Ribosomal Protein uS5 and Friends: Protein–Protein Interactions Involved in Ribosome Assembly and Beyond
Abstract
:1. Introduction
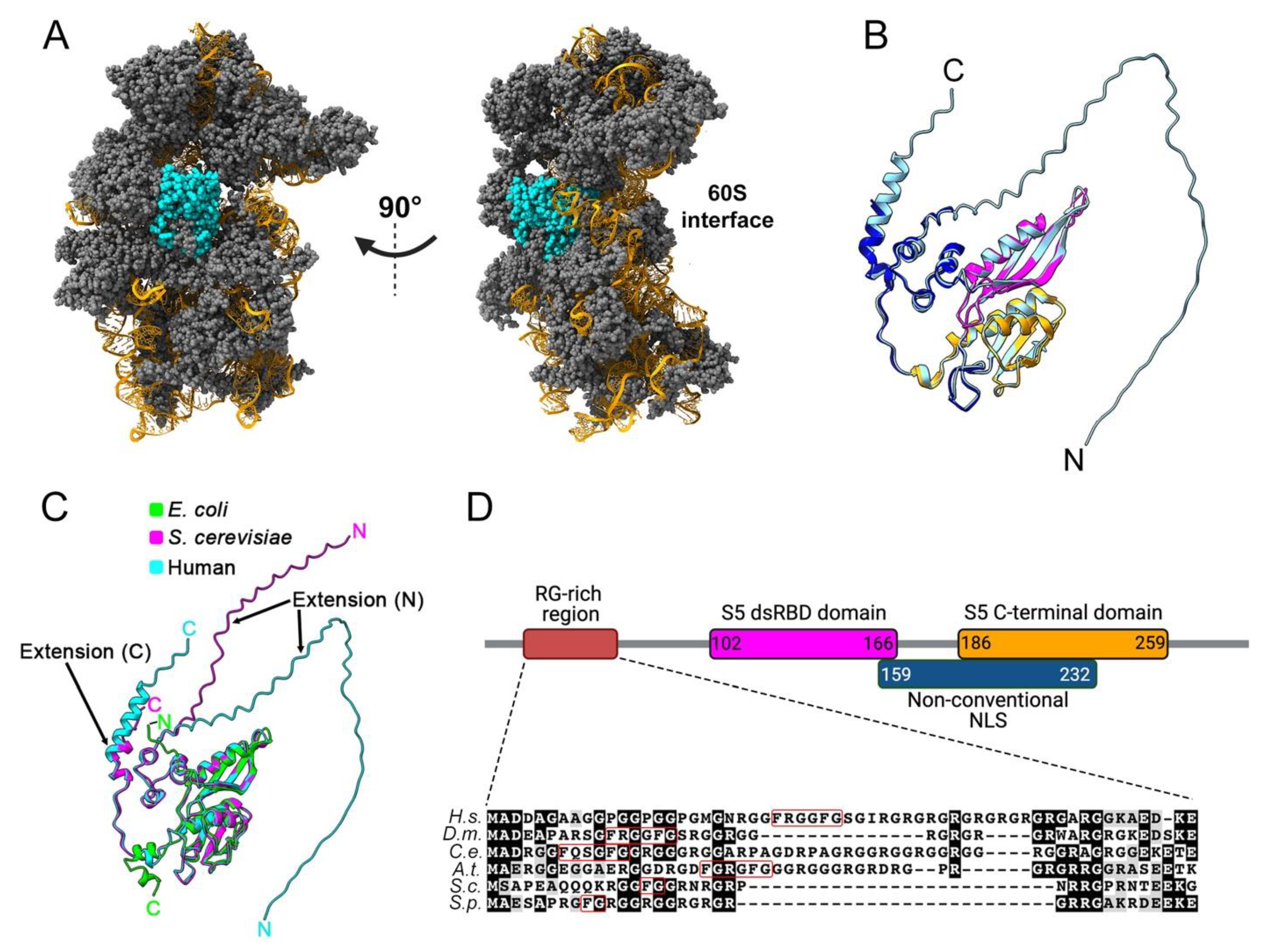
2. Structural Features of Eukaryotic uS5 and Role in Translation
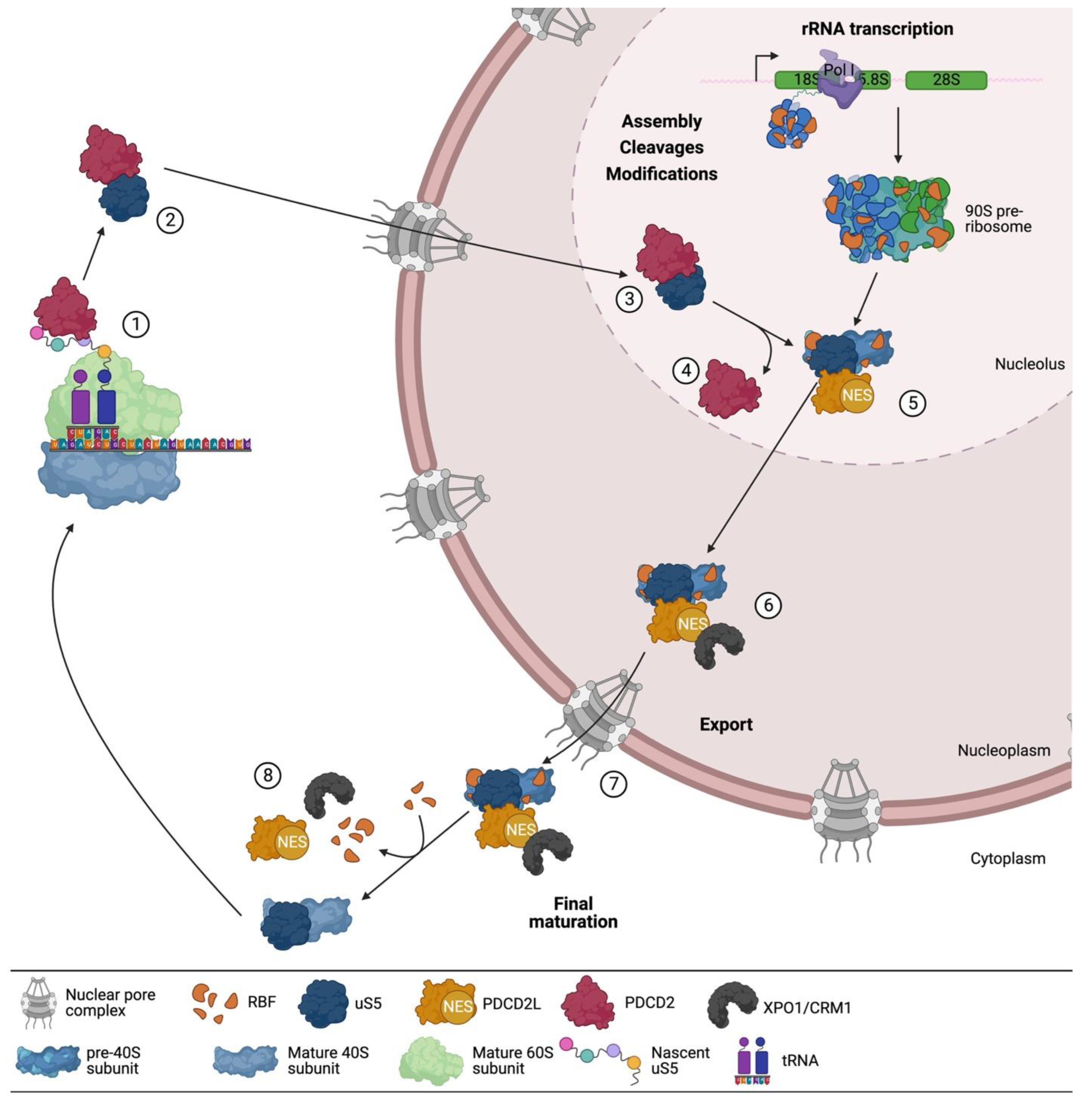
3. uS5 Is an Essential Protein Required for 40s Ribosomal Subunit Production
A Multifaceted Network of uS5-Associated Proteins
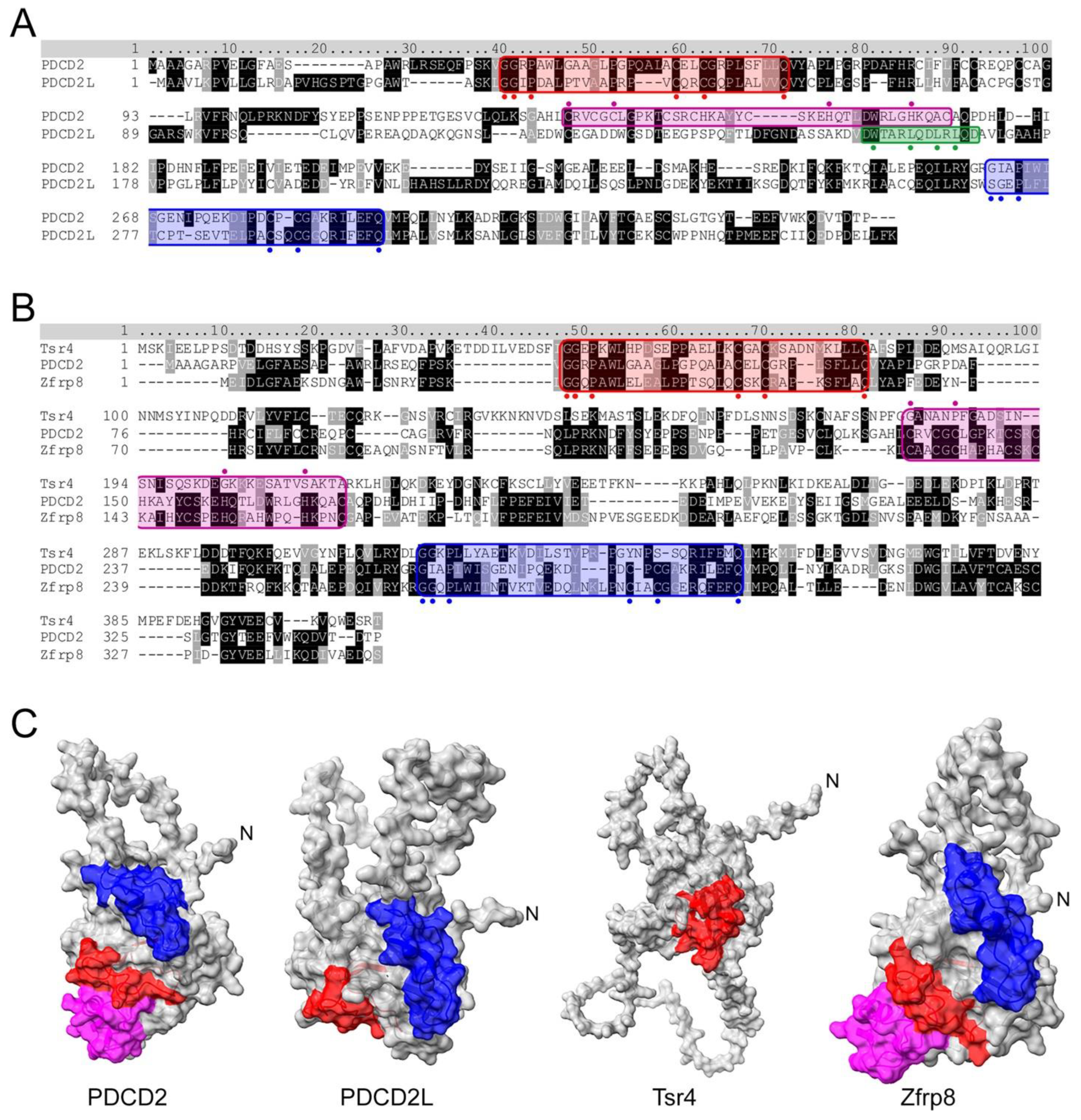
4. PDCD2 and PDCD2L: uS5-Associated Paralogs That Take Part in Human Ribosome Assembly
5. PDCD2 Is a Conserved Dedicated Chaperone for uS5
6. Conserved Role of PDCD2 in Stem Cell Biology and Embryonic Development
7. PDCD2L: A Paralog of Human PDCD2 That Associates with uS5
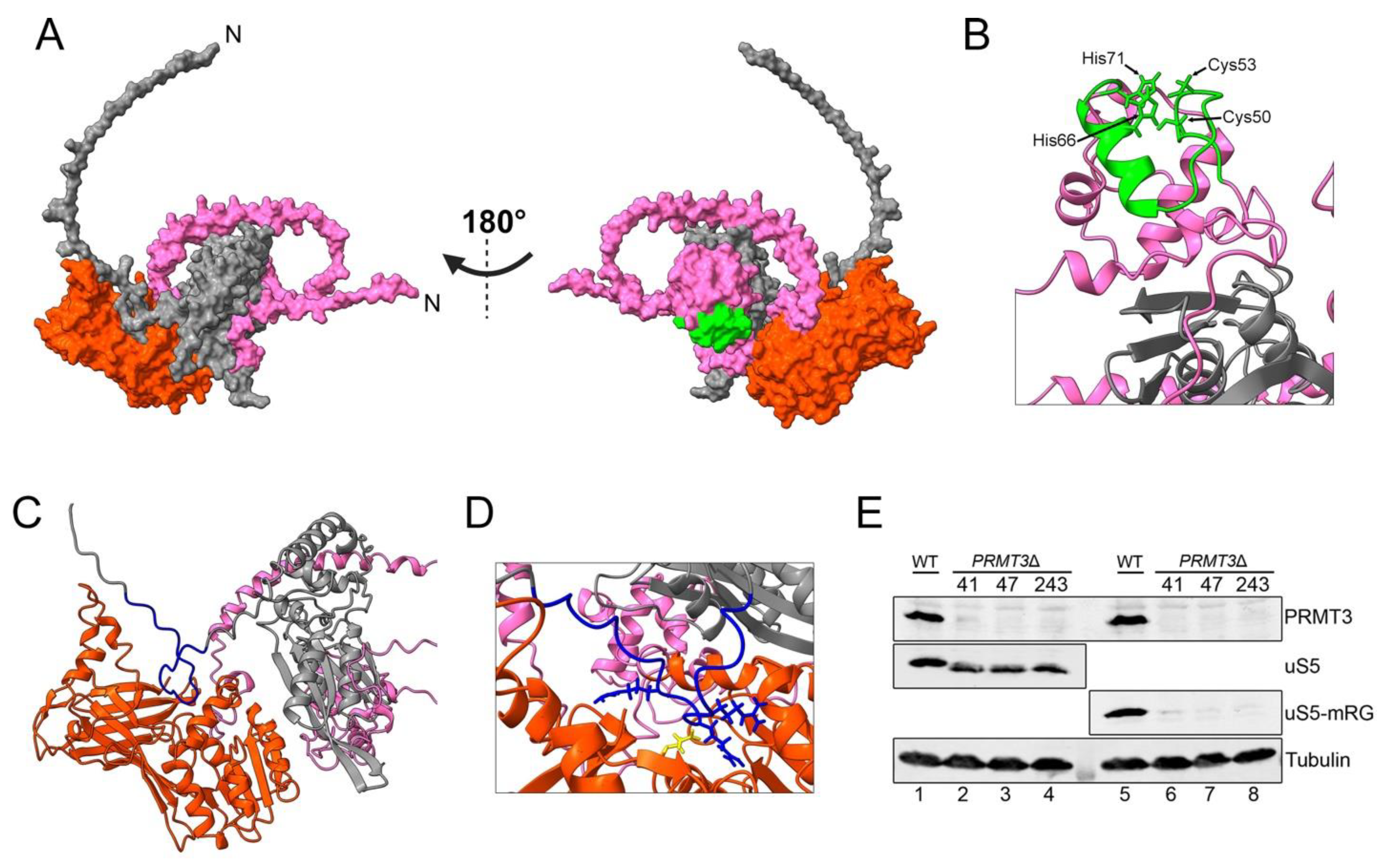
8. uS5 Arginine Methylation and uS5–PRMT3 Complex
9. ZNF277: The Newest Member among Conserved uS5-Associated Proteins
10. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anger, A.M.; Armache, J.P.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85. [Google Scholar] [CrossRef]
- Petrov, A.S.; Bernier, C.R.; Hsiao, C.; Norris, A.M.; Kovacs, N.A.; Waterbury, C.C.; Stepanov, V.G.; Harvey, S.C.; Fox, G.E.; Wartell, R.M.; et al. Evolution of the ribosome at atomic resolution. Proc. Natl. Acad. Sci. USA 2014, 111, 10251–10256. [Google Scholar] [CrossRef] [PubMed]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, S.; Manakongtreecheep, K.; Soll, D. Revising the Structural Diversity of Ribosomal Proteins Across the Three Domains of Life. Mol. Biol. Evol. 2018, 35, 1588–1598. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Thomas, G.; Volarevic, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef]
- Kos, M.; Tollervey, D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 2010, 37, 809–820. [Google Scholar] [CrossRef]
- Dorner, K.; Ruggeri, C.; Zemp, I.; Kutay, U. Ribosome biogenesis factors-from names to functions. EMBO J. 2023, 42, e112699. [Google Scholar] [CrossRef]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.F.; Chakraborty, A.; Gleizes, P.E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef]
- Ferreira-Cerca, S.; Poll, G.; Gleizes, P.E.; Tschochner, H.; Milkereit, P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell 2005, 20, 263–275. [Google Scholar] [CrossRef]
- Ferreira-Cerca, S.; Poll, G.; Kuhn, H.; Neueder, A.; Jakob, S.; Tschochner, H.; Milkereit, P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell 2007, 28, 446–457. [Google Scholar] [CrossRef]
- O’Donohue, M.F.; Choesmel, V.; Faubladier, M.; Fichant, G.; Gleizes, P.E. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 2010, 190, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Poll, G.; Braun, T.; Jakovljevic, J.; Neueder, A.; Jakob, S.; Woolford, J.L., Jr.; Tschochner, H.; Milkereit, P. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS ONE 2009, 4, e8249. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Behrmann, E.; Loerke, J.; Budkevich, T.V.; Yamamoto, K.; Schmidt, A.; Penczek, P.A.; Vos, M.R.; Burger, J.; Mielke, T.; Scheerer, P.; et al. Structural snapshots of actively translating human ribosomes. Cell 2015, 161, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Chowdhury, M.N.; Jin, H. The RGG motif proteins: Interactions, functions, and regulations. Wiley Interdiscip. Rev. RNA 2023, 14, e1748. [Google Scholar] [CrossRef]
- Bachand, F.; Silver, P.A. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 2004, 23, 2641–2650. [Google Scholar] [CrossRef]
- Lipson, R.S.; Webb, K.J.; Clarke, S.G. Rmt1 catalyzes zinc-finger independent arginine methylation of ribosomal protein Rps2 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2010, 391, 1658–1662. [Google Scholar] [CrossRef]
- Swiercz, R.; Person, M.D.; Bedford, M.T. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 2005, 386, 85–91. [Google Scholar] [CrossRef]
- Antoine, M.; Reimers, K.; Wirz, W.; Gressner, A.M.; Muller, R.; Kiefer, P. Identification of an unconventional nuclear localization signal in human ribosomal protein S2. Biochem. Biophys. Res. Commun. 2005, 335, 146–153. [Google Scholar] [CrossRef]
- Agarwal, D.; Kamath, D.; Gregory, S.T.; O’Connor, M. Modulation of decoding fidelity by ribosomal proteins S4 and S5. J. Bacteriol. 2015, 197, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Piepersberg, W.; Bock, A.; Wittmann, H.G. Effect of different mutations in ribosomal protein S5 of Escherichia coli on translational fidelity. Mol. Gen. Genet. 1975, 140, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Piepersberg, W.; Bock, A.; Yaguchi, M.; Wittmann, H.G. Genetic position and amino acid replacements of several mutations in ribosomal protein S5 from Escherichia coli. Mol. Gen. Genet. 1975, 143, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Rosset, R.; Gorini, L. A ribosomal ambiguity mutation. J. Mol. Biol. 1969, 39, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Culver, G.M. Meanderings of the mRNA through the ribosome. Structure 2001, 9, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Kurkcuoglu, O.; Doruker, P.; Sen, T.Z.; Kloczkowski, A.; Jernigan, R.L. The ribosome structure controls and directs mRNA entry, translocation and exit dynamics. Phys. Biol. 2008, 5, 046005. [Google Scholar] [CrossRef]
- Llacer, J.L.; Hussain, T.; Saini, A.K.; Nanda, J.S.; Kaur, S.; Gordiyenko, Y.; Kumar, R.; Hinnebusch, A.G.; Lorsch, J.R.; Ramakrishnan, V. Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. eLife 2018, 7, e39273. [Google Scholar] [CrossRef]
- Dong, J.; Hinnebusch, A.G. uS5/Rps2 residues at the 40S ribosome entry channel enhance initiation at suboptimal start codons in vivo. Genetics 2022, 220, iyab176. [Google Scholar] [CrossRef]
- Shcherbakov, D.; Teo, Y.; Boukari, H.; Cortes-Sanchon, A.; Mantovani, M.; Osinnii, I.; Moore, J.; Juskeviciene, R.; Brilkova, M.; Duscha, S.; et al. Ribosomal mistranslation leads to silencing of the unfolded protein response and increased mitochondrial biogenesis. Commun. Biol. 2019, 2, 381. [Google Scholar] [CrossRef]
- Moore, J.; Akbergenov, R.; Nigri, M.; Isnard-Petit, P.; Grimm, A.; Seebeck, P.; Restelli, L.; Frank, S.; Eckert, A.; Thiam, K.; et al. Random errors in protein synthesis activate an age-dependent program of muscle atrophy in mice. Commun. Biol. 2021, 4, 703. [Google Scholar] [CrossRef]
- Steffen, K.K.; McCormick, M.A.; Pham, K.M.; MacKay, V.L.; Delaney, J.R.; Murakami, C.J.; Kaeberlein, M.; Kennedy, B.K. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 2012, 191, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Perreault, A.; Bellemer, C.; Bachand, F. Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2. Nucleic Acids Res. 2008, 36, 6132–6142. [Google Scholar] [CrossRef] [PubMed]
- Cramton, S.E.; Laski, F.A. String of pearls encodes Drosophila ribosomal protein S2, has Minute-like characteristics, and is required during oogenesis. Genetics 1994, 137, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Marygold, S.J.; Roote, J.; Reuter, G.; Lambertsson, A.; Ashburner, M.; Millburn, G.H.; Harrison, P.M.; Yu, Z.; Kenmochi, N.; Kaufman, T.C.; et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007, 8, R216. [Google Scholar] [CrossRef]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e516. [Google Scholar] [CrossRef]
- Wang, M.; Hu, Y.; Stearns, M.E. RPS2: A novel therapeutic target in prostate cancer. J. Exp. Clin. Cancer Res. CR 2009, 28, 6. [Google Scholar] [CrossRef]
- Cheng, J.; Lau, B.; Thoms, M.; Ameismeier, M.; Berninghausen, O.; Hurt, E.; Beckmann, R. The nucleoplasmic phase of pre-40S formation prior to nuclear export. Nucleic Acids Res. 2022, 50, 11924–11937. [Google Scholar] [CrossRef]
- Landry-Voyer, A.M.; Bergeron, D.; Yague-Sanz, C.; Baker, B.; Bachand, F. PDCD2 functions as an evolutionarily conserved chaperone dedicated for the 40S ribosomal protein uS5 (RPS2). Nucleic Acids Res. 2020, 48, 12900–12916. [Google Scholar] [CrossRef]
- Landry-Voyer, A.M.; Bilodeau, S.; Bergeron, D.; Dionne, K.L.; Port, S.A.; Rouleau, C.; Boisvert, F.M.; Kehlenbach, R.H.; Bachand, F. Human PDCD2L Is an Export Substrate of CRM1 That Associates with 40S Ribosomal Subunit Precursors. Mol. Cell. Biol. 2016, 36, 3019–3032. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Aravind, L. Analysis of two domains with novel RNA-processing activities throws light on the complex evolution of ribosomal RNA biogenesis. Front. Genet. 2014, 5, 424. [Google Scholar] [CrossRef]
- Lutterbach, B.; Sun, D.; Schuetz, J.; Hiebert, S.W. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol. Cell. Biol. 1998, 18, 3604–3611. [Google Scholar] [CrossRef] [PubMed]
- Melnick, A.M.; Westendorf, J.J.; Polinger, A.; Carlile, G.W.; Arai, S.; Ball, H.J.; Lutterbach, B.; Hiebert, S.W.; Licht, J.D. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 2000, 20, 2075–2086. [Google Scholar] [CrossRef] [PubMed]
- Owens, G.P.; Hahn, W.E.; Cohen, J.J. Identification of mRNAs associated with programmed cell death in immature thymocytes. Mol. Cell. Biol. 1991, 11, 4177–4188. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.W.; Chan, C.C.; Chao, C.C.; Fan, H.A.; Sheu, D.L.; Chan, E.C. Expression patterns of cell cycle and apoptosis-related genes in a multidrug-resistant human colon carcinoma cell line. Scand. J. Gastroenterol. 2004, 39, 464–469. [Google Scholar] [CrossRef]
- Kawakami, T.; Furukawa, Y.; Sudo, K.; Saito, H.; Takami, S.; Takahashi, E.; Nakamura, Y. Isolation and mapping of a human gene (PDCD2) that is highly homologous to Rp8, a rat gene associated with programmed cell death. Cytogenet. Cell Genet. 1995, 71, 41–43. [Google Scholar] [CrossRef]
- Baron, B.W.; Zeleznik-Le, N.; Baron, M.J.; Theisler, C.; Huo, D.; Krasowski, M.D.; Thirman, M.J.; Baron, R.M.; Baron, J.M. Repression of the PDCD2 gene by BCL6 and the implications for the pathogenesis of human B and T cell lymphomas. Proc. Natl. Acad. Sci. USA 2007, 104, 7449–7454. [Google Scholar] [CrossRef]
- Kusam, S.; Munugalavadla, V.; Sawant, D.; Dent, A. BCL6 cooperates with CD40 stimulation and loss of p53 function to rapidly transform primary B cells. Int. J. Cancer J. Int. Cancer 2009, 125, 977–981. [Google Scholar] [CrossRef]
- Liu, H.; Wang, M.; Liang, N.; Guan, L. PDCD2 sensitizes HepG2 cells to sorafenib by suppressing epithelial-mesenchymal transition. Mol. Med. Rep. 2019, 19, 2173–2179. [Google Scholar] [CrossRef]
- Steinemann, D.; Gesk, S.; Zhang, Y.; Harder, L.; Pilarsky, C.; Hinzmann, B.; Martin-Subero, J.I.; Calasanz, M.J.; Mungall, A.; Rosenthal, A.; et al. Identification of candidate tumor-suppressor genes in 6q27 by combined deletion mapping and electronic expression profiling in lymphoid neoplasms. Genes Chromosomes Cancer 2003, 37, 421–426. [Google Scholar] [CrossRef]
- Wang, W.; Song, X.W.; Bu, X.M.; Zhang, N.; Zhao, C.H. PDCD2 and NCoR1 as putative tumor suppressors in gastric gastrointestinal stromal tumors. Cell. Oncol. 2016, 39, 129–137. [Google Scholar] [CrossRef]
- Yang, X.; Lee, Y.; Fan, H.; Sun, X.; Lussier, Y.A. Identification of common microRNA-mRNA regulatory biomodules in human epithelial cancers. Chin. Sci. Bull. 2010, 55, 3576–3589. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jin, Y.; Du, W. Programmed cell death 2 functions as a tumor suppressor in osteosarcoma. Int. J. Clin. Exp. Pathol. 2015, 8, 10894–10900. [Google Scholar] [PubMed]
- Fukae, J.; Sato, S.; Shiba, K.; Sato, K.; Mori, H.; Sharp, P.A.; Mizuno, Y.; Hattori, N. Programmed cell death-2 isoform1 is ubiquitinated by parkin and increased in the substantia nigra of patients with autosomal recessive Parkinson’s disease. FEBS Lett. 2009, 583, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Schauder, C.; Naryshkina, T.; Minakhina, S.; Steward, R. Zfrp8 forms a complex with fragile-X mental retardation protein and regulates its localization and function. Dev. Biol. 2016, 410, 202–212. [Google Scholar] [CrossRef]
- Mu, W.; Munroe, R.J.; Barker, A.K.; Schimenti, J.C. PDCD2 is essential for inner cell mass development and embryonic stem cell maintenance. Dev. Biol. 2010, 347, 279–288. [Google Scholar] [CrossRef]
- Black, J.J.; Musalgaonkar, S.; Johnson, A.W. Tsr4 Is a Cytoplasmic Chaperone for the Ribosomal Protein Rps2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2019, 39, e00019–e00094. [Google Scholar] [CrossRef]
- Rossler, I.; Embacher, J.; Pillet, B.; Murat, G.; Liesinger, L.; Hafner, J.; Unterluggauer, J.J.; Birner-Gruenberger, R.; Kressler, D.; Pertschy, B. Tsr4 and Nap1, two novel members of the ribosomal protein chaperOME. Nucleic Acids Res. 2019, 47, 6984–7002. [Google Scholar] [CrossRef]
- Li, Z.; Lee, I.; Moradi, E.; Hung, N.J.; Johnson, A.W.; Marcotte, E.M. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009, 7, e1000213. [Google Scholar] [CrossRef]
- Minakhina, S.; Naryshkina, T.; Changela, N.; Tan, W.; Steward, R. Zfrp8/PDCD2 Interacts with RpS2 Connecting Ribosome Maturation and Gene-Specific Translation. PLoS ONE 2016, 11, e0147631. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. BioRxiv 2022. [Google Scholar] [CrossRef]
- Ben-Shem, A.; Garreau de Loubresse, N.; Melnikov, S.; Jenner, L.; Yusupova, G.; Yusupov, M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 2011, 334, 1524–1529. [Google Scholar] [CrossRef]
- Minakhina, S.; Changela, N.; Steward, R. Zfrp8/PDCD2 is required in ovarian stem cells and interacts with the piRNA pathway machinery. Development 2014, 141, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Minakhina, S.; Steward, R. Hematopoietic stem cells in Drosophila. Development 2010, 137, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Kokorina, N.A.; Granier, C.J.; Zakharkin, S.O.; Davis, S.; Rabson, A.B.; Sabaawy, H.E. PDCD2 knockdown inhibits erythroid but not megakaryocytic lineage differentiation of human hematopoietic stem/progenitor cells. Exp. Hematol. 2012, 40, 1028–1042.e1023. [Google Scholar] [CrossRef] [PubMed]
- Kramer, J.; Granier, C.J.; Davis, S.; Piso, K.; Hand, J.; Rabson, A.B.; Sabaawy, H.E. PDCD2 controls hematopoietic stem cell differentiation during development. Stem Cells Dev. 2013, 22, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Minakhina, S.; Druzhinina, M.; Steward, R. Zfrp8, the Drosophila ortholog of PDCD2, functions in lymph gland development and controls cell proliferation. Development 2007, 134, 2387–2396. [Google Scholar] [CrossRef]
- Abbas, M.N.; Liang, H.; Kausar, S.; Dong, Z.; Cui, H. Zinc finger protein RP-8, the Bombyx mori ortholog of programmed cell death 2, regulates cell proliferation. Dev. Comp. Immunol. 2020, 104, 103542. [Google Scholar] [CrossRef]
- Dionne, K.L.; Bergeron, D.; Landry-Voyer, A.M.; Bachand, F. The 40S ribosomal protein uS5 (RPS2) assembles into an extra-ribosomal complex with human ZNF277 that competes with the PRMT3-uS5 interaction. J. Biol. Chem. 2019, 294, 1944–1955. [Google Scholar] [CrossRef]
- Houston, B.J.; Oud, M.S.; Aguirre, D.M.; Merriner, D.J.; O’Connor, A.E.; Okutman, O.; Viville, S.; Burke, R.; Veltman, J.A.; O’Bryan, M.K. Programmed Cell Death 2-Like (Pdcd2l) Is Required for Mouse Embryonic Development. G3 2020, 10, 4449–4457. [Google Scholar] [CrossRef]
- Lee, S.W.; Berger, S.J.; Martinovic, S.; Pasa-Tolic, L.; Anderson, G.A.; Shen, Y.; Zhao, R.; Smith, R.D. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc. Natl. Acad. Sci. USA 2002, 99, 5942–5947. [Google Scholar] [CrossRef]
- Louie, D.F.; Resing, K.A.; Lewis, T.S.; Ahn, N.G. Mass spectrometric analysis of 40 S ribosomal proteins from Rat-1 fibroblasts. J. Biol. Chem. 1996, 271, 28189–28198. [Google Scholar] [CrossRef] [PubMed]
- Odintsova, T.I.; Muller, E.C.; Ivanov, A.V.; Egorov, T.A.; Bienert, R.; Vladimirov, S.N.; Kostka, S.; Otto, A.; Wittmann-Liebold, B.; Karpova, G.G. Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J. Protein Chem. 2003, 22, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Barna, M. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012, 13, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.N.; Navickas, I.J.; Chang, C.N.; Dancis, B.M. Methylation of ribosomal proteins in HeLa cells. Arch. Biochem. Biophys. 1976, 172, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Kruiswijk, T.; Kunst, A.; Planta, R.J.; Mager, W.H. Modification of yeast ribosomal proteins. Methylation. Biochem. J. 1978, 175, 221–225. [Google Scholar] [CrossRef]
- Lhoest, J.; Lobet, Y.; Costers, E.; Colson, C. Methylated proteins and amino acids in the ribosomes of Saccharomyces cerevisiae. Eur. J. Biochem. 1984, 141, 585–590. [Google Scholar] [CrossRef]
- Ramagopal, S. Covalent modifications of ribosomal proteins in growing and aggregation-competent dictyostelium discoideum: Phosphorylation and methylation. Biochem. Cell Biol. 1991, 69, 263–268. [Google Scholar] [CrossRef]
- Chang, F.N.; Navickas, I.J.; Au, C.; Budzilowicz, C. Identification of the methylated ribosomal proteins in HeLa cells and the fluctuation of methylation during the cell cycle. Biochim. Biophys. Acta 1978, 518, 89–94. [Google Scholar] [CrossRef]
- Xu, J.; Richard, S. Cellular pathways influenced by protein arginine methylation: Implications for cancer. Mol. Cell 2021, 81, 4357–4368. [Google Scholar] [CrossRef]
- Bachand, F. Protein Arginine Methyltransferases: From Unicellular Eukaryotes to Humans. Eukaryot. Cell 2007, 6, 889–898. [Google Scholar] [CrossRef]
- Swiercz, R.; Cheng, D.; Kim, D.; Bedford, M.T. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J. Biol. Chem. 2007, 282, 16917–16923. [Google Scholar] [CrossRef]
- Ladror, D.T.; Frey, B.L.; Scalf, M.; Levenstein, M.E.; Artymiuk, J.M.; Smith, L.M. Methylation of yeast ribosomal protein S2 is elevated during stationary phase growth conditions. Biochem. Biophys. Res. Commun. 2014, 445, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Perreault, A.; Gascon, S.; D’Amours, A.; Aletta, J.M.; Bachand, F. A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis. J. Biol. Chem. 2009, 284, 15026–15037. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Gary, J.D.; Clarke, S.; Herschman, H.R. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J. Biol. Chem. 1998, 273, 16935–16945. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.; Clarke, S. PRMT3 is a distinct member of the protein arginine N-methyltransferase family. Conferral of substrate specificity by a zinc-finger domain. J. Biol. Chem. 2000, 275, 32974–32982. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, M.B.; Karbstein, K. Does functional specialization of ribosomes really exist? RNA 2019, 25, 521–538. [Google Scholar] [CrossRef] [PubMed]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef]
- Choi, S.; Jung, C.R.; Kim, J.Y.; Im, D.S. PRMT3 inhibits ubiquitination of ribosomal protein S2 and together forms an active enzyme complex. Biochim. Biophys. Acta 2008, 1780, 1062–1069. [Google Scholar] [CrossRef]
- Hang, R.; Liu, C.; Ahmad, A.; Zhang, Y.; Lu, F.; Cao, X. Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc. Natl. Acad. Sci. USA 2014, 111, 16190–16195. [Google Scholar] [CrossRef]
- Hang, R.; Wang, Z.; Yang, C.; Luo, L.; Mo, B.; Chen, X.; Sun, J.; Liu, C.; Cao, X. Protein arginine methyltransferase 3 fine-tunes the assembly/disassembly of pre-ribosomes to repress nucleolar stress by interacting with RPS2B in arabidopsis. Mol. Plant 2021, 14, 223–236. [Google Scholar] [CrossRef]
- Pausch, P.; Singh, U.; Ahmed, Y.L.; Pillet, B.; Murat, G.; Altegoer, F.; Stier, G.; Thoms, M.; Hurt, E.; Sinning, I.; et al. Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat. Commun. 2015, 6, 7494. [Google Scholar] [CrossRef] [PubMed]
- Pillet, B.; Mitterer, V.; Kressler, D.; Pertschy, B. Hold on to your friends: Dedicated chaperones of ribosomal proteins: Dedicated chaperones mediate the safe transfer of ribosomal proteins to their site of pre-ribosome incorporation. Bioessays 2017, 39, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Giot, L.; Bader, J.S.; Brouwer, C.; Chaudhuri, A.; Kuang, B.; Li, Y.; Hao, Y.L.; Ooi, C.E.; Godwin, B.; Vitols, E.; et al. A Protein Interaction Map of Drosophila melanogaster. Science 2003, 302, 1727–1736. [Google Scholar] [CrossRef]
- Xu, T.; Liao, S.; Huang, M.; Zhu, C.; Huang, X.; Jin, Q.; Xu, D.; Fu, C.; Chen, X.; Feng, X.; et al. A ZTF-7/RPS-2 complex mediates the cold-warm response in C. elegans. PLoS Genet. 2023, 19, e1010628. [Google Scholar] [CrossRef]
- Negishi, M.; Saraya, A.; Mochizuki, S.; Helin, K.; Koseki, H.; Iwama, A. A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1. PLoS ONE 2010, 5, e12373. [Google Scholar] [CrossRef]
- Xie, G.; Peng, Z.; Liang, J.; Larabee, S.M.; Drachenberg, C.B.; Yfantis, H.; Raufman, J.P. Zinc finger protein 277 is an intestinal transit-amplifying cell marker and colon cancer oncogene. JCI Insight 2022, 7, e150894. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Yang, Y.M.; Karbstein, K. The chaperone Tsr2 regulates Rps26 release and reincorporation from mature ribosomes to enable a reversible, ribosome-mediated response to stress. Sci. Adv. 2022, 8, eabl4386. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landry-Voyer, A.-M.; Mir Hassani, Z.; Avino, M.; Bachand, F. Ribosomal Protein uS5 and Friends: Protein–Protein Interactions Involved in Ribosome Assembly and Beyond. Biomolecules 2023, 13, 853. https://doi.org/10.3390/biom13050853
Landry-Voyer A-M, Mir Hassani Z, Avino M, Bachand F. Ribosomal Protein uS5 and Friends: Protein–Protein Interactions Involved in Ribosome Assembly and Beyond. Biomolecules. 2023; 13(5):853. https://doi.org/10.3390/biom13050853
Chicago/Turabian StyleLandry-Voyer, Anne-Marie, Zabih Mir Hassani, Mariano Avino, and François Bachand. 2023. "Ribosomal Protein uS5 and Friends: Protein–Protein Interactions Involved in Ribosome Assembly and Beyond" Biomolecules 13, no. 5: 853. https://doi.org/10.3390/biom13050853
APA StyleLandry-Voyer, A.-M., Mir Hassani, Z., Avino, M., & Bachand, F. (2023). Ribosomal Protein uS5 and Friends: Protein–Protein Interactions Involved in Ribosome Assembly and Beyond. Biomolecules, 13(5), 853. https://doi.org/10.3390/biom13050853





