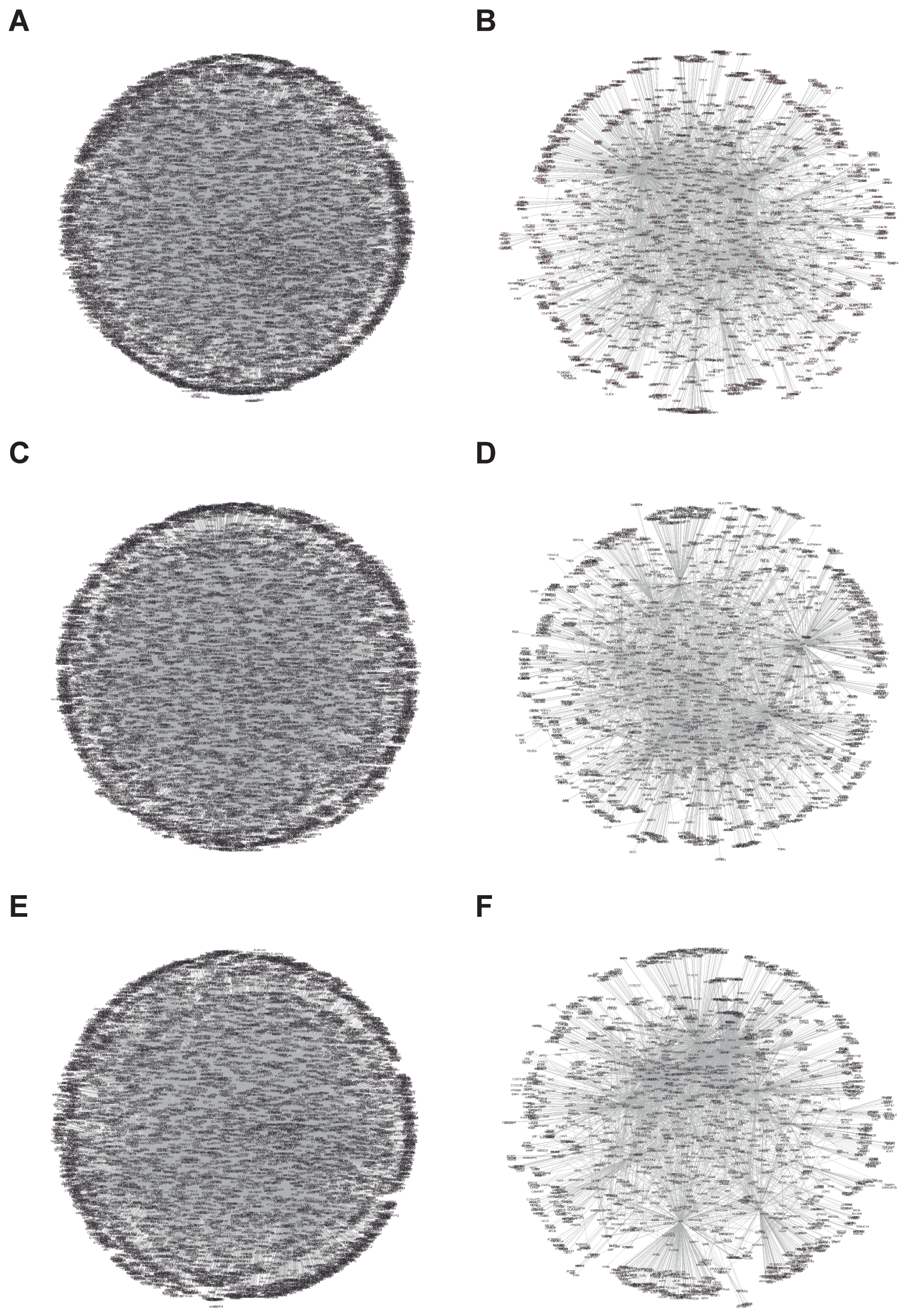

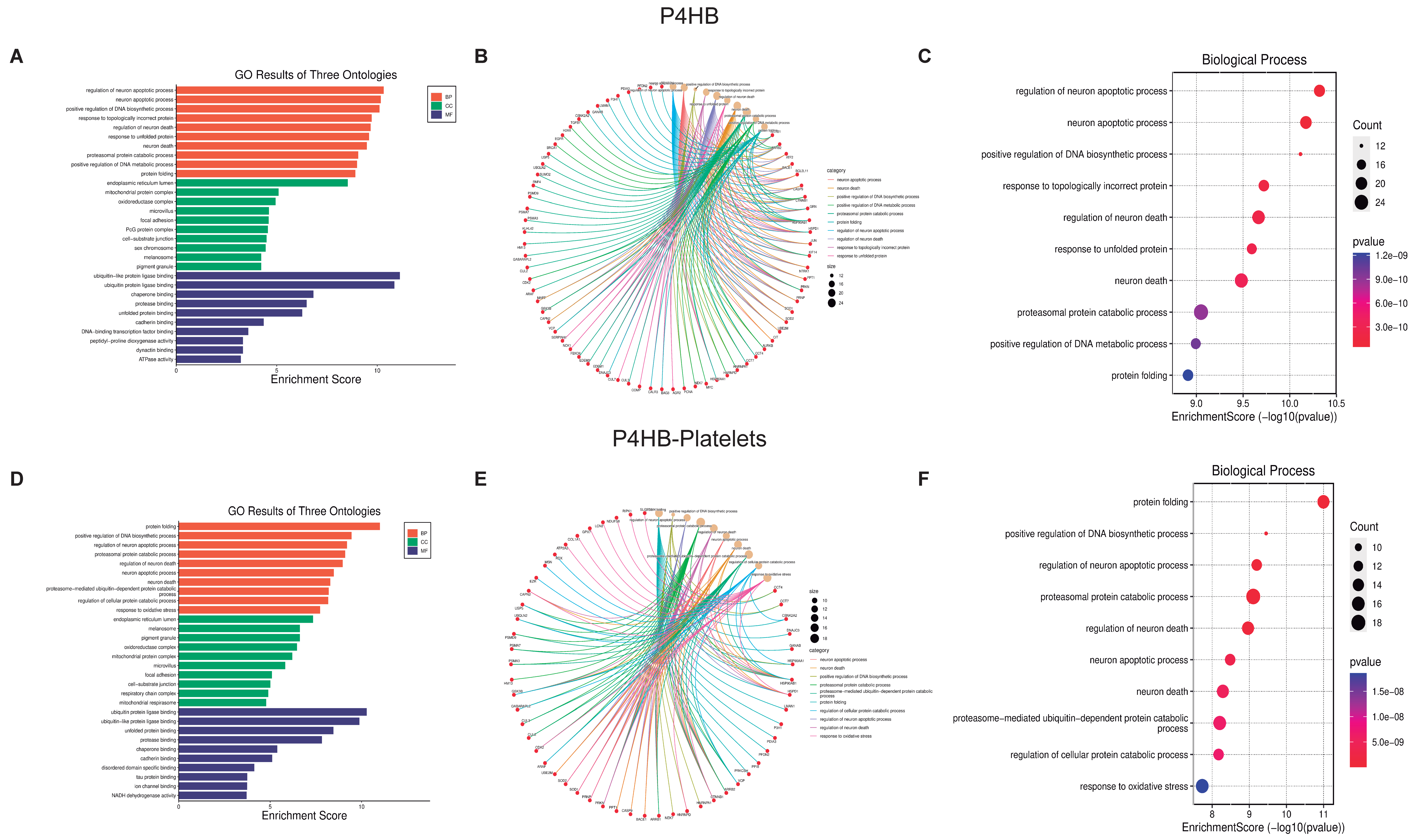
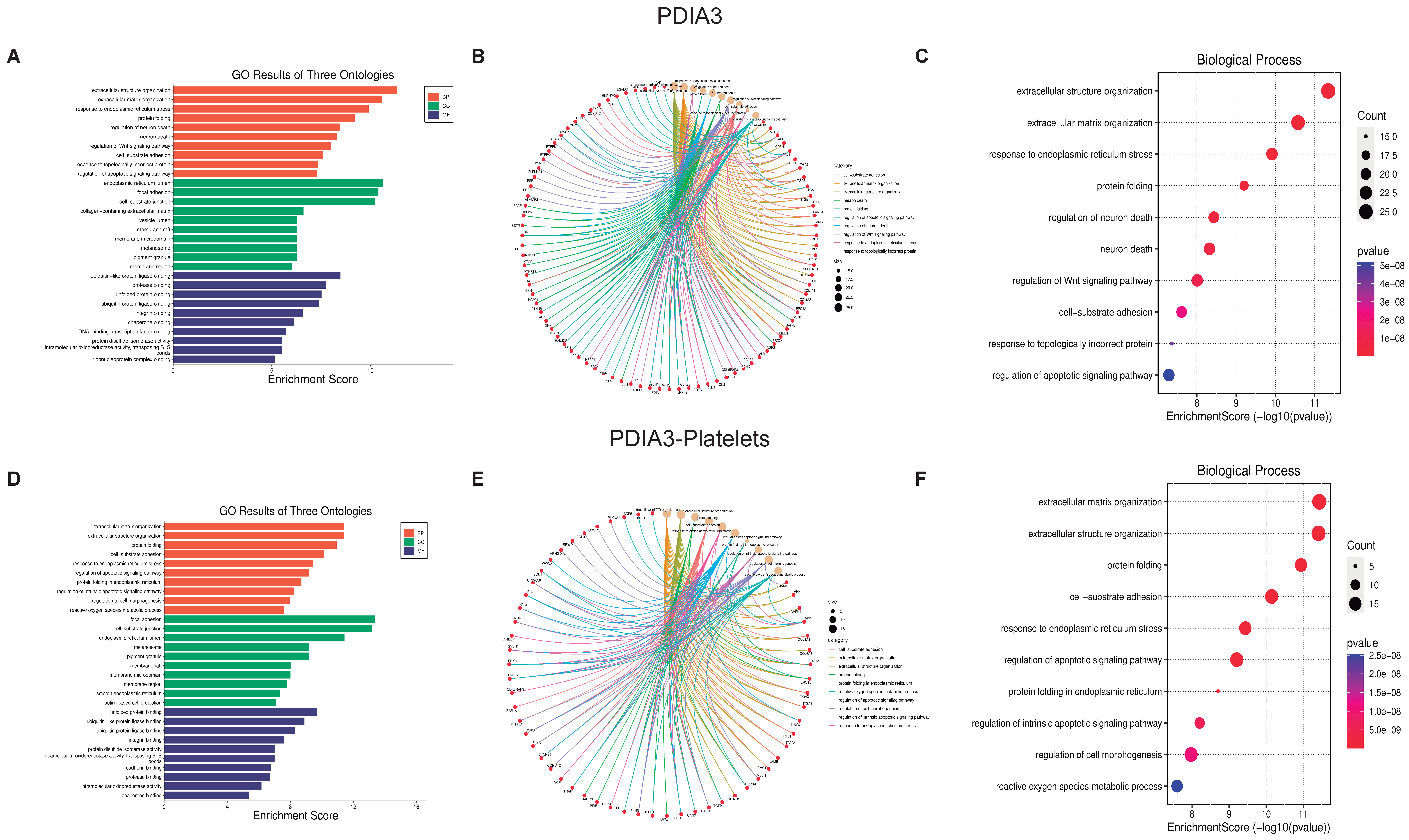
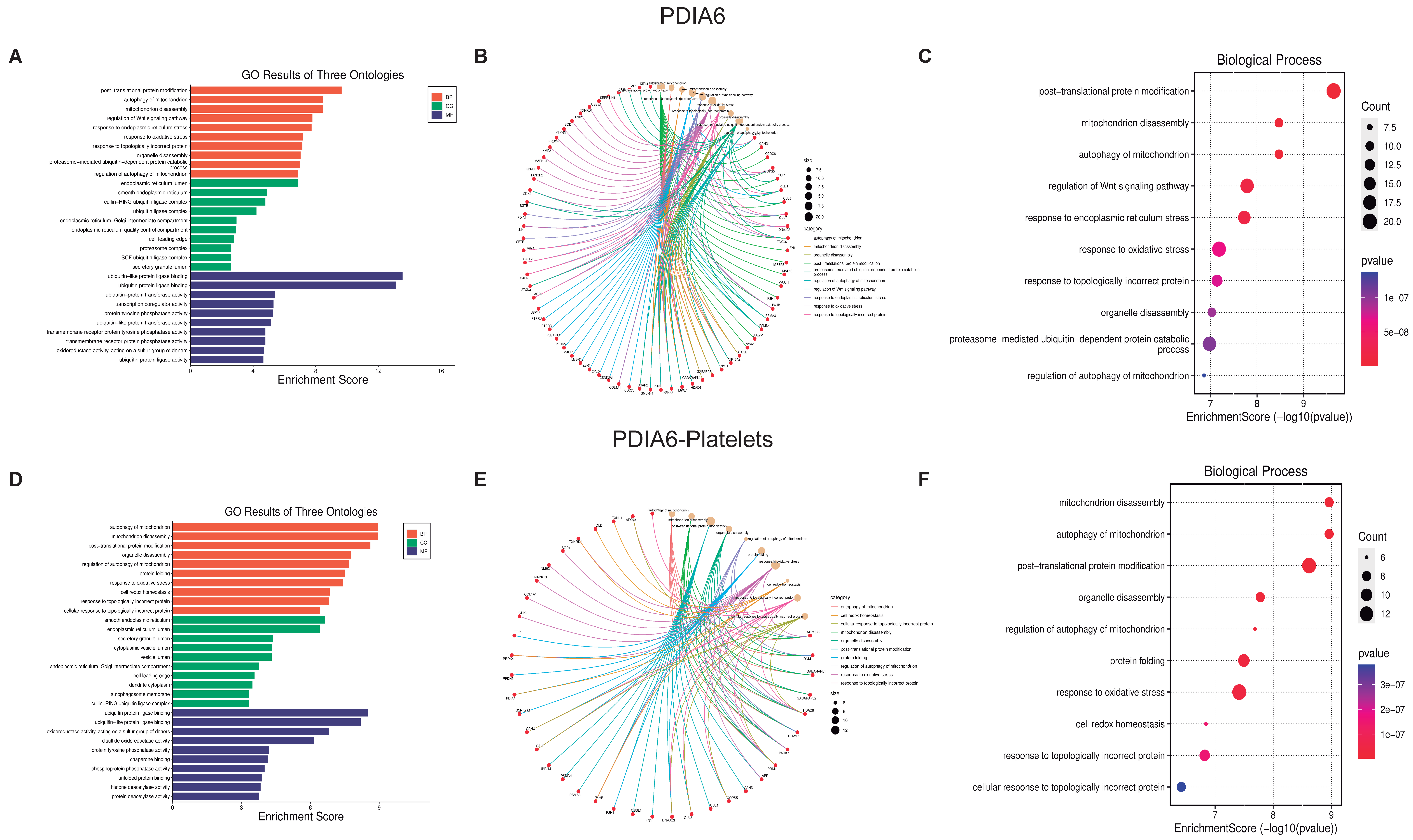
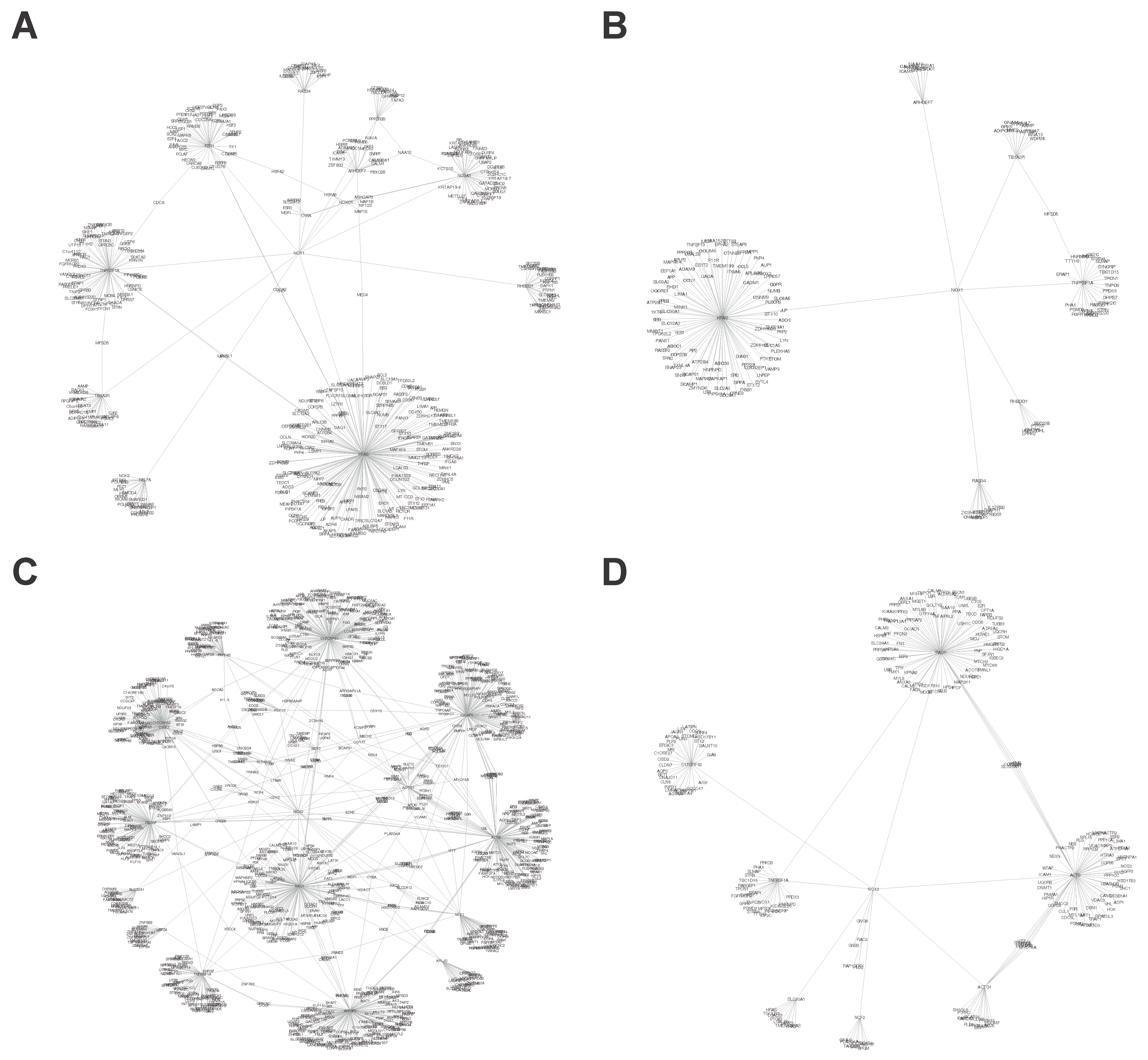
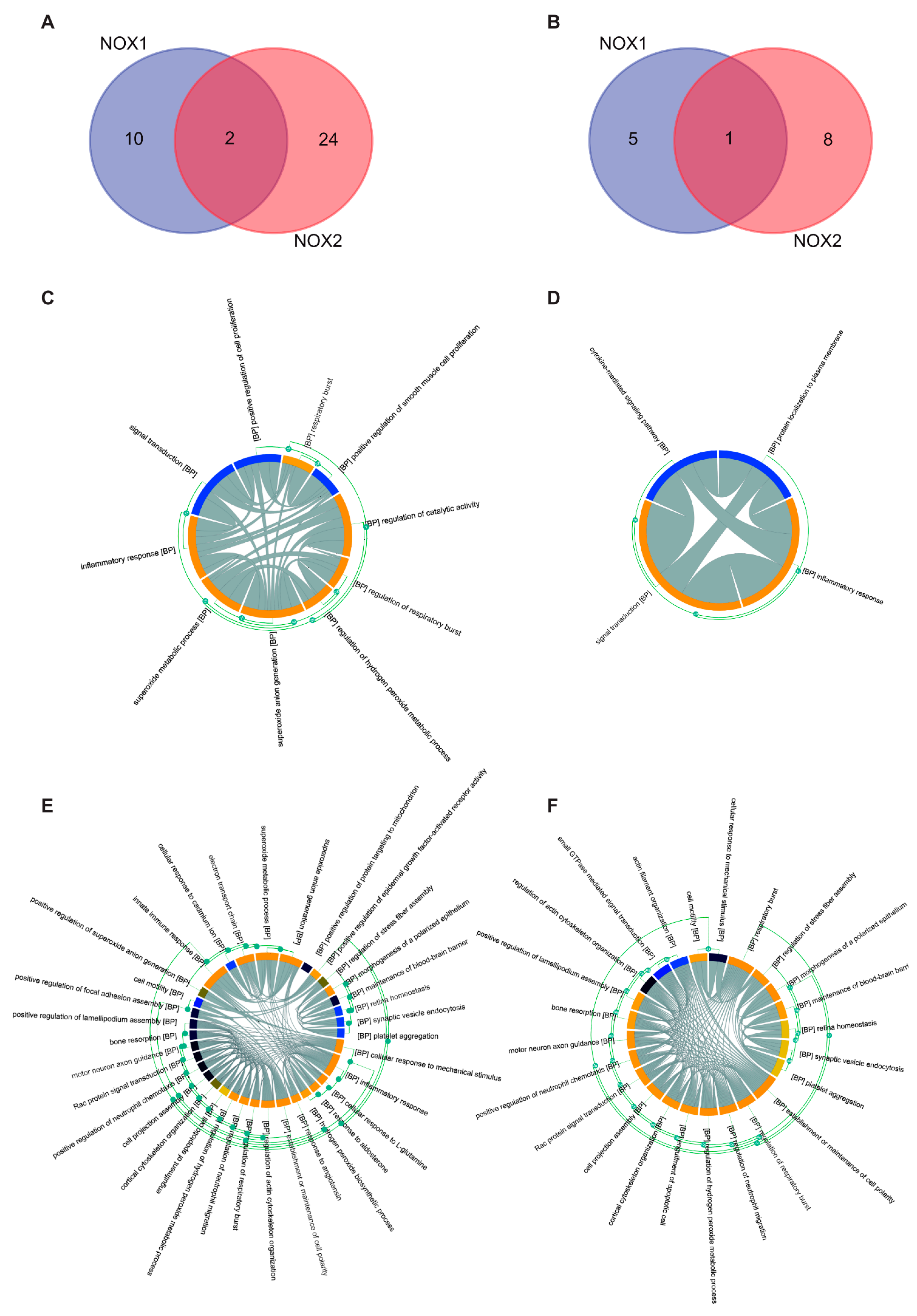
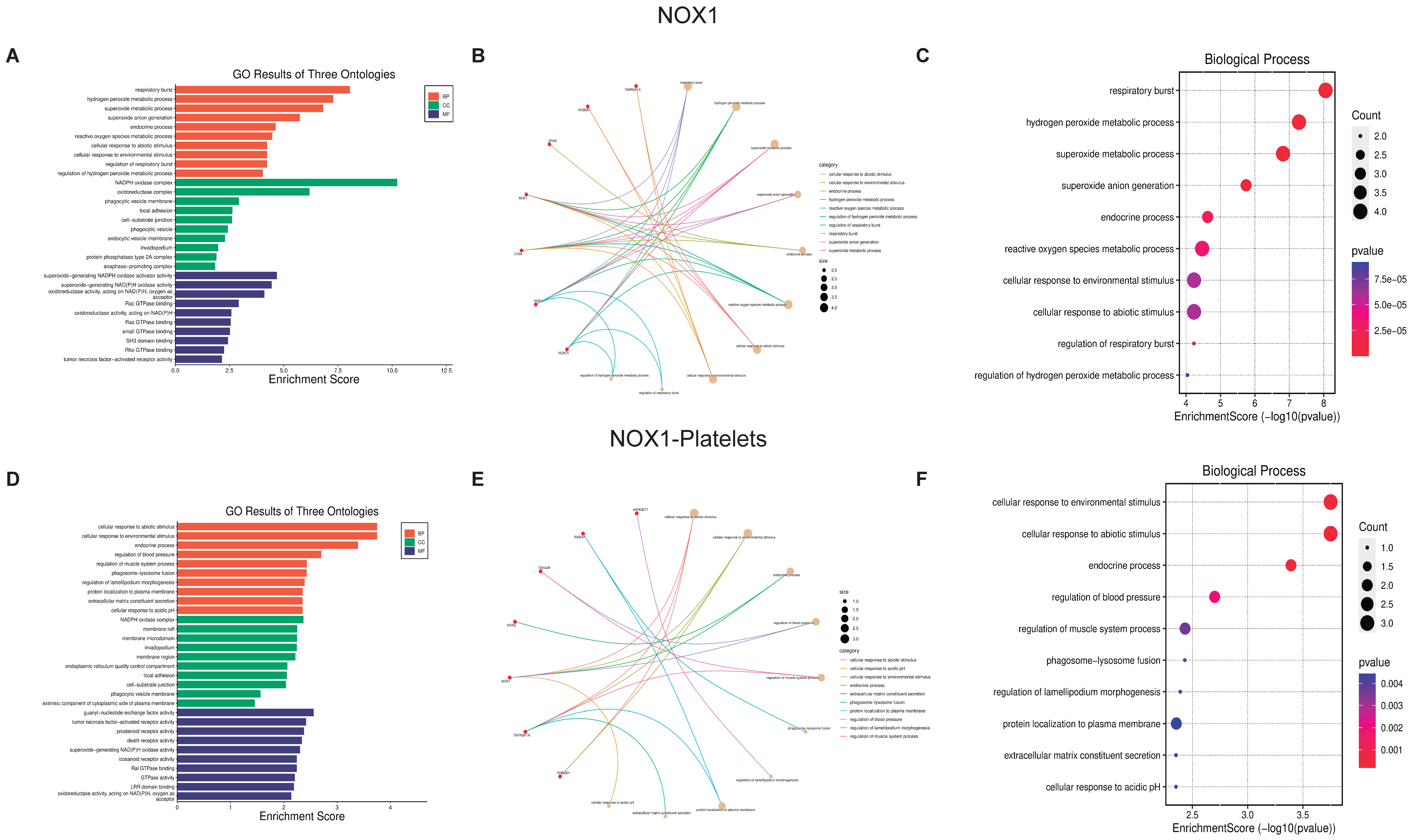
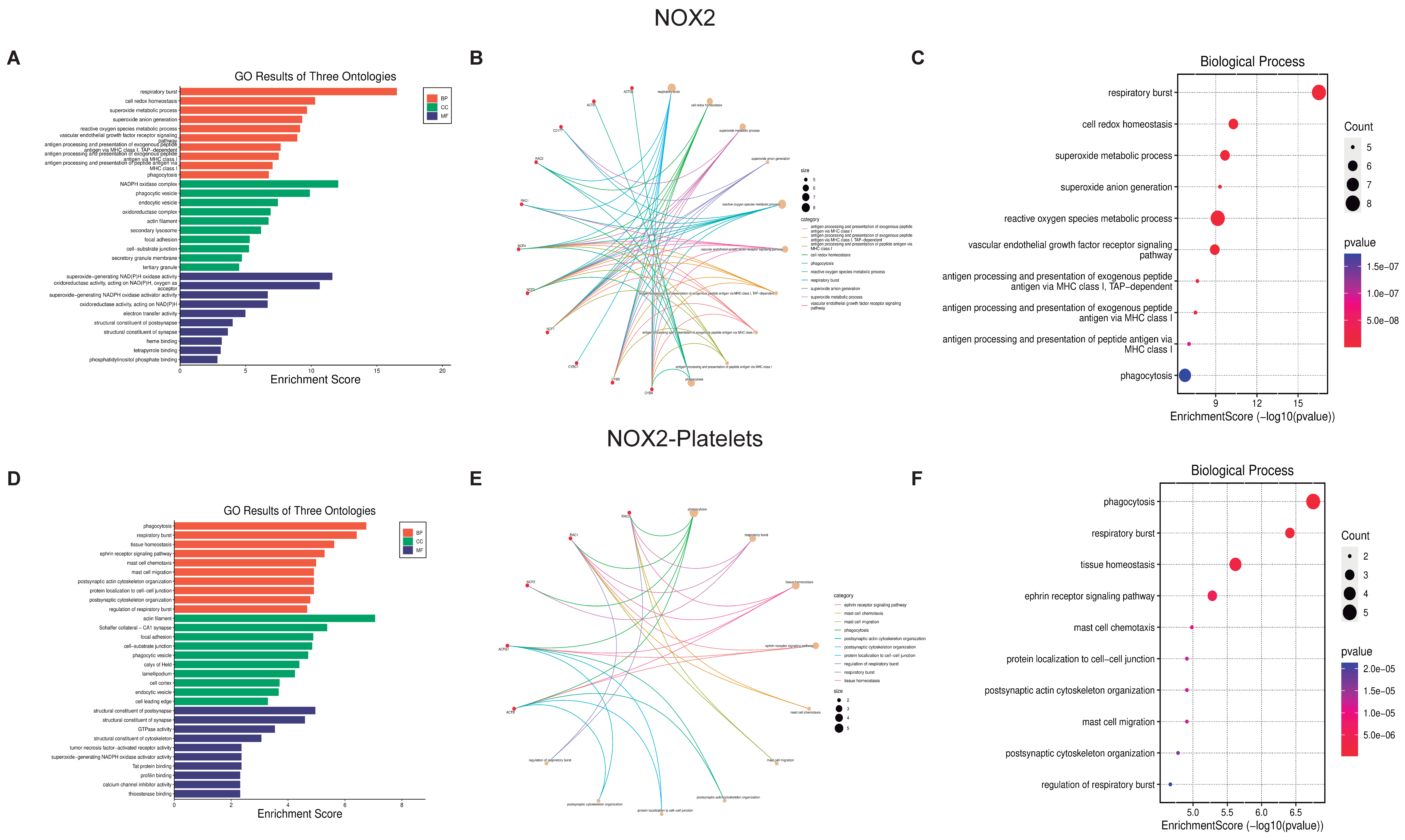

| Protein | Interaction | Node | Edges |
|---|---|---|---|
| P4HB | All | 8185 | 20,975 |
| Platelet | 1713 | 3842 | |
| PDIA3 | All | 7988 | 18,831 |
| Platelet | 1859 | 4001 | |
| PDIA6 | All | 7542 | 17,855 |
| Platelet | 1868 | 4299 | |
| NOX1 | All | 480 | 499 |
| Platelet | 158 | 161 | |
| NOX2 | All | 1181 | 1349 |
| Platelet | 242 | 254 |
| Protein | Interaction | Count |
|---|---|---|
| P4HB | All | 284 |
| Platelet | 181 | |
| PDIA3 | All | 309 |
| Platelet | 207 | |
| PDIA6 | All | 177 |
| Platelet | 105 | |
| NOX1 | All | 14 |
| Platelet | 4 | |
| NOX2 | All | 40 |
| Platelet | 27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trostchansky, A.; Alarcon, M. An Overview of Two Old Friends Associated with Platelet Redox Signaling, the Protein Disulfide Isomerase and NADPH Oxidase. Biomolecules 2023, 13, 848. https://doi.org/10.3390/biom13050848
Trostchansky A, Alarcon M. An Overview of Two Old Friends Associated with Platelet Redox Signaling, the Protein Disulfide Isomerase and NADPH Oxidase. Biomolecules. 2023; 13(5):848. https://doi.org/10.3390/biom13050848
Chicago/Turabian StyleTrostchansky, Andrés, and Marcelo Alarcon. 2023. "An Overview of Two Old Friends Associated with Platelet Redox Signaling, the Protein Disulfide Isomerase and NADPH Oxidase" Biomolecules 13, no. 5: 848. https://doi.org/10.3390/biom13050848
APA StyleTrostchansky, A., & Alarcon, M. (2023). An Overview of Two Old Friends Associated with Platelet Redox Signaling, the Protein Disulfide Isomerase and NADPH Oxidase. Biomolecules, 13(5), 848. https://doi.org/10.3390/biom13050848









