Orchestration of Mitochondrial Function and Remodeling by Post-Translational Modifications Provide Insight into Mechanisms of Viral Infection
Abstract
1. Introduction
2. Mitochondrial PTMs Remodeling Mitochondrial Structure
3. Mitochondrial PTMs Regulating Cellular Processes
3.1. Metabolism
3.2. Apoptosis
3.3. Immune Signaling
4. Enzymatic and Nonenzymatic Regulation of Mitochondrial PTMs
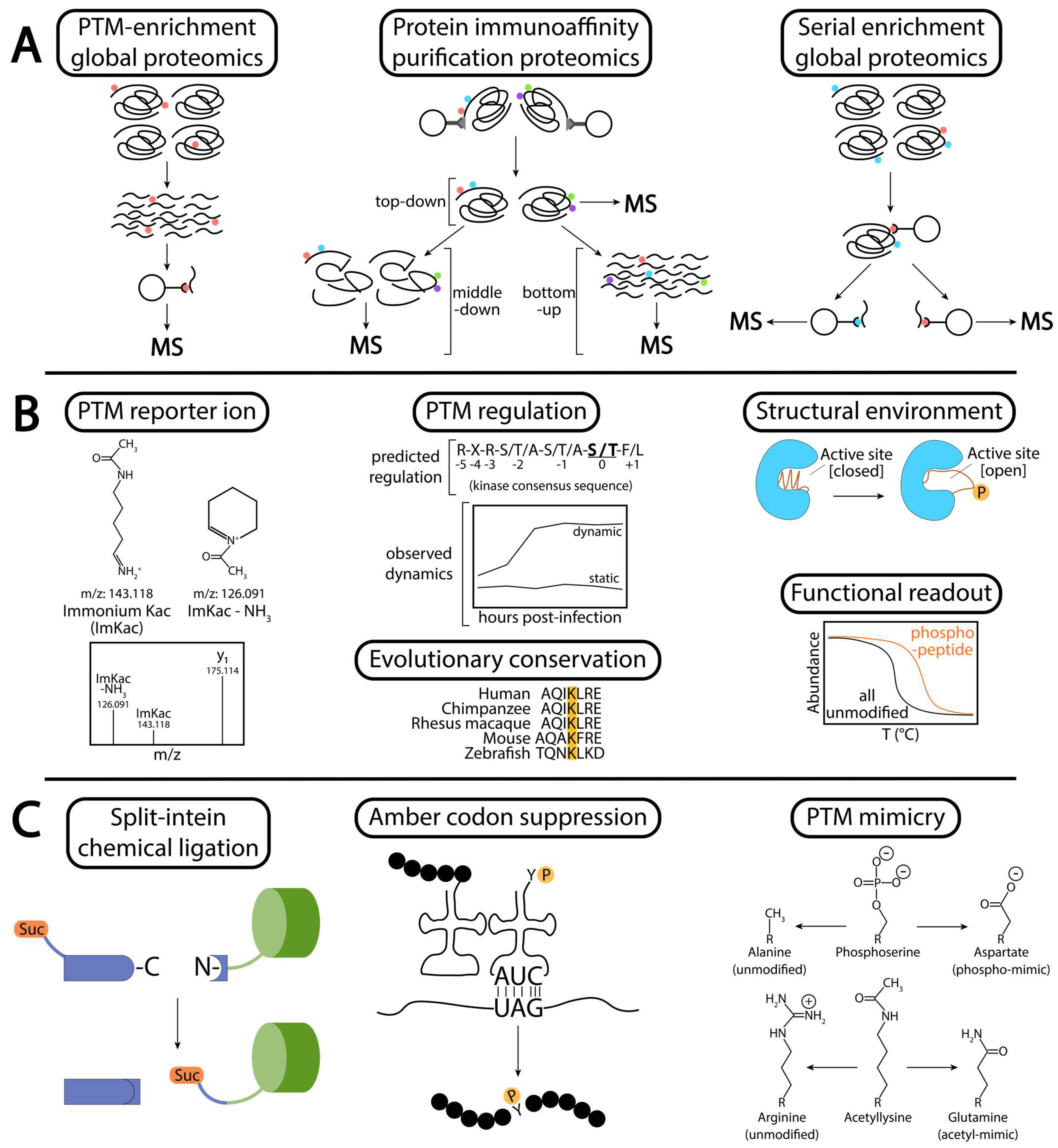
5. Methods for Identifying and Determining the Mechanism of PTMs
6. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pernas, L.; Scorrano, L. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu. Rev. Physiol. 2016, 78, 505–531. [Google Scholar] [CrossRef]
- Kasahara, A.; Scorrano, L. Mitochondria: From Cell Death Executioners to Regulators of Cell Differentiation. Trends Cell Biol. 2014, 24, 761–770. [Google Scholar] [CrossRef]
- Rath, S.; Sharma, R.; Gupta, R.; Ast, T.; Chan, C.; Durham, T.J.; Goodman, R.P.; Grabarek, Z.; Haas, M.E.; Hung, W.H.W.; et al. MitoCarta3.0: An Updated Mitochondrial Proteome Now with Sub-Organelle Localization and Pathway Annotations. Nucleic Acids Res. 2021, 49, D1541–D1547. [Google Scholar] [CrossRef]
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial Proteins: From Biogenesis to Functional Networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284. [Google Scholar] [CrossRef]
- Dougherty, S.E.; Maduka, A.O.; Inada, T.; Silva, G.M. Expanding Role of Ubiquitin in Translational Control. Int. J. Mol. Sci. 2020, 21, 1151. [Google Scholar] [CrossRef]
- Calnan, D.R.; Brunet, A. The FoxO Code. Oncogene 2008, 27, 2276–2288. [Google Scholar] [CrossRef] [PubMed]
- Doulias, P.T.; Greene, J.L.; Greco, T.M.; Tenopoulou, M.; Seeholzer, S.H.; Dunbrack, R.L.; Ischiropoulos, H. Structural Profiling of Endogenous S-Nitrosocysteine Residues Reveals Unique Features That Accommodate Diverse Mechanisms for Protein S-Nitrosylation. Proc. Natl. Acad. Sci. USA 2010, 107, 16958–16963. [Google Scholar] [CrossRef] [PubMed]
- Chastain, C.J.; Failing, C.J.; Manandhar, L.; Zimmerman, M.A.; Lakner, M.M.; Nguyen, T.H.T. Functional Evolution of C(4) Pyruvate, Orthophosphate Dikinase. J. Exp. Bot. 2011, 62, 3083–3091. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Tao, W.A.; Wollscheid, B.; O’Brien, R.; Eng, J.K.; Li, X.J.; Bodenmiller, B.; Watts, J.D.; Hood, L.; Aebersold, R. Quantitative Phosphoproteome Analysis Using a Dendrimer Conjugation Chemistry and Tandem Mass Spectrometry. Nat. Methods 2005, 2, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.I.; Kane, L.A.; Uhrigshardt, H.; Wang, S.B.; Van Eyk, J.E. Site-Mapping of in Vitro S-Nitrosation in Cardiac Mitochondria: Implications for Cardioprotection. Mol. Cell. Proteom. 2011, 10, M110.004721. [Google Scholar] [CrossRef]
- Deng, N.; Zhang, J.; Zong, C.; Wang, Y.; Lu, H.; Yang, P.; Wang, W.; Young, G.W.; Wang, Y.; Korge, P.; et al. Phosphoproteome Analysis Reveals Regulatory Sites in Major Pathways of Cardiac Mitochondria. Mol. Cell. Proteom. 2011, 10, M110.000117. [Google Scholar] [CrossRef] [PubMed]
- Griffante, G.; Gugliesi, F.; Pasquero, S.; Dell’Oste, V.; Biolatti, M.; Salinger, A.J.; Mondal, S.; Thompson, P.R.; Weerapana, E.; Lebbink, R.J.; et al. Human Cytomegalovirus-Induced Host Protein Citrullination Is Crucial for Viral Replication. Nat. Commun. 2021, 12, 3910. [Google Scholar] [CrossRef]
- Sheng, X.; Cristea, I.M. The Antiviral Sirtuin 3 Bridges Protein Acetylation to Mitochondrial Integrity and Metabolism during Human Cytomegalovirus Infection. PLoS Pathog. 2021, 17, e1009506. [Google Scholar] [CrossRef] [PubMed]
- Oberstein, A.; Perlman, D.H.; Shenk, T.; Terry, L.J. Human Cytomegalovirus PUL97 Kinase Induces Global Changes in the Infected Cell Phosphoproteome. Proteomics 2015, 15, 2006–2022. [Google Scholar] [CrossRef] [PubMed]
- Murray, L.A.; Sheng, X.; Cristea, I.M. Orchestration of Protein Acetylation as a Toggle for Cellular Defense and Virus Replication. Nat. Commun. 2018, 9, 4967. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, H.; Zhu, J.; Dong, Q.; Wang, J.; Fan, H.; Chen, Y.; Zhang, X.; Han, X.; Li, Q.; et al. Ubiquitin-Modified Proteome of SARS-CoV-2-Infected Host Cells Reveals Insights into Virus-Host Interaction and Pathogenesis. J. Proteome Res. 2021, 20, 2224–2239. [Google Scholar] [CrossRef]
- Sloan, E.; Tatham, M.H.; Groslambert, M.; Glass, M.; Orr, A.; Hay, R.T.; Everett, R.D. Analysis of the SUMO2 Proteome during HSV-1 Infection. PLoS Pathog. 2015, 11, e1005059. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712.e19. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, H.; Zhang, H.; Sui, L.; Li, L.; Xu, W.; Du, S.; Hao, P.; Jiang, Y.; Chen, J.; et al. The Global Succinylation of SARS-CoV-2-Infected Host Cells Reveals Drug Targets. Proc. Natl. Acad. Sci. USA 2022, 119, e2123065119. [Google Scholar] [CrossRef]
- Ramazi, S.; Zahiri, J. Posttranslational Modifications in Proteins: Resources, Tools and Prediction Methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, H.; Liu, J.; Long, J. Post-Translational Modifications on Mitochondrial Metabolic Enzymes in Cancer. Free Radic. Biol. Med. 2022, 179, 11–23. [Google Scholar] [CrossRef]
- Guo, J.; Cheng, J.; North, B.J.; Wei, W. Functional Analyses of Major Cancer-Related Signaling Pathways in Alzheimer’s Disease Etiology. Biochim. Biophys. Acta (BBA) Rev. Cancer 2017, 1868, 341–358. [Google Scholar] [CrossRef]
- Yang, Y.; Gibson, G.E. Succinylation Links Metabolism to Protein Functions. Neurochem. Res. 2019, 44, 2346–2359. [Google Scholar] [CrossRef]
- Refolo, G.; Vescovo, T.; Piacentini, M.; Fimia, G.M.; Ciccosanti, F. Mitochondrial Interactome: A Focus on Antiviral Signaling Pathways. Front. Cell Dev. Biol. 2020, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Munger, J.; Bajad, S.U.; Coller, H.A.; Shenk, T.; Rabinowitz, J.D. Dynamics of the Cellular Metabolome during Human Cytomegalovirus Infection. PLoS Pathog. 2006, 2, 1165–1175. [Google Scholar] [CrossRef]
- Munger, J.; Bennett, B.D.; Parikh, A.; Feng, X.J.; McArdle, J.; Rabitz, H.A.; Shenk, T.; Rabinowitz, J.D. Systems-Level Metabolic Flux Profiling Identifies Fatty Acid Synthesis as a Target for Antiviral Therapy. Nat. Biotechnol. 2008, 26, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Betsinger, C.N.; Jankowski, C.S.R.; Hofstadter, W.A.; Federspiel, J.D.; Otter, C.J.; Jean Beltran, P.M.; Cristea, I.M. The Human Cytomegalovirus Protein PUL13 Targets Mitochondrial Cristae Architecture to Increase Cellular Respiration during Infection. Proc. Natl. Acad. Sci. USA 2021, 118, e2101675118. [Google Scholar] [CrossRef] [PubMed]
- Saunier, E.; Benelli, C.; Bortoli, S. The Pyruvate Dehydrogenase Complex in Cancer: An Old Metabolic Gatekeeper Regulated by New Pathways and Pharmacological Agents. Int. J. Cancer 2016, 138, 809–817. [Google Scholar] [CrossRef]
- Ampomah, P.B.; Lim, L.H.K. Influenza A Virus-Induced Apoptosis and Virus Propagation. Apoptosis 2020, 25, 1–11. [Google Scholar] [CrossRef]
- Turpin, J.; El Safadi, D.; Lebeau, G.; Krejbich, M.; Chatelain, C.; Desprès, P.; Viranaïcken, W.; Krejbich-Trotot, P. Apoptosis during ZIKA Virus Infection: Too Soon or Too Late? Int. J. Mol. Sci. 2022, 23, 1287. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Brenner, C.; Morselli, E.; Touat, Z.; Kroemer, G. Viral Control of Mitochondrial Apoptosis. PLoS Pathog. 2008, 4, e1000018. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.T.; Grishin, N.V.; et al. Phosphorylation of Innate Immune Adaptor Proteins MAVS, STING, and TRIF Induces IRF3 Activation. Science 2015, 347, aaa2630. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.C.; Cristea, I.M. Location Is Everything: Protein Translocations as a Viral Infection Strategy. Curr. Opin. Chem. Biol. 2019, 48, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Paulus, C.; Krauss, S.; Nevels, M. A Human Cytomegalovirus Antagonist of Type I IFN-Dependent Signal Transducer and Activator of Transcription Signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 3840–3845. [Google Scholar] [CrossRef]
- Ma, Z.; Jacobs, S.R.; West, J.A.; Stopford, C.; Zhang, Z.; Davis, Z.; Barber, G.N.; Glaunsinger, B.A.; Dittmer, D.P.; Damania, B. Modulation of the CGAS-STING DNA Sensing Pathway by Gammaherpesviruses. Proc. Natl. Acad. Sci. USA 2015, 112, E4306–E4315. [Google Scholar] [CrossRef]
- Choi, H.J.; Park, A.; Kang, S.; Lee, E.; Lee, T.A.; Ra, E.A.; Lee, J.; Lee, S.; Park, B. Human Cytomegalovirus-Encoded US9 Targets MAVS and STING Signaling to Evade Type i Interferon Immune Responses. Nat. Commun. 2018, 9, 125. [Google Scholar] [CrossRef]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.S.; Huang, I.C.; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; García-Sastre, A. Influenza A Virus NS1 Targets the Ubiquitin Ligase TRIM25 to Evade Recognition by the Host Viral RNA Sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar] [CrossRef]
- Huang, Z.F.; Zou, H.M.; Liao, B.W.; Zhang, H.Y.; Yang, Y.; Fu, Y.Z.; Wang, S.Y.; Luo, M.H.; Wang, Y.Y. Human Cytomegalovirus Protein UL31 Inhibits DNA Sensing of CGAS to Mediate Immune Evasion. Cell Host Microbe 2018, 24, 69–80.e4. [Google Scholar] [CrossRef]
- Li, T.; Chen, J.; Cristea, I.M. Human Cytomegalovirus Tegument Protein PUL83 Inhibits IFI16-Mediated DNA Sensing for Immune Evasion. Cell Host Microbe 2013, 14, 591–599. [Google Scholar] [CrossRef]
- Jean Beltran, P.M.; Cook, K.C.; Cristea, I.M. Exploring and Exploiting Proteome Organization during Viral Infection. J. Virol. 2017, 91, e00268-17. [Google Scholar] [CrossRef] [PubMed]
- Bagshaw, O.R.M.; Balardo, C.J.; Bland, N.A.; Pardiwalla, N.; Samuel, I.A.J.; Zoso, S.L.S.; Stuart, J.A. Impaired Mitochondrial Dynamics in Disease. In Mitochondrial Dysfunction and Nanotherapeutics: Aging, Diseases, and Nanotechnology-Related Strategies in Mitochondrial Medicine; Elsevier: Amsterdam, The Netherlands, 2021; pp. 57–90. ISBN 9780323856669. [Google Scholar]
- Westermann, B. Bioenergetic Role of Mitochondrial Fusion and Fission. Biochim. Biophys. Acta 2012, 1817, 1833–1838. [Google Scholar] [CrossRef] [PubMed]
- Tait, S.W.G.; Green, D.R. Mitochondria and Cell Signalling. J. Cell Sci. 2012, 125, 807–815. [Google Scholar] [CrossRef]
- Castanier, C.; Garcin, D.; Vazquez, A.; Arnoult, D. Mitochondrial Dynamics Regulate the RIG-I-like Receptor Antiviral Pathway. EMBO Rep. 2010, 11, 133–138. [Google Scholar] [CrossRef]
- Cipolat, S.; De Brito, O.M.; Dal Zilio, B.; Scorrano, L. OPA1 Requires Mitofusin 1 to Promote Mitochondrial Fusion. Proc. Natl. Acad. Sci. USA 2004, 101, 15927–15932. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.D.; Wagner, J.A.; Gorsich, S.W.; McCaffery, J.M.; Shaw, J.M.; Nunnari, J. The Dynamin-Related GTPase, Mgm1p, Is an Intermembrane Space Protein Required for Maintenance of Fusion Competent Mitochondria. J. Cell Biol. 2000, 151, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Frezza, C.; Cipolat, S.; Martins de Brito, O.; Micaroni, M.; Beznoussenko, G.V.; Rudka, T.; Bartoli, D.; Polishuck, R.S.; Danial, N.N.; De Strooper, B.; et al. OPA1 Controls Apoptotic Cristae Remodeling Independently from Mitochondrial Fusion. Cell 2006, 126, 177–189. [Google Scholar] [CrossRef]
- Samant, S.A.; Zhang, H.J.; Hong, Z.; Pillai, V.B.; Sundaresan, N.R.; Wolfgeher, D.; Archer, S.L.; Chan, D.C.; Gupta, M.P. SIRT3 Deacetylates and Activates OPA1 To Regulate Mitochondrial Dynamics during Stress. Mol. Cell. Biol. 2014, 34, 807–819. [Google Scholar] [CrossRef]
- Tondera, D.; Grandemange, S.; Jourdain, A.; Karbowski, M.; Mattenberger, Y.; Herzig, S.; Da Cruz, S.; Clerc, P.; Raschke, I.; Merkwirth, C.; et al. SLP-2 Is Required for Stress-Induced Mitochondrial Hyperfusion. EMBO J. 2009, 28, 1589–1600. [Google Scholar] [CrossRef]
- Barbier, V.; Lang, D.; Valois, S.; Rothman, A.L.; Medin, C.L. Dengue Virus Induces Mitochondrial Elongation through Impairment of Drp1-Triggered Mitochondrial Fission. Virology 2017, 500, 149–160. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Qian, D.; Wang, Z.; Qin, Z.; Liu, X.; Liu, T.; Wang, B. HCMV-Encoded IE2 Promotes NAFLD Progression by up-Regulation of SREBP1c Expression in UL122 Genetically Modified Mice. Int. J. Clin. Exp. Pathol. 2018, 11, 4213–4220. [Google Scholar] [PubMed]
- Yan, K.; Wang, K.; Li, P. The Role of Post-Translational Modifications in Cardiac Hypertrophy. J. Cell. Mol. Med. 2019, 23, 3795–3807. [Google Scholar] [CrossRef]
- Kashatus, J.A.; Nascimento, A.; Myers, L.J.; Sher, A.; Byrne, F.L.; Hoehn, K.L.; Counter, C.M.; Kashatus, D.F. Erk2 Phosphorylation of Drp1 Promotes Mitochondrial Fission and MAPK-Driven Tumor Growth. Mol. Cell 2015, 57, 537–551. [Google Scholar] [CrossRef]
- Jung, J.U.; Ravi, S.; Lee, D.W.; McFadden, K.; Kamradt, M.L.; Toussaint, L.G.; Sitcheran, R. NIK/MAP3K14 Regulates Mitochondrial Dynamics and Trafficking to Promote Cell Invasion. Curr. Biol. 2016, 26, 3288–3302. [Google Scholar] [CrossRef]
- Kim, S.J.; Khan, M.; Quan, J.; Till, A.; Subramani, S.; Siddiqui, A. Hepatitis B Virus Disrupts Mitochondrial Dynamics: Induces Fission and Mitophagy to Attenuate Apoptosis. PLoS Pathog. 2013, 9, e1003722. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Syed, G.H.; Khan, M.; Chiu, W.W.; Sohail, M.A.; Gish, R.G.; Siddiqui, A. Hepatitis C Virus Triggers Mitochondrial Fission and Attenuates Apoptosis to Promote Viral Persistence. Proc. Natl. Acad. Sci. USA 2014, 111, 6413–6418. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, N.; Ishihara, N.; Jofuku, A.; Oka, T.; Mihara, K. Mitotic Phosphorylation of Dynamin-Related GTPase Drp1 Participates in Mitochondrial Fission. J. Biol. Chem. 2007, 282, 11521–11529. [Google Scholar] [CrossRef]
- Valera-Alberni, M.; Joffraud, M.; Miro-Blanch, J.; Capellades, J.; Junza, A.; Dayon, L.; Núñez Galindo, A.; Sanchez-Garcia, J.L.; Valsesia, A.; Cercillieux, A.; et al. Crosstalk between Drp1 Phosphorylation Sites during Mitochondrial Remodeling and Their Impact on Metabolic Adaptation. Cell Rep. 2021, 36, 109565. [Google Scholar] [CrossRef]
- Han, X.J.; Lu, Y.F.; Li, S.A.; Kaitsuka, T.; Sato, Y.; Tomizawa, K.; Nairn, A.C.; Takei, K.; Matsui, H.; Matsushita, M. CaM Kinase Iα–Induced Phosphorylation of Drp1 Regulates Mitochondrial Morphology. J. Cell Biol. 2008, 182, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Cereghetti, G.M.; Stangherlin, A.; Martins De Brito, O.; Chang, C.R.; Blackstone, C.; Bernardi, P.; Scorrano, L. Dephosphorylation by Calcineurin Regulates Translocation of Drp1 to Mitochondria. Proc. Natl. Acad. Sci. USA 2008, 105, 15803–15808. [Google Scholar] [CrossRef]
- Cho, D.H.; Nakamura, T.; Fang, J.; Cieplak, P.; Godzik, A.; Gu, Z.; Lipton, S.A. S-Nitrosylation of Drp1 Mediates Beta-Amyloid-Related Mitochondrial Fission and Neuronal Injury. Science 2009, 324, 102–105. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, J.M.; Yan, J.; Zhang, D.L.; Liu, B.Q.; Jiang, J.Y.; Li, C.; Li, S.; Meng, X.N.; Wang, H.Q. BAG3 Promotes Autophagy and Glutaminolysis via Stabilizing Glutaminase. Cell Death Dis. 2019, 10, 284. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Wang, H.L.; Xu, J.; Tan, J.; Fu, L.N.; Wang, J.L.; Zou, T.H.; Sun, D.F.; Gao, Q.Y.; Chen, Y.X.; et al. Sirtuin 5 Contributes to Colorectal Carcinogenesis by Enhancing Glutaminolysis in a Deglutarylation-Dependent Manner. Nat. Commun. 2018, 9, 545. [Google Scholar] [CrossRef]
- Yu, W.; Dittenhafer-Reed, K.E.; Denu, J.M. SIRT3 Protein Deacetylates Isocitrate Dehydrogenase 2 (IDH2) and Regulates Mitochondrial Redox Status. J. Biol. Chem. 2012, 287, 14078–14086. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, F.; Sun, R.; Chen, X.; Zhang, M.; Xu, Q.; Wang, Y.; Wang, S.; Xiong, Y.; Guan, K. SIRT5 Promotes IDH2 Desuccinylation and G6PD Deglutarylation to Enhance Cellular Antioxidant Defense. EMBO Rep. 2016, 17, 811–822. [Google Scholar] [CrossRef]
- Hirschey, M.D.; Shimazu, T.; Huang, J.Y.; Schwer, B.; Verdin, E. SIRT3 Regulates Mitochondrial Protein Acetylation and Intermediary Metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 267–277. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, M.; Chu, H.; Zhang, H.; Wu, H.; Song, G.; Wang, P.; Zhao, K.; Hou, J.; Wang, X.; et al. The Ubiquitin E3 Ligase TRIM31 Promotes Aggregation and Activation of the Signaling Adaptor MAVS through Lys63-Linked Polyubiquitination. Nat. Immunol. 2016, 18, 214–224. [Google Scholar] [CrossRef]
- Liu, X.; Zhu, C.; Zha, H.; Tang, J.; Rong, F.; Chen, X.; Fan, S.; Xu, C.; Du, J.; Zhu, J.; et al. SIRT5 Impairs Aggregation and Activation of the Signaling Adaptor MAVS through Catalyzing Lysine Desuccinylation. EMBO J. 2020, 39, e103285. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-Z.; Shu, Q.-P.; Song, Y.; Zhang, H.-H.; Liu, Y.; Jin, B.-X.; Liuyu, T.-Z.; Li, C.; Huang, X.-C.; Du, R.-L.; et al. Phosphorylation of MAVS/VISA by Nemo-like Kinase (NLK) for Degradation Regulates the Antiviral Innate Immune Response. Nat. Commun. 2019, 10, 3233. [Google Scholar] [CrossRef] [PubMed]
- Laurent, G.; German, N.J.; Saha, A.K.; de Boer, V.C.J.; Davies, M.; Koves, T.R.; Dephoure, N.; Fischer, F.; Boanca, G.; Vaitheesvaran, B.; et al. SIRT4 Coordinates the Balance between Lipid Synthesis and Catabolism by Repressing Malonyl CoA Decarboxylase. Mol. Cell 2013, 50, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Pyakurel, A.; Savoia, C.; Hess, D.; Scorrano, L. Extracellular Regulated Kinase Phosphorylates Mitofusin 1 to Control Mitochondrial Morphology and Apoptosis. Mol. Cell 2015, 58, 244–254. [Google Scholar] [CrossRef] [PubMed]
- McLelland, G.L.; Goiran, T.; Yi, W.; Dorval, G.; Chen, C.X.; Lauinger, N.D.; Krahn, A.I.; Valimehr, S.; Rakovic, A.; Rouiller, I.; et al. Mfn2 Ubiquitination by PINK1/Parkin Gates the P97-Dependent Release of ER from Mitochondria to Drive Mitophagy. Elife 2018, 7, e32866. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dorn, G.W. PINK1-Phosphorylated Mitofusin 2 Is a Parkin Receptor for Culling Damaged Mitochondria. Science 2013, 340, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Korotchkina, L.G.; Patel, M.S. Mutagenesis Studies of the Phosphorylation Sites of Recombinant Human Pyruvate Dehydrogenase. Site-Specific Regulation. J. Biol. Chem. 1995, 270, 14297–14304. [Google Scholar] [CrossRef] [PubMed]
- Korotchkina, L.G.; Patel, M.S. Probing the Mechanism of Inactivation of Human Pyruvate Dehydrogenase by Phosphorylation of Three Sites. J. Biol. Chem. 2001, 276, 5731–5738. [Google Scholar] [CrossRef]
- Korotchkina, L.G.; Patel, M.S. Site Specificity of Four Pyruvate Dehydrogenase Kinase Isoenzymes toward the Three Phosphorylation Sites of Human Pyruvate Dehydrogenase. J. Biol. Chem. 2001, 276, 37223–37229. [Google Scholar] [CrossRef]
- Kolobova, E.; Tuganova, A.; Boulatnikov, I.; Popov, K.M. Regulation of Pyruvate Dehydrogenase Activity through Phosphorylation at Multiple Sites. Biochem. J. 2001, 358, 69–77. [Google Scholar] [CrossRef]
- Park, J.; Chen, Y.; Tishkoff, D.X.; Peng, C.; Tan, M.; Dai, L.; Xie, Z.; Zhang, Y.; Zwaans, B.M.M.; Skinner, M.E.; et al. SIRT5-Mediated Lysine Desuccinylation Impacts Diverse Metabolic Pathways. Mol. Cell 2013, 50, 919–930. [Google Scholar] [CrossRef]
- Mathias, R.A.; Greco, T.M.; Oberstein, A.; Budayeva, H.G.; Chakrabarti, R.; Rowland, E.A.; Kang, Y.; Shenk, T.; Cristea, I.M. Sirtuin 4 Is a Lipoamidase Regulating Pyruvate Dehydrogenase Complex Activity. Cell 2014, 159, 1615–1625. [Google Scholar] [CrossRef]
- Du, Z.G.; Liu, X.J.; Chen, T.; Gao, W.C.; Wu, Z.M.; Hu, Z.Q.; Wei, D.; Gao, C.F.; Li, Q.Q. Targeting a Sirt5-Positive Subpopulation Overcomes Multidrug Resistance in Wild-Type Kras Colorectal Carcinomas. Cell Rep. 2018, 22, 2677–2689. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, Y.; Song, Z.; Zhao, R. Pan-Cancer Analysis of Voltage-Dependent Anion Channel (VDAC1) as a Cancer Therapeutic Target or Diagnostic Biomarker. Dis. Markers 2022, 2022, 5946110. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Fu, Y.; Wang, X.; Shi, H.; Huang, Y.; Song, X.; Li, L.; Song, N.; Luo, Y. Voltage-Dependent Anion Channel 1 Is Involved in Endostatin-Induced Endothelial Cell Apoptosis. FASEB J. 2008, 22, 2809–2820. [Google Scholar] [CrossRef]
- Chen, Y.; Gaczynska, M.; Osmulski, P.; Polci, R.; Riley, D.J. Phosphorylation by Nek1 Regulates Opening and Closing of Voltage Dependent Anion Channel 1. Biochem. Biophys. Res. Commun. 2010, 394, 798–803. [Google Scholar] [CrossRef]
- Carr, E.L.; Kelman, A.; Wu, G.S.; Gopaul, R.; Senkevitch, E.; Aghvanyan, A.; Turay, A.M.; Frauwirth, K.A. Glutamine Uptake and Metabolism Are Coordinately Regulated by ERK/MAPK during T Lymphocyte Activation. J. Immunol. 2010, 185, 1037–1044. [Google Scholar] [CrossRef]
- Li, W.; Ji, L.; Tian, J.; Tang, W.; Shan, X.; Zhao, P.; Chen, H.; Zhang, C.; Xu, M.; Lu, R.; et al. Ophiopogonin D Alleviates Diabetic Myocardial Injuries by Regulating Mitochondrial Dynamics. J. Ethnopharmacol. 2021, 271, 113853. [Google Scholar] [CrossRef]
- Lapek, J.D.; Lewinski, M.K.; Wozniak, J.M.; Guatelli, J.; Gonzalez, D.J. Quantitative Temporal Viromics of an Inducible HIV-1 Model Yields Insight to Global Host Targets and Phospho-Dynamics Associated with Protein Vpr. Mol. Cell. Proteom. 2017, 16, 1447–1461. [Google Scholar] [CrossRef]
- Huang, C.Y.; Chiang, S.F.; Lin, T.Y.; Chiou, S.H.; Chow, K.C. HIV-1 Vpr Triggers Mitochondrial Destruction by Impairing Mfn2-Mediated ER-Mitochondria Interaction. PLoS ONE 2012, 7, e33657. [Google Scholar] [CrossRef]
- Giacomello, M.; Pellegrini, L. The Coming of Age of the Mitochondria-ER Contact: A Matter of Thickness. Cell Death Differ. 2016, 23, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Scorrano, L.; De Matteis, M.A.; Emr, S.; Giordano, F.; Hajnóczky, G.; Kornmann, B.; Lackner, L.L.; Levine, T.P.; Pellegrini, L.; Reinisch, K.; et al. Coming Together to Define Membrane Contact Sites. Nat. Commun. 2019, 10, 1287. [Google Scholar] [CrossRef] [PubMed]
- Flis, V.V.; Daum, G. Lipid Transport between the Endoplasmic Reticulum and Mitochondria. Cold Spring Harb. Perspect. Biol. 2013, 5, a013235. [Google Scholar] [CrossRef]
- Csordás, G.; Thomas, A.P.; Hajnóczky, G. Quasi-Synaptic Calcium Signal Transmission between Endoplasmic Reticulum and Mitochondria. EMBO J. 1999, 18, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Booth, D.M.; Enyedi, B.; Geiszt, M.; Várnai, P.; Hajnóczky, G. Redox Nanodomains Are Induced by and Control Calcium Signaling at the ER-Mitochondrial Interface. Mol. Cell 2016, 63, 240–248. [Google Scholar] [CrossRef]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes Form at ER-Mitochondria Contact Sites. Nature 2013, 495, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Perrone, M.; Caroccia, N.; Genovese, I.; Missiroli, S.; Modesti, L.; Pedriali, G.; Vezzani, B.; Vitto, V.A.M.; Antenori, M.; Lebiedzinska-Arciszewska, M.; et al. The Role of Mitochondria-Associated Membranes in Cellular Homeostasis and Diseases. Int. Rev. Cell. Mol. Biol. 2020, 350, 119–196. [Google Scholar] [CrossRef]
- Jean Beltran, P.M.; Mathias, R.A.; Cristea, I.M. A Portrait of the Human Organelle Proteome In Space and Time during Cytomegalovirus Infection. Cell Syst. 2016, 3, 361–373.e6. [Google Scholar] [CrossRef]
- Cook, K.C.; Tsopurashvili, E.; Needham, J.M.; Thompson, S.R.; Cristea, I.M. Restructured Membrane Contacts Rewire Organelles for Human Cytomegalovirus Infection. Nat. Commun. 2022, 13, 4720. [Google Scholar] [CrossRef] [PubMed]
- Jean Beltran, P.M.; Cook, K.C.; Hashimoto, Y.; Galitzine, C.; Murray, L.A.; Vitek, O.; Cristea, I.M. Infection-Induced Peroxisome Biogenesis Is a Metabolic Strategy for Herpesvirus Replication. Cell Host Microbe 2018, 24, 526–541.e7. [Google Scholar] [CrossRef]
- Knoops, K.; Kikkert, M.; Van Den Worm, S.H.E.; Zevenhoven-Dobbe, J.C.; Van Der Meer, Y.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum. PLoS Biol. 2008, 6, 1957–1974. [Google Scholar] [CrossRef]
- Brobeil, A.; Bobrich, M.; Tag, C.; Wimmer, M. PTPIP51 in Protein Interactions: Regulation and In Situ Interacting Partners. Cell Biochem. Biophys. 2012, 63, 211–222. [Google Scholar] [CrossRef]
- Brobeil, A.; Koch, P.; Eiber, M.; Tag, C.; Wimmer, M. The Known Interactome of PTPIP51 in HaCaT Cells—Inhibition of Kinases and Receptors. Int. J. Biochem. Cell Biol. 2014, 46, 19–31. [Google Scholar] [CrossRef]
- Brobeil, A.; Graf, M.; Eiber, M.; Wimmer, M. Interaction of PTPIP51 with Tubulin, CGI-99 and Nuf2 During Cell Cycle Progression. Biomolecules 2012, 2, 122–142. [Google Scholar] [CrossRef] [PubMed]
- Mattia, T.D.; Martinet, A.; Ikhlef, S.; McEwen, A.G.; Nominé, Y.; Wendling, C.; Poussin-Courmontagne, P.; Voilquin, L.; Eberling, P.; Ruffenach, F.; et al. FFAT Motif Phosphorylation Controls Formation and Lipid Transfer Function of Inter-Organelle Contacts. EMBO J. 2020, 39, e104369. [Google Scholar] [CrossRef]
- Delgado, T.; Carroll, P.A.; Punjabi, A.S.; Margineantu, D.; Hockenbery, D.M.; Lagunoff, M. Induction of the Warburg Effect by Kaposi’s Sarcoma Herpesvirus Is Required for the Maintenance of Latently Infected Endothelial Cells. Proc. Natl. Acad. Sci. USA 2010, 107, 10696–10701. [Google Scholar] [CrossRef]
- Ritter, J.B.; Wahl, A.S.; Freund, S.; Genzel, Y.; Reichl, U. Metabolic Effects of Influenza Virus Infection in Cultured Animal Cells: Intra- and Extracellular Metabolite Profiling. BMC Syst. Biol. 2010, 4, 61. [Google Scholar] [CrossRef] [PubMed]
- Hollenbaugh, J.A.; Munger, J.; Kim, B. Metabolite Profiles of Human Immunodeficiency Virus Infected CD4+ T Cells and Macrophages Using LC-MS/MS Analysis. Virology 2011, 415, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, K.A.; Sanchez, E.L.; Camarda, R.; Lagunoff, M. Dengue Virus Induces and Requires Glycolysis for Optimal Replication. J. Virol. 2015, 89, 2358–2366. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, R.; Kim, S.H.; Shah, H.; Zhang, S.; Liang, J.H.; Fang, Y.; Gentili, M.; Leary, C.N.O.; Elledge, S.J.; et al. SARS-CoV-2 Hijacks Folate and One-Carbon Metabolism for Viral Replication. Nat. Commun. 2021, 12, 1676. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, G.; Xu, Z.G.; Tu, H.; Hu, F.; Dai, J.; Chang, Y.; Chen, Y.; Lu, Y.; Zeng, H.; et al. Lactate Is a Natural Suppressor of RLR Signaling by Targeting MAVS. Cell 2019, 178, 176–189.e15. [Google Scholar] [CrossRef]
- Errea, A.; Cayet, D.; Marchetti, P.; Tang, C.; Kluza, J.; Offermanns, S.; Sirard, J.-C.; Rumbo, M. Lactate Inhibits the Pro-Inflammatory Response and Metabolic Reprogramming in Murine Macrophages in a GPR81-Independent Manner. PLoS ONE 2016, 11, e0163694. [Google Scholar] [CrossRef]
- Watson, M.L.J.; Vignali, P.D.A.; Mullett, S.J.; Overacre-Delgoffe, A.E.; Peralta, R.M.; Grebinoski, S.; Menk, A.V.; Rittenhouse, N.L.; DePeaux, K.; Whetstone, R.D.; et al. Metabolic Support of Tumour-Infiltrating Regulatory T Cells by Lactic Acid. Nature 2021, 591, 645–651. [Google Scholar] [CrossRef]
- Colegio, O.R.; Chu, N.Q.; Szabo, A.L.; Chu, T.; Rhebergen, A.M.; Jairam, V.; Cyrus, N.; Brokowski, C.E.; Eisenbarth, S.C.; Phillips, G.M.; et al. Functional Polarization of Tumour-Associated Macrophages by Tumour-Derived Lactic Acid. Nature 2014, 513, 559–563. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic Regulation of Gene Expression by Histone Lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Gu, J.; Zhou, J.; Chen, Q.; Xu, X.; Gao, J.; Li, X.; Shao, Q.; Zhou, B.; Zhou, H.; Wei, S.; et al. Tumor Metabolite Lactate Promotes Tumorigenesis by Modulating MOESIN Lactylation and Enhancing TGF-β Signaling in Regulatory T Cells. Cell Rep. 2022, 39, 110986. [Google Scholar] [CrossRef]
- Yu, Y.; Clippinger, A.J.; Alwine, J.C. Viral Effects on Metabolism: Changes in Glucose and Glutamine Utilization during Human Cytomegalovirus Infection. Trends Microbiol. 2011, 19, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Vastag, L.; Koyuncu, E.; Grady, S.L.; Shenk, T.E.; Rabinowitz, J.D. Divergent Effects of Human Cytomegalovirus and Herpes Simplex Virus-1 on Cellular Metabolism. PLoS Pathog. 2011, 7, e1002124. [Google Scholar] [CrossRef] [PubMed]
- Mullen, P.J.; Garcia, G.; Purkayastha, A.; Matulionis, N.; Schmid, E.W.; Momcilovic, M.; Sen, C.; Langerman, J.; Ramaiah, A.; Shackelford, D.B.; et al. SARS-CoV-2 Infection Rewires Host Cell Metabolism and Is Potentially Susceptible to MTORC1 Inhibition. Nat. Commun. 2021, 12, 1876. [Google Scholar] [CrossRef]
- Kaarbø, M.; Ager-Wick, E.; Osenbroch, P.Ø.; Kilander, A.; Skinnes, R.; Müller, F.; Eide, L. Human Cytomegalovirus Infection Increases Mitochondrial Biogenesis. Mitochondrion 2011, 11, 935–945. [Google Scholar] [CrossRef]
- Chu, J.; Xing, C.; Du, Y.; Duan, T.; Liu, S.; Zhang, P.; Cheng, C.; Henley, J.; Liu, X.; Qian, C.; et al. Pharmacological Inhibition of Fatty Acid Synthesis Blocks SARS-CoV-2 Replication. Nat. Metab. 2021, 3, 1466–1475. [Google Scholar] [CrossRef] [PubMed]
- Moniruzzaman, M.; Martinez-Gutierrez, C.A.; Weinheimer, A.R.; Aylward, F.O. Dynamic Genome Evolution and Complex Virocell Metabolism of Globally-Distributed Giant Viruses. Nat. Commun. 2020, 11, 1710. [Google Scholar] [CrossRef]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A Role for Mitochondria in NLRP3 Inflammasome Activation. Nature 2011, 469, 221–226. [Google Scholar] [CrossRef]
- Stram, A.R.; Payne, R.M. Post-Translational Modifications in Mitochondria: Protein Signaling in the Powerhouse. Cell. Mol. Life Sci. 2016, 73, 4063–4073. [Google Scholar] [CrossRef] [PubMed]
- Kulej, K.; Avgousti, D.C.; Sidoli, S.; Herrmann, C.; Della Fera, A.N.; Kim, E.T.; Garcia, B.A.; Weitzman, M.D. Time-Resolved Global and Chromatin Proteomics during Herpes Simplex Virus Type 1 (HSV-1) Infection. Mol. Cell. Proteom. 2017, 16, S92–S107. [Google Scholar] [CrossRef]
- Soderholm, S.; Kainov, D.E.; Ohman, T.; Denisova, O.V.; Schepens, B.; Kulesskiy, E.; Imanishi, S.Y.; Corthals, G.; Hintsanen, P.; Aittokallio, T.; et al. Phosphoproteomics to Characterize Host Response During Influenza A Virus Infection of Human Macrophages. Mol. Cell. Proteom. 2016, 15, 3203–3219. [Google Scholar] [CrossRef]
- Miao, M.; Yu, F.; Wang, D.; Tong, Y.; Yang, L.; Xu, J.; Qiu, Y.; Zhou, X.; Zhao, X. Proteomics Profiling of Host Cell Response via Protein Expression and Phosphorylation upon Dengue Virus Infection. Virol. Sin. 2019, 34, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Wynn, R.M.; Chuang, J.L.; Tso, S.C.; Machius, M.; Li, J.; Chuang, D.T. Structural Basis for Inactivation of the Human Pyruvate Dehydrogenase Complex by Phosphorylation: Role of Disordered Phosphorylation Loops. Structure 2008, 16, 1849–1859. [Google Scholar] [CrossRef] [PubMed]
- Ciszak, E.M.; Korotchkina, L.G.; Dominiak, P.M.; Sidhu, S.; Patel, M.S. Structural Basis for Flip-Flop Action of Thiamin Pyrophosphate-Dependent Enzymes Revealed by Human Pyruvate Dehydrogenase. J. Biol. Chem. 2003, 278, 21240–21246. [Google Scholar] [CrossRef]
- Ciszak, E.M.; Makal, A.; Hong, Y.S.; Vettaikkorumakankauv, A.K.; Korotchkina, L.G.; Patel, M.S. How Dihydrolipoamide Dehydrogenase-Binding Protein Binds Dihydrolipoamide Dehydrogenase in the Human Pyruvate Dehydrogenase Complex. J. Biol. Chem. 2006, 281, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Indalao, I.L.; Chida, J.; Yamamoto, Y.; Hanawa, M.; Kido, H. Diisopropylamine Dichloroacetate, a Novel Pyruvate Dehydrogenase Kinase 4 Inhibitor, as a Potential Therapeutic Agent for Metabolic Disorders and Multiorgan Failure in Severe Influenza. PLoS ONE 2014, 9, e98032. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Xu, W.; Jiang, W.; Yu, W.; Lin, Y.; Zhang, T.; Yao, J.; Zhou, L.; Zeng, Y.; Li, H.; et al. Regulation of Cellular Metabolism by Protein Lysine Acetylation. Science 2010, 327, 1000–1004. [Google Scholar] [CrossRef]
- Sawant Dessai, A.; Kalhotra, P.; Novickis, A.T.; Dasgupta, S. Regulation of Tumor Metabolism by Post Translational Modifications on Metabolic Enzymes. Cancer Gene Ther. 2023, 30, 548–558. [Google Scholar] [CrossRef]
- Cimen, H.; Han, M.J.; Yang, Y.; Tong, Q.; Koc, H.; Koc, E.C. Regulation of Succinate Dehydrogenase Activity by SIRT3 in Mammalian Mitochondria. Biochemistry 2010, 49, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Finley, L.W.S.; Haas, W.; Desquiret-Dumas, V.; Wallace, D.C.; Procaccio, V.; Gygi, S.P.; Haigis, M.C. Succinate Dehydrogenase Is a Direct Target of Sirtuin 3 Deacetylase Activity. PLoS ONE 2011, 6, e23295. [Google Scholar] [CrossRef] [PubMed]
- Ogura, M.; Yamaki, J.; Homma, M.K.; Homma, Y. Mitochondrial C-Src Regulates Cell Survival through Phosphorylation of Respiratory Chain Components. Biochem. J. 2012, 447, 281–289. [Google Scholar] [CrossRef]
- Röhrig, F.; Schulze, A. The Multifaceted Roles of Fatty Acid Synthesis in Cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V. Cellular Fatty Acid Metabolism and Cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef]
- Hunkeler, M.; Hagmann, A.; Stuttfeld, E.; Chami, M.; Guri, Y.; Stahlberg, H.; Maier, T. Structural Basis for Regulation of Human Acetyl-CoA Carboxylase. Nature 2018, 558, 470–474. [Google Scholar] [CrossRef] [PubMed]
- German, N.J.; Yoon, H.; Yusuf, R.Z.; Murphy, J.P.; Finley, L.W.S.; Laurent, G.; Haas, W.; Satterstrom, F.K.; Guarnerio, J.; Zaganjor, E.; et al. PHD3 Loss in Cancer Enables Metabolic Reliance on Fatty Acid Oxidation via Deactivation of ACC2. Mol. Cell 2016, 63, 1006–1020. [Google Scholar] [CrossRef]
- Bock, F.J.; Tait, S.W.G. Mitochondria as Multifaceted Regulators of Cell Death. Nat. Rev. Mol. Cell. Biol. 2020, 21, 85–100. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Seol, D.W. The Role of Mitochondria in Apoptosis. BMB Rep. 2008, 41, 11–22. [Google Scholar] [CrossRef]
- Adams, J.M. Ways of Dying: Multiple Pathways to Apoptosis. Genes Dev. 2003, 17, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Ferri, K.F.; Kroemer, G. Organelle-Specific Initiation of Cell Death Pathways. Nat. Cell Biol. 2001, 3, E255–E263. [Google Scholar] [CrossRef]
- Xiang, J.; Chao, D.T.; Korsmeyer, S.J. BAX-Induced Cell Death May Not Require Interleukin 1-Converting Enzyme-like Proteases (Apoptosiscysteine ProteaseFASmitochondriareactive Oxygen Species). Proc. Natl. Acad. Sci. USA 1996, 93, 14559–14563. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, N.J.; Whyte, M.K.; Gilbert, C.S.; Evan, G.I. Inhibition of Ced-3/ICE-Related Proteases Does Not Prevent Cell Death Induced by Oncogenes, DNA Damage, or the Bcl-2 Homologue Bak. J. Cell Biol. 1997, 136, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Amarante-Mendes, G.P.; Finucane, D.M.; Martin, S.J.; Cotter, T.G.; Salvesen, G.S.; Green, D.R. Anti-Apoptotic Oncogenes Prevent Caspase-Dependent and Independent Commitment for Cell Death. Cell Death Differ. 1998, 5, 298–306. [Google Scholar] [CrossRef]
- Julien, O.; Wells, J.A. Caspases and Their Substrates. Cell Death Differ. 2017, 24, 1380–1389. [Google Scholar] [CrossRef]
- Grinberg, M.; Sarig, R.; Zaltsman, Y.; Frumkin, D.; Grammatikakis, N.; Reuveny, E.; Gross, A. TBID Homooligomerizes in the Mitochondrial Membrane to Induce Apoptosis. J. Biol. Chem. 2002, 277, 12237–12245. [Google Scholar] [CrossRef]
- Dix, M.M.; Simon, G.M.; Wang, C.; Okerberg, E.; Patricelli, M.P.; Cravatt, B.F. Functional Interplay between Caspase Cleavage and Phosphorylation Sculpts the Apoptotic Proteome. Cell 2012, 150, 426–440. [Google Scholar] [CrossRef]
- Kurokawa, M.; Kornbluth, S. Caspases and Kinases in a Death Grip. Cell 2009, 138, 838–854. [Google Scholar] [CrossRef]
- Voss, O.H.; Kim, S.; Wewers, M.D.; Doseff, A.I. Regulation of Monocyte Apoptosis by the Protein Kinase Cdelta-Dependent Phosphorylation of Caspase-3. J. Biol. Chem. 2005, 280, 17371–17379. [Google Scholar] [CrossRef]
- Cardone, M.H.; Roy, N.; Stennicke, H.R.; Salvesen, G.S.; Franke, T.F.; Stanbridge, E.; Frisch, S.; Reed, J.C. Regulation of Cell Death Protease Caspase-9 by Phosphorylation. Science 1998, 282, 1318–1321. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.L.; Sahai, E.A.; Yeo, M.; Bosch, M.; Dewar, A.; Olson, M.F. Membrane Blebbing during Apoptosis Results from Caspase-Mediated Activation of ROCK I. Nat. Cell Biol. 2001, 3, 339–345. [Google Scholar] [CrossRef]
- Peng, R.; Zhu, J.; Deng, S.; Shi, H.; Xu, S.; Wu, H.; Zou, F. Targeting BAX Ubiquitin-Binding Sites Reveals That BAX Activation Is Essential for Its Ubiquitin-Dependent Degradation. J. Cell. Biochem. 2020, 121, 2802–2810. [Google Scholar] [CrossRef]
- Bazylianska, V.; Kalpage, H.A.; Wan, J.; Vaishnav, A.; Mahapatra, G.; Turner, A.A.; Chowdhury, D.D.; Kim, K.; Morse, P.T.; Lee, I.; et al. Lysine 53 Acetylation of Cytochrome c in Prostate Cancer: Warburg Metabolism and Evasion of Apoptosis. Cells 2021, 10, 802. [Google Scholar] [CrossRef] [PubMed]
- Wasiak, S.; Zunino, R.; McBride, H.M. Bax/Bak Promote Sumoylation of DRP1 and Its Stable Association with Mitochondria during Apoptotic Cell Death. J. Cell Biol. 2007, 177, 439–450. [Google Scholar] [CrossRef]
- Harder, Z.; Zunino, R.; McBride, H. Sumo1 Conjugates Mitochondrial Substrates and Participates in Mitochondrial Fission. Curr. Biol. 2004, 14, 340–345. [Google Scholar] [CrossRef]
- Prudent, J.; Zunino, R.; Sugiura, A.; Mattie, S.; Shore, G.C.; McBride, H.M. MAPL SUMOylation of Drp1 Stabilizes an ER/Mitochondrial Platform Required for Cell Death. Mol. Cell 2015, 59, 941–955. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zou, Q.; Zhao, L. VPTMdb: A Viral Posttranslational Modification Database. Brief. Bioinform. 2021, 22, bbaa251. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Mukherjee, A.; Patra, U.; Bhowmick, R.; Basak, T.; Sengupta, S.; Chawla-Sarkar, M. Tyrosine Phosphorylation Modulates Mitochondrial Chaperonin Hsp60 and Delays Rotavirus NSP4-Mediated Apoptotic Signaling in Host Cells. Cell. Microbiol. 2017, 19, e12670. [Google Scholar] [CrossRef]
- Yoo, Y.S.; Park, Y.J.; Lee, H.S.; Oanh, N.T.K.; Cho, M.Y.; Heo, J.; Lee, E.S.; Cho, H.; Park, Y.Y.; Cho, H. Mitochondria Ubiquitin Ligase, MARCH5 Resolves Hepatitis B Virus X Protein Aggregates in the Liver Pathogenesis. Cell Death Dis. 2019, 10, 938. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Shteinfer-Kuzmine, A.; Verma, A. VDAC1 at the Intersection of Cell Metabolism, Apoptosis, and Diseases. Biomolecules 2020, 10, 1485. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Ide, T.; Yanagida, T.; Tsujimoto, Y. Electrophysiological Study of a Novel Large Pore Formed by Bax and the Voltage-Dependent Anion Channel That Is Permeable to Cytochrome c. J. Biol. Chem. 2000, 275, 12321–12325. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Zakar, M.; Rosenthal, K.; Abu-Hamad, S. Key Regions of VDAC1 Functioning in Apoptosis Induction and Regulation by Hexokinase. Biochim. Biophys. Acta (BBA) Bioenerg. 2009, 1787, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Schein, S.J.; Colombini, M.; Finkelstein, A. Reconstitution in Planar Lipid Bilayers of a Voltage-Dependent Anion-Selective Channel Obtained from Paramecium Mitochondria. J. Membr. Biol. 1976, 30, 99–120. [Google Scholar] [CrossRef]
- Shimizu, S.; Narita, M.; Tsujimoto, Y. Bcl-2 Family Proteins Regulate the Release of Apoptogenic Cytochrome c by the Mitochondrial Channel VDAC. Nature 1999, 399, 483–487. [Google Scholar] [CrossRef]
- Ujwal, R.; Cascio, D.; Colletier, J.P.; Faham, S.; Zhang, J.; Toro, L.; Ping, P.; Abramson, J. The Crystal Structure of Mouse VDAC1 at 2.3 A Resolution Reveals Mechanistic Insights into Metabolite Gating. Proc. Natl. Acad. Sci. USA 2008, 105, 17742–17747. [Google Scholar] [CrossRef]
- Young, M.J.; Bay, D.C.; Hausner, G.; Court, D.A. The Evolutionary History of Mitochondrial Porins. BMC Evol. Biol. 2007, 7, 31. [Google Scholar] [CrossRef]
- Pittalà, M.G.G.; Conti Nibali, S.; Reina, S.; Cunsolo, V.; Di Francesco, A.; De Pinto, V.; Messina, A.; Foti, S.; Saletti, R. Vdacs Post-Translational Modifications Discovery by Mass Spectrometry: Impact on Their Hub Function. Int. J. Mol. Sci. 2021, 22, 12833. [Google Scholar] [CrossRef]
- Kerner, J.; Lee, K.; Tandler, B.; Hoppel, C.L. VDAC Proteomics: Post-Translation Modifications. Biochim. Biophys. Acta Biomembr. 2012, 1818, 1520–1525. [Google Scholar] [CrossRef]
- Ramírez, C.M.; González, M.; Díaz, M.; Alonso, R.; Ferrer, I.; Santpere, G.; Puig, B.; Meyer, G.; Marin, R. VDAC and ERα Interaction in Caveolae from Human Cortex Is Altered in Alzheimer’s Disease. Mol. Cell. Neurosci. 2009, 42, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.C.; Fountoulakis, M.; Cairns, N.; Lubec, G. Changes of Voltage-Dependent Anion-Selective Channel Proteins VDAC1 and VDAC2 Brain Levels in Patients with Alzheimer’s Disease and Down Syndrome. Electrophoresis 2001, 22, 172–179. [Google Scholar] [CrossRef]
- Fukada, K.; Zhang, F.; Vien, A.; Cashman, N.R.; Zhu, H. Mitochondrial Proteomic Analysis of a Cell Line Model of Familial Amyotrophic Lateral Sclerosis. Mol. Cell. Proteom. 2004, 3, 1211–1223. [Google Scholar] [CrossRef] [PubMed]
- Israelson, A.; Arbel, N.; Da Cruz, S.; Ilieva, H.; Yamanaka, K.; Shoshan-Barmatz, V.; Cleveland, D.W. Misfolded Mutant SOD1 Directly Inhibits VDAC1 Conductance in a Mouse Model of Inherited ALS. Neuron 2010, 67, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Pittalà, M.G.G.; Reina, S.; Cubisino, S.A.M.; Cucina, A.; Formicola, B.; Cunsolo, V.; Foti, S.; Saletti, R.; Messina, A. Post-Translational Modification Analysis of VDAC1 in ALS-SOD1 Model Cells Reveals Specific Asparagine and Glutamine Deamidation. Antioxidants 2020, 9, 1218. [Google Scholar] [CrossRef]
- Pittalà, M.G.G.; Reina, S.; Nibali, S.C.; Cucina, A.; Cubisino, S.A.M.; Cunsolo, V.; Amodeo, G.F.; Foti, S.; De Pinto, V.; Saletti, R.; et al. Specific Post-Translational Modifications of VDAC3 in ALS-SOD1 Model Cells Identified by High-Resolution Mass Spectrometry. Int. J. Mol. Sci. 2022, 23, 15853. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, J.G.; Hoek, J.B.; Shulga, N. Activation of Glycogen Synthase Kinase 3β Disrupts the Binding of Hexokinase II to Mitochondria by Phosphorylating Voltage-Dependent Anion Channel and Potentiates Chemotherapy-Induced Cytotoxicity. Cancer Res. 2005, 65, 10545–10554. [Google Scholar] [CrossRef]
- Rahmani, Z.; Huh, K.-W.; Lasher, R.; Siddiqui, A. Hepatitis B Virus X Protein Colocalizes to Mitochondria with a Human Voltage-Dependent Anion Channel, HVDAC3, and Alters Its Transmembrane Potential. J. Virol. 2000, 74, 2840–2846. [Google Scholar] [CrossRef] [PubMed]
- Waris, G.; Huh, K.-W.; Siddiqui, A. Mitochondrially Associated Hepatitis B Virus X Protein Constitutively Activates Transcription Factors STAT-3 and NF-ΚB via Oxidative Stress. Mol. Cell. Biol. 2001, 21, 7721–7730. [Google Scholar] [CrossRef] [PubMed]
- Pittalà, M.G.G.; Saletti, R.; Reina, S.; Cunsolo, V.; De Pinto, V.; Foti, S. A High Resolution Mass Spectrometry Study Reveals the Potential of Disulfide Formation in Human Mitochondrial Voltage-Dependent Anion Selective Channel Isoforms (HVDACs). Int. J. Mol. Sci. 2020, 21, 1468. [Google Scholar] [CrossRef]
- Saletti, R.; Reina, S.; Pittalà, M.G.G.; Belfiore, R.; Cunsolo, V.; Messina, A.; De Pinto, V.; Foti, S. High Resolution Mass Spectrometry Characterization of the Oxidation Pattern of Methionine and Cysteine Residues in Rat Liver Mitochondria Voltage-Dependent Anion Selective Channel 3 (VDAC3). Biochim. Biophys. Acta Biomembr. 2017, 1859, 301–311. [Google Scholar] [CrossRef]
- Reina, S.; Checchetto, V.; Saletti, R.; Gupta, A.; Chaturvedi, D.; Guardiani, C.; Guarino, F.; Scorciapino, M.A.; Magrì, A.; Foti, S.; et al. VDAC3 as a Sensor of Oxidative State of the Intermembrane Space of Mitochondria: The Putative Role of Cysteine Residue Modifications. Oncotarget 2016, 7, 2249–2268. [Google Scholar] [CrossRef]
- Baines, C.P.; Kaiser, R.A.; Sheiko, T.; Craigen, W.J.; Molkentin, J.D. Voltage-Dependent Anion Channels Are Dispensable for Mitochondrial-Dependent Cell Death. Nat. Cell Biol. 2007, 9, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Gupta, R.; Blanco, L.P.; Yang, S.; Shteinfer-Kuzmine, A.; Wang, K.; Zhu, J.; Yoon, H.E.; Wang, X.; Kerkhofs, M.; et al. Mitochondrial Biology VDAC Oligomers Form Mitochondrial Pores to Release MtDNA Fragments and Promote Lupus-like Disease. Science 2019, 366, 1531–1536. [Google Scholar] [CrossRef]
- Ham, S.J.; Lee, D.; Yoo, H.; Jun, K.; Shin, H.; Chung, J. Decision between Mitophagy and Apoptosis by Parkin via VDAC1 Ubiquitination. Proc. Natl. Acad. Sci. USA 2020, 117, 4281–4291. [Google Scholar] [CrossRef]
- Le, A.; Lane, A.N.; Hamaker, M.; Bose, S.; Gouw, A.; Barbi, J.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Zhang, H.; et al. Glucose-Independent Glutamine Metabolism via TCA Cycling for Proliferation and Survival in b Cells. Cell Metab. 2012, 15, 110–121. [Google Scholar] [CrossRef]
- Torres, C.-R.; Hart, G.W. Topography and Polypeptide Distribution of Terminal N-Acetylglucosamine Residues on the Surfaces of Intact Lymphocytes EVIDENCE FOR O-LINKED GlcNAc. J. Biol. Chem. 1984, 259, 3308–3317. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.S.; Ma, J.; Hart, G.W. Diabetes-Associated Dysregulation of O-GlcNAcylation in Rat Cardiac Mitochondria. Proc. Natl. Acad. Sci. USA 2015, 112, 6050–6055. [Google Scholar] [CrossRef]
- Love, D.C.; Kochran, J.; Cathey, R.L.; Shin, S.H.; Hanover, J.A. Mitochondrial and Nucleocytoplasmic Targeting of O-Linked GlcNAc Transferase. J. Cell Sci. 2003, 116, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, T.; Wei, A.C.; Banerjee, P.; O’Rourke, B.; Hart, G.W. O-GlcNAcomic Profiling Identifies Widespread O-Linked β-N-Acetylglucosamine Modification (O-GlcNAcylation) in Oxidative Phosphorylation System Regulating Cardiac Mitochondrial Function. J. Biol. Chem. 2015, 290, 29141–29153. [Google Scholar] [CrossRef]
- Ma, J.; Banerjee, P.; Whelan, S.A.; Liu, T.; Wei, A.C.; Ramirez-Correa, G.; McComb, M.E.; Costello, C.E.; O’Rourke, B.; Murphy, A.; et al. Comparative Proteomics Reveals Dysregulated Mitochondrial O-GlcNAcylation in Diabetic Hearts. J. Proteome Res. 2016, 15, 2254–2264. [Google Scholar] [CrossRef]
- Golks, A.; Tran, T.T.T.; Goetschy, J.F.; Guerini, D. Requirement for O-Linked N-Acetylglucosaminyltransferase in Lymphocytes Activation. EMBO J. 2007, 26, 4368–4379. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.; Pathak, S.; Grzes, K.M.; Damerow, S.; Sinclair, L.V.; Van Aalten, D.M.F.; Cantrell, D.A. Glucose and Glutamine Fuel Protein O-GlcNAcylation to Control T Cell Self-Renewal and Malignancy. Nat. Immunol. 2016, 17, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Kneass, Z.T.; Marchase, R.B. Protein O-GlcNAc Modulates Motility-Associated Signaling Intermediates in Neutrophils. J. Biol. Chem. 2005, 280, 14579–14585. [Google Scholar] [CrossRef] [PubMed]
- Wallace, C.; Keast, D. Glutamine and Macrophage Function. Metabolism 1992, 41, 1016–1020. [Google Scholar] [CrossRef]
- Murphy, C.; Newsholme, P. Importance of Glutamine Metabolism in Murine Macrophages and Human Monocytes to L-Arginine Biosynthesis and Rates of Nitrite or Urea Production. Clin. Sci. 1998, 95, 397–407. [Google Scholar] [CrossRef]
- Bellows, C.F.; Jaffe, B.M. Glutamine Is Essential for Nitric Oxide Synthesis by Murine Macrophages. J. Surg. Res. 1999, 86, 213–219. [Google Scholar] [CrossRef]
- Xu, L.G.; Wang, Y.Y.; Han, K.J.; Li, L.Y.; Zhai, Z.; Shu, H.B. VISA Is an Adapter Protein Required for Virus-Triggered IFN-β Signaling. Mol. Cell 2005, 19, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.B.; Sun, L.; Ea, C.K.; Chen, Z.J. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein That Activates NF-ΚB and IRF3. Cell 2005, 122, 669–682. [Google Scholar] [CrossRef]
- Meylan, E.; Curran, J.; Hofmann, K.; Moradpour, D.; Binder, M.; Bartenschlager, R.; Tschopp, J. Cardif Is an Adaptor Protein in the RIG-I Antiviral Pathway and Is Targeted by Hepatitis C Virus. Nature 2005, 437, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an Adaptor Triggering RIG-I- and Mda5-Mediated Type I Interferon Induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef]
- Peisley, A.; Wu, B.; Yao, H.; Walz, T.; Hur, S. RIG-I Forms Signaling-Competent Filaments in an ATP-Dependent, Ubiquitin-Independent Manner. Mol. Cell 2013, 51, 573–583. [Google Scholar] [CrossRef]
- Wu, B.; Peisley, A.; Richards, C.; Yao, H.; Zeng, X.; Lin, C.; Chu, F.; Walz, T.; Hur, S. Structural Basis for DsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5. Cell 2013, 152, 276–289. [Google Scholar] [CrossRef]
- Hou, F.; Sun, L.; Zheng, H.; Skaug, B.; Jiang, Q.X.; Chen, Z.J. MAVS Forms Functional Prion-like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell 2011, 146, 448–461. [Google Scholar] [CrossRef]
- Zhong, B.; Yang, Y.; Li, S.; Wang, Y.Y.; Li, Y.; Diao, F.; Lei, C.; He, X.; Zhang, L.; Tien, P.; et al. The Adaptor Protein MITA Links Virus-Sensing Receptors to IRF3 Transcription Factor Activation. Immunity 2008, 29, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Barber, G.N. STING Is an Endoplasmic Reticulum Adaptor That Facilitates Innate Immune Signalling. Nature 2008, 455, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.H.; MacMillan, J.B.; Chen, Z.J. RNA Polymerase III Detects Cytosolic DNA and Induces Type I Interferons through the RIG-I Pathway. Cell 2009, 138, 576–591. [Google Scholar] [CrossRef] [PubMed]
- Nazmi, A.; Mukhopadhyay, R.; Dutta, K.; Basu, A. STING Mediates Neuronal Innate Immune Response Following Japanese Encephalitis Virus Infection. Sci. Rep. 2012, 2, 347. [Google Scholar] [CrossRef]
- Holm, C.K.; Rahbek, S.H.; Gad, H.H.; Bak, R.O.; Jakobsen, M.R.; Jiang, Z.; Hansen, A.L.; Jensen, S.K.; Sun, C.; Thomsen, M.K.; et al. Influenza A Virus Targets a CGAS-Independent STING Pathway That Controls Enveloped RNA Viruses. Nat. Commun. 2016, 7, 10680. [Google Scholar] [CrossRef]
- Lum, K.K.; Song, B.; Federspiel, J.D.; Diner, B.A.; Howard, T.; Cristea, I.M. Interactome and Proteome Dynamics Uncover Immune Modulatory Associations of the Pathogen Sensing Factor CGAS. Cell Syst. 2018, 7, 627–642.e6. [Google Scholar] [CrossRef]
- Vitour, D.; Dabo, S.; Pour, M.A.; Vilasco, M.; Vidalain, P.O.; Jacob, Y.; Mezel-Lemoine, M.; Paz, S.; Arguello, M.; Lin, R.; et al. Polo-like Kinase 1 (PLK1) Regulates Interferon (IFN) Induction by MAVS. J. Biol. Chem. 2009, 284, 21797–21809. [Google Scholar] [CrossRef]
- Cheng, J.; Liao, Y.; Xiao, L.; Wu, R.; Zhao, S.; Chen, H.; Hou, B.; Zhang, X.; Liang, C.; Xu, Y.; et al. Autophagy Regulates MAVS Signaling Activation in a Phosphorylation-Dependent Manner in Microglia. Cell Death Differ. 2016, 24, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Arimoto, K.I.; Takahashi, H.; Hishiki, T.; Konishi, H.; Fujita, T.; Shimotohno, K. Negative Regulation of the RIG-I Signaling by the Ubiquitin Ligase RNF125. Proc. Natl. Acad. Sci. USA 2007, 104, 7500–7505. [Google Scholar] [CrossRef] [PubMed]
- Tanzer, M.C.; Bludau, I.; Stafford, C.A.; Hornung, V.; Mann, M. Phosphoproteome Profiling Uncovers a Key Role for CDKs in TNF Signaling. Nat. Commun. 2021, 12, 6053. [Google Scholar] [CrossRef] [PubMed]
- Erber, L.N.; Luo, A.; Gong, Y.; Beeson, M.; Tu, M.; Tran, P.; Chen, Y. Iron Deficiency Reprograms Phosphorylation Signaling and Reduces O-Glcnac Pathways in Neuronal Cells. Nutrients 2021, 13, 179. [Google Scholar] [CrossRef]
- Rinaldi, L.; Senatore, E.; Iannucci, R.; Chiuso, F.; Feliciello, A. Control of Mitochondrial Activity by the Ubiquitin Code in Health and Cancer. Cells 2023, 12, 234. [Google Scholar] [CrossRef] [PubMed]
- Linn, T.C.; Pettit, F.H.; Reed, L.J. Alpha-Keto Acid Dehydrogenase Complexes. X. Regulation of the Activity of the Pyruvate Dehydrogenase Complex from Beef Kidney Mitochondria by Phosphorylation and Dephosphorylation. Proc. Natl. Acad. Sci. USA 1969, 62, 234–241. [Google Scholar] [CrossRef]
- Teague, W.M.; Pettit, F.H.; Wu, T.-L.; Silberman, S.R.; Reed, L.J. Purification and Properties of Pyruvate Dehydrogenase Phosphatase from Bovine Heart and Kidney. Biochemistry 1982, 22, 5585–5592. [Google Scholar] [CrossRef]
- Niemi, N.M.; Pagliarini, D.J. The Extensive and Functionally Uncharacterized Mitochondrial Phosphoproteome. J. Biol. Chem. 2021, 297, 100880. [Google Scholar] [CrossRef]
- Lennicke, C.; Cochemé, H.M. Redox Metabolism: ROS as Specific Molecular Regulators of Cell Signaling and Function. Mol. Cell 2021, 81, 3691–3707. [Google Scholar] [CrossRef]
- Foo, J.; Bellot, G.; Pervaiz, S.; Alonso, S. Mitochondria-Mediated Oxidative Stress during Viral Infection. Trends Microbiol. 2022, 30, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.R.; Hirschey, M.D. Nonenzymatic Protein Acylation as a Carbon Stress Regulated by Sirtuin Deacylases. Mol. Cell 2014, 54, 5–16. [Google Scholar] [CrossRef]
- Muoio, D.M.; Williams, A.S.; Grimsrud, P.A. Mitochondrial Lysine Acylation and Cardiometabolic Stress: Truth or Consequence? Curr. Opin. Physiol. 2022, 27, 100551. [Google Scholar] [CrossRef]
- Wagner, G.R.; Payne, R.M. Widespread and Enzyme-Independent Nε-Acetylation and Nε-Succinylation of Proteins in the Chemical Conditions of the Mitochondrial Matrix. J. Biol. Chem. 2013, 288, 29036–29045. [Google Scholar] [CrossRef]
- James, A.M.; Hoogewijs, K.; Logan, A.; Hall, A.R.; Ding, S.; Fearnley, I.M.; Murphy, M.P. Non-Enzymatic N-Acetylation of Lysine Residues by AcetylCoA Often Occurs via a Proximal S-Acetylated Thiol Intermediate Sensitive to Glyoxalase II. Cell Rep. 2017, 18, 2105–2112. [Google Scholar] [CrossRef]
- Weinert, B.T.; Moustafa, T.; Iesmantavicius, V.; Zechner, R.; Choudhary, C. Analysis of Acetylation Stoichiometry Suggests That SIRT3 Repairs Nonenzymatic Acetylation Lesions. EMBO J. 2015, 34, 2620–2632. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.K.; Gupta, R.; Baldus, L.; Lyon, D.; Narita, T.; Lammers, M.; Choudhary, C.; Weinert, B.T. Analysis of Human Acetylation Stoichiometry Defines Mechanistic Constraints on Protein Regulation. Nat. Commun. 2019, 10, 1055. [Google Scholar] [CrossRef]
- Weinert, B.T.; Schölz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine Succinylation Is a Frequently Occurring Modification in Prokaryotes and Eukaryotes and Extensively Overlaps with Acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Frye, R.A. Characterization of Five Human CDNAs with Homology to the Yeast SIR2 Gene: Sir2-like Proteins (Sirtuins) Metabolize NAD and May Have Protein ADP-Ribosyltransferase Activity. Biochem. Biophys. Res. Commun. 1999, 260, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Frye, R.A. Phylogenetic Classification of Prokaryotic and Eukaryotic Sir2-like Proteins. Biochem. Biophys. Res. Commun. 2000, 273, 793–798. [Google Scholar] [CrossRef]
- Feldman, J.L.; Baeza, J.; Denu, J.M. Activation of the Protein Deacetylase SIRT6 by Long-Chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. J. Biol. Chem. 2013, 288, 31350–31356. [Google Scholar] [CrossRef]
- Budayeva, H.G.; Rowland, E.A.; Cristea, I.M. Intricate Roles of Mammalian Sirtuins in Defense against Viral Pathogens. J. Virol. 2016, 90, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Betsinger, C.N.; Cristea, I.M. Mitochondrial Function, Metabolic Regulation, and Human Disease Viewed through the Prism of Sirtuin 4 (SIRT4) Functions. J. Proteome Res. 2019, 18, 1929–1938. [Google Scholar] [CrossRef] [PubMed]
- Lombard, D.B.; Alt, F.W.; Cheng, H.-L.; Bunkenborg, J.; Streeper, R.S.; Mostoslavsky, R.; Kim, J.; Yancopoulos, G.; Valenzuela, D.; Murphy, A.; et al. Mammalian Sir2 Homolog SIRT3 Regulates Global Mitochondrial Lysine Acetylation. Mol. Cell. Biol. 2007, 27, 8807–8814. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Lu, Z.; Xie, Z.; Cheng, Z.; Chen, Y.; Tan, M.; Luo, H.; Zhang, Y.; He, W.; Yang, K.; et al. The First Identification of Lysine Malonylation Substrates and Its Regulatory Enzyme. Mol. Cell. Proteom. 2011, 10, M111.012658. [Google Scholar] [CrossRef]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Sirt5 Is an NAD-Dependent Protein Lysine Demalonylase and Desuccinylase. Science 2012, 334, 806–809. [Google Scholar] [CrossRef]
- Tan, M.; Peng, C.; Anderson, K.A.; Chhoy, P.; Xie, Z.; Dai, L.; Park, J.; Chen, Y.; Huang, H.; Zhang, Y.; et al. Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5. Cell Metab. 2014, 19, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.A.; Huynh, F.K.; Fisher-Wellman, K.; Stuart, J.D.; Peterson, B.S.; Douros, J.D.; Wagner, G.R.; Thompson, J.W.; Madsen, A.S.; Green, M.F.; et al. SIRT4 Is a Lysine Deacylase That Controls Leucine Metabolism and Insulin Secretion. Cell Metab. 2017, 25, 838–855.e15. [Google Scholar] [CrossRef]
- Hallows, W.C.; Lee, S.; Denu, J.M. Sirtuins Deacetylate and Activate Mammalian Acetyl-CoA Synthetases. Proc. Natl. Acad. Sci. USA 2006, 103, 10230–10235. [Google Scholar] [CrossRef]
- Schwer, B.; Bunkenborg, J.; Verdin, R.O.; Andersen, J.S.; Verdin, E. Reversible Lysine Acetylation Controls the Activity of the Mitochondrial Enzyme Acetyl-CoA Synthetase 2. Proc. Natl. Acad. Sci. USA 2006, 103, 10224–10229. [Google Scholar] [CrossRef]
- Hirschey, M.D.; Shimazu, T.; Goetzman, E.; Jing, E.; Schwer, B.; Lombard, D.B.; Grueter, C.A.; Harris, C.; Biddinger, S.; Ilkayeva, O.R.; et al. SIRT3 Regulates Mitochondrial Fatty-Acid Oxidation by Reversible Enzyme Deacetylation. Nature 2010, 464, 121–125. [Google Scholar] [CrossRef]
- Someya, S.; Yu, W.; Hallows, W.C.; Xu, J.; Vann, J.M.; Leeuwenburgh, C.; Tanokura, M.; Denu, J.M.; Prolla, T.A. Sirt3 Mediates Reduction of Oxidative Damage and Prevention of Age-Related Hearing Loss under Caloric Restriction. Cell 2010, 143, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Coleman, M.C.; Pennington, J.D.; Ozden, O.; Park, S.H.; Jiang, H.; Kim, H.S.; Flynn, C.R.; Hill, S.; McDonald, W.H.; et al. Sirt3-Mediated Deacetylation of Evolutionarily Conserved Lysine 122 Regulates MnSOD Activity in Response to Stress. Mol. Cell 2010, 40, 893–904. [Google Scholar] [CrossRef]
- Qiu, X.; Brown, K.; Hirschey, M.D.; Verdin, E.; Chen, D. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metab. 2010, 12, 662–667. [Google Scholar] [CrossRef]
- Dyck, J.R.B.; Cheng, J.F.; Stanley, W.C.; Barr, R.; Chandler, M.P.; Brown, S.; Wallace, D.; Arrhenius, T.; Harmon, C.; Yang, G.; et al. Malonyl Coenzyme a Decarboxylase Inhibition Protects the Ischemic Heart by Inhibiting Fatty Acid Oxidation and Stimulating Glucose Oxidation. Circ. Res. 2004, 94, e78–e84. [Google Scholar] [CrossRef]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 Is a Leucine Sensor for the MTORC1 Pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Rardin, M.J.; He, W.; Nishida, Y.; Newman, J.C.; Carrico, C.; Danielson, S.R.; Guo, A.; Gut, P.; Sahu, A.K.; Li, B.; et al. SIRT5 Regulates the Mitochondrial Lysine Succinylome and Metabolic Networks. Cell Metab. 2014, 18, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Zur Hausen, H. Viruses in Human Cancers. Science 1991, 254, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Patel, K.; Muldoon-Jacobs, K.; Bisht, K.S.; Aykin-Burns, N.; Pennington, J.D.; van der Meer, R.; Nguyen, P.; Savage, J.; Owens, K.M.; et al. SIRT3 Is a Mitochondria-Localized Tumor Suppressor Required for Maintenance of Mitochondrial Integrity and Metabolism during Stress. Cancer Cell 2010, 17, 41–52. [Google Scholar] [CrossRef]
- Finley, L.W.S.; Carracedo, A.; Lee, J.; Souza, A.; Egia, A.; Zhang, J.; Teruya-Feldstein, J.; Moreira, P.I.; Cardoso, S.M.; Clish, C.B.; et al. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1α Destabilization. Cancer Cell 2011, 19, 416–428. [Google Scholar] [CrossRef]
- Bell, E.L.; Emerling, B.M.; Ricoult, S.J.H.; Guarente, L. SirT3 Suppresses Hypoxia Inducible Factor 1α and Tumor Growth by Inhibiting Mitochondrial ROS Production. Oncogene 2011, 30, 2986–2996. [Google Scholar] [CrossRef]
- Alhazzazi, T.Y.; Kamarajan, P.; Joo, N.; Huang, J.Y.; Verdin, E.; D’Silva, N.J.; Kapila, Y.L. Sirtuin-3 (SIRT3), a Novel Potential Therapeutic Target for Oral Cancer. Cancer 2011, 117, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Greene, K.S.; Lukey, M.J.; Wang, X.; Blank, B.; Druso, J.E.; Lin, M.C.J.; Stalnecker, C.A.; Zhang, C.; Abril, Y.N.; Erickson, J.W.; et al. SIRT5 Stabilizes Mitochondrial Glutaminase and Supports Breast Cancer Tumorigenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 26625–26632. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; He, X.; Ye, D.; Lin, Y.; Yu, H.; Yao, C.; Huang, L.; Zhang, J.; Wang, F.; Xu, S.; et al. NADP+-IDH Mutations Promote Hypersuccinylation That Impairs Mitochondria Respiration and Induces Apoptosis Resistance. Mol. Cell 2015, 60, 661–675. [Google Scholar] [CrossRef] [PubMed]
- Csibi, A.; Fendt, S.M.; Li, C.; Poulogiannis, G.; Choo, A.Y.; Chapski, D.J.; Jeong, S.M.; Dempsey, J.M.; Parkhitko, A.; Morrison, T.; et al. The MTORC1 Pathway Stimulates Glutamine Metabolism and Cell Proliferation by Repressing SIRT4. Cell 2013, 153, 840–854. [Google Scholar] [CrossRef]
- Jeong, S.M.; Xiao, C.; Finley, L.W.S.; Lahusen, T.; Souza, A.L.; Pierce, K.; Li, Y.H.; Wang, X.; Laurent, G.; German, N.J.; et al. SIRT4 Has Tumor-Suppressive Activity and Regulates the Cellular Metabolic Response to DNA Damage by Inhibiting Mitochondrial Glutamine Metabolism. Cancer Cell 2013, 23, 450–463. [Google Scholar] [CrossRef]
- Gassen, N.C.; Papies, J.; Bajaj, T.; Emanuel, J.; Dethloff, F.; Chua, R.L.; Trimpert, J.; Heinemann, N.; Niemeyer, C.; Weege, F.; et al. SARS-CoV-2-Mediated Dysregulation of Metabolism and Autophagy Uncovers Host-Targeting Antivirals. Nat. Commun. 2021, 12, 3818. [Google Scholar] [CrossRef]
- Koyuncu, E.; Budayeva, H.G.; Miteva, Y.V.; Ricci, D.P.; Silhavy, T.J.; Shenk, T.; Cristea, I.M. Sirtuins Are Evolutionarily Conserved Viral Restriction Factors. mBio 2014, 5, e02249-14. [Google Scholar] [CrossRef]
- Walter, M.; Chen, I.P.; Vallejo-Gracia, A.; Kim, I.J.; Bielska, O.; Lam, V.L.; Hayashi, J.M.; Cruz, A.; Shah, S.; Soveg, F.W.; et al. SIRT5 Is a Proviral Factor That Interacts with SARS-CoV-2 Nsp14 Protein. PLoS Pathog. 2022, 18, e1010811. [Google Scholar] [CrossRef]
- Gong, Y.; Tang, N.; Liu, P.; Sun, Y.; Lu, S.; Liu, W.; Tan, L.; Song, C.; Qiu, X.; Liao, Y.; et al. Newcastle Disease Virus Degrades SIRT3 via PINK1-PRKN-Dependent Mitophagy to Reprogram Energy Metabolism in Infected Cells. Autophagy 2022, 18, 1503–1521. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-Spectrometric Exploration of Proteome Structure and Function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef]
- Thygesen, C.; Boll, I.; Finsen, B.; Modzel, M.; Larsen, M.R. Characterizing Disease-Associated Changes in Post-Translational Modifications by Mass Spectrometry. Expert Rev. Proteom. 2018, 15, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Geiszler, D.J.; Kong, A.T.; Avtonomov, D.M.; Yu, F.; da Veiga Leprevost, F.; Nesvizhskii, A.I. PTM-Shepherd: Analysis and Summarization of Post-Translational and Chemical Modifications from Open Search Results. Mol. Cell. Proteom. 2021, 20, 100018. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.R.; Thingholm, T.E.; Jensen, O.N.; Roepstorff, P.; Jørgensen, T.J.D. Highly Selective Enrichment of Phosphorylated Peptides from Peptide Mixtures Using Titanium Dioxide Microcolumns. Mol. Cell. Proteom. 2005, 4, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Sprung, R.; Chen, Y.; Xu, Y.; Ball, H.; Pei, J.; Cheng, T.; Kho, Y.; Xiao, H.; Xiao, L.; et al. Substrate and Functional Diversity of Lysine Acetylation Revealed by a Proteomics Survey. Mol. Cell 2006, 23, 607–618. [Google Scholar] [CrossRef]
- Thingholm, T.E.; Jørgensen, T.J.D.; Jensen, O.N.; Larsen, M.R. Highly Selective Enrichment of Phosphorylated Peptides Using Titanium Dioxide. Nat. Protoc. 2006, 1, 1929–1935. [Google Scholar] [CrossRef]
- Udeshi, N.D.; Mani, D.C.; Satpathy, S.; Fereshetian, S.; Gasser, J.A.; Svinkina, T.; Olive, M.E.; Ebert, B.L.; Mertins, P.; Carr, S.A. Rapid and Deep-Scale Ubiquitylation Profiling for Biology and Translational Research. Nat. Commun. 2020, 11, 359. [Google Scholar] [CrossRef]
- Svinkina, T.; Gu, H.; Silva, J.C.; Mertins, P.; Qiao, J.; Fereshetian, S.; Jaffe, J.D.; Kuhn, E.; Udeshi, N.D.; Carr, S.A. Deep, Quantitative Coverage of the Lysine Acetylome Using Novel Anti-Acetyl-Lysine Antibodies and an Optimized Proteomic Workflow. Mol. Cell. Proteom. 2015, 14, 2429–2440. [Google Scholar] [CrossRef]
- Herrmann, C.; Dybas, J.M.; Liddle, J.C.; Price, A.M.; Hayer, K.E.; Lauman, R.; Purman, C.E.; Charman, M.; Kim, E.T.; Garcia, B.A.; et al. Adenovirus-Mediated Ubiquitination Alters Protein-RNA Binding and Aids Viral RNA Processing. Nat. Microbiol. 2020, 5, 1217–1231. [Google Scholar] [CrossRef]
- Song, B.; Greco, T.M.; Lum, K.K.; Taber, C.; Cristea, I.M. The DNA Sensor CGAS Is Decorated by Acetylation and Phosphorylation Modifications in the Context of Immune Signaling. Mol. Cell. Proteom. 2020, 19, 1193–1208. [Google Scholar] [CrossRef]
- Garcia, B.A.; Pesavento, J.J.; Mizzen, C.A.; Kelleher, N.L. Pervasive Combinatorial Modification of Histone H3 in Human Cells. Nat. Methods 2007, 4, 487–489. [Google Scholar] [CrossRef]
- Wu, C.; Tran, J.C.; Zamdborg, L.; Durbin, K.R.; Li, M.; Ahlf, D.R.; Early, B.P.; Thomas, P.M.; Sweedler, J.V.; Kelleher, N.L. A Protease for “middle-down” Proteomics. Nat. Methods 2012, 9, 822–824. [Google Scholar] [CrossRef]
- Donnelly, D.P.; Rawlins, C.M.; DeHart, C.J.; Fornelli, L.; Schachner, L.F.; Lin, Z.; Lippens, J.L.; Aluri, K.C.; Sarin, R.; Chen, B.; et al. Best Practices and Benchmarks for Intact Protein Analysis for Top-down Mass Spectrometry. Nat. Methods 2019, 16, 587–594. [Google Scholar] [CrossRef]
- Brown, K.A.; Chen, B.; Guardado-Alvarez, T.M.; Lin, Z.; Hwang, L.; Ayaz-Guner, S.; Jin, S.; Ge, Y. A Photocleavable Surfactant for Top-down Proteomics. Nat. Methods 2019, 16, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Swaney, D.L.; Beltrao, P.; Starita, L.; Guo, A.; Rush, J.; Fields, S.; Krogan, N.J.; Villén, J. Global Analysis of Phosphorylation and Ubiquitylation Cross-Talk in Protein Degradation. Nat. Methods 2013, 10, 676–682. [Google Scholar] [CrossRef]
- Mertins, P.; Qiao, J.W.; Patel, J.; Udeshi, N.D.; Clauser, K.R.; Mani, D.R.; Burgess, M.W.; Gillette, M.A.; Jaffe, J.D.; Carr, S.A. Integrated Proteomic Analysis of Post-Translational Modifications by Serial Enrichment. Nat. Methods 2013, 10, 634–637. [Google Scholar] [CrossRef]
- Leutert, M.; Entwisle, S.W.; Villén, J. Decoding Post-Translational Modification Crosstalk with Proteomics. Mol. Cell. Proteom. 2021, 20, 100129. [Google Scholar] [CrossRef]
- Ochoa, D.; Jarnuczak, A.F.; Viéitez, C.; Gehre, M.; Soucheray, M.; Mateus, A.; Kleefeldt, A.A.; Hill, A.; Garcia-Alonso, L.; Stein, F.; et al. The Functional Landscape of the Human Phosphoproteome. Nat. Biotechnol. 2020, 38, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Steen, H.; Kuster, B.; Fernandez, M.; Pandey, A.; Mann, M. Detection of Tyrosine Phosphorylated Peptides by Precursor Ion Scanning Quadrupole TOF Mass Spectrometry in Positive Ion Mode. Anal. Chem. 2001, 73, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Trelle, M.B.; Jensen, O.N. Utility of Immonium Ions for Assignment of ε-N-Acetyllysine-Containing Peptides by Tandem Mass Spectrometry. Anal. Chem. 2008, 80, 3422–3430. [Google Scholar] [CrossRef]
- Deng, W.; Wang, C.; Zhang, Y.; Xu, Y.; Zhang, S.; Liu, Z.; Xue, Y. GPS-PAIL: Prediction of Lysine Acetyltransferase-Specific Modification Sites from Protein Sequences. Sci. Rep. 2016, 6, 39787. [Google Scholar] [CrossRef]
- Song, J.; Wang, H.; Wang, J.; Leier, A.; Marquez-Lago, T.; Yang, B.; Zhang, Z.; Akutsu, T.; Webb, G.I.; Daly, R.J. PhosphoPredict: A Bioinformatics Tool for Prediction of Human Kinase-Specific Phosphorylation Substrates and Sites by Integrating Heterogeneous Feature Selection. Sci. Rep. 2017, 7, 6862. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, D.; Jonikas, M.; Lawrence, R.T.; El Debs, B.; Selkrig, J.; Typas, A.; Villén, J.; Santos, S.D.M.; Beltrao, P. An Atlas of Human Kinase Regulation. Mol. Syst. Biol. 2016, 12, 888. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Bludau, I.; Willems, S.; Zeng, W.F.; Strauss, M.T.; Hansen, F.M.; Tanzer, M.C.; Karayel, O.; Schulman, B.A.; Mann, M. The Structural Context of Posttranslational Modifications at a Proteome-Wide Scale. PLoS Biol. 2022, 20, e3001636. [Google Scholar] [CrossRef]
- Beltrao, P.; Albanèse, V.; Kenner, L.R.; Swaney, D.L.; Burlingame, A.; Villén, J.; Lim, W.A.; Fraser, J.S.; Frydman, J.; Krogan, N.J. Systematic Functional Prioritization of Protein Posttranslational Modifications. Cell 2012, 150, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Weinert, B.T.; Wagner, S.A.; Horn, H.; Henriksen, P.; Liu, W.R.; Olsen, J.V.; Jensen, L.J.; Choudhary, C. Proteome-Wide Mapping of the Drosophila Acetylome Demonstrates a High Degree of Conservation of Lysine Acetylation. Sci. Signal. 2011, 4, ra48. [Google Scholar] [CrossRef]
- Tan, C.S.H.; Go, K.D.; Bisteau, X.; Dai, L.; Yong, C.H.; Prabhu, N.; Ozturk, M.B.; Lim, Y.T.; Sreekumar, L.; Lengqvist, J.; et al. Thermal Proximity Coaggregation for System-Wide Profiling of Protein Complex Dynamics in Cells. Science 2018, 359, 1170–1177. [Google Scholar] [CrossRef]
- Justice, J.L.; Kennedy, M.A.; Hutton, J.E.; Liu, D.; Song, B.; Phelan, B.; Cristea, I.M. Systematic Profiling of Protein Complex Dynamics Reveals DNA-PK Phosphorylation of IFI16 En Route to Herpesvirus Immunity. Sci. Adv. 2021, 7, eabg6680. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Sheng, X.; Murray-Nerger, L.A.; Cristea, I.M. Temporal Dynamics of Protein Complex Formation and Dissociation during Human Cytomegalovirus Infection. Nat. Commun. 2020, 11, 806. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.X.; Lee, G.; Cavanaugh, K.E.; Chang, J.W.; Gardel, M.L.; Moellering, R.E. High Throughput Discovery of Functional Protein Modifications by Hotspot Thermal Profiling. Nat. Methods 2019, 16, 894–901. [Google Scholar] [CrossRef]
- Potel, C.M.; Kurzawa, N.; Becher, I.; Typas, A.; Mateus, A.; Savitski, M.M. Impact of Phosphorylation on Thermal Stability of Proteins. Nat. Methods 2021, 18, 757–759. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Bauman, D.; Davis, J.S.; Loyola, A.; Nishioka, K.; Gronlund, J.L.; Reinberg, D.; Meng, F.; Kelleher, N.; McCafferty, D.G. Facile Synthesis of Site-Specifically Acetylated and Methylated Histone Proteins: Reagents for Evaluation of the Histone Code Hypothesis. Proc. Natl. Acad. Sci. USA 2003, 100, 12033–12038. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.D.; Chu, F.; Racki, L.R.; de la Cruz, C.C.; Burlingame, A.L.; Panning, B.; Narlikar, G.J.; Shokat, K.M. The Site-Specific Installation of Methyl-Lysine Analogs into Recombinant Histones. Cell 2007, 128, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Burton, A.J.; Haugbro, M.; Parisi, E.; Muir, T.W. Live-Cell Protein Engineering with an Ultra-Short Split Intein. Proc. Natl. Acad. Sci. USA 2020, 117, 12041–12049. [Google Scholar] [CrossRef] [PubMed]
- Hoppmann, C.; Wong, A.; Yang, B.; Li, S.; Hunter, T.; Shokat, K.M.; Wang, L. Site-Specific Incorporation of Phosphotyrosine Using an Expanded Genetic Code. Nat. Chem. Biol. 2017, 13, 842–844. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Fu, G.; Wang, R.E.; Zhu, X.; Zambaldo, C.; Liu, R.; Liu, T.; Lyu, X.; Du, J.; Xuan, W.; et al. Genetically Encoding Phosphotyrosine and Its Nonhydrolyzable Analog in Bacteria. Nat. Chem. Biol. 2017, 13, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Yang, A.; Lee, S.; Lee, H.W.; Park, C.B.; Park, H.S. Expanding the Genetic Code of Mus Musculus. Nat. Commun. 2017, 8, 14568. [Google Scholar] [CrossRef]
- Uttamapinant, C.; Howe, J.D.; Lang, K.; Beránek, V.; Davis, L.; Mahesh, M.; Barry, N.P.; Chin, J.W. Genetic Code Expansion Enables Live-Cell and Super-Resolution Imaging of Site-Specifically Labeled Cellular Proteins. J. Am. Chem. Soc. 2015, 137, 4602–4605. [Google Scholar] [CrossRef]
- Mideksa, Y.G.; Fottner, M.; Braus, S.; Weiß, C.A.M.; Nguyen, T.A.; Meier, S.; Lang, K.; Feige, M.J. Site-Specific Protein Labeling with Fluorophores as a Tool To Monitor Protein Turnover. ChemBioChem 2020, 21, 1861–1867. [Google Scholar] [CrossRef]
- Duvic, M.; Vu, J. Vorinostat in Cutaneous T-Cell Lymphoma. Drugs Today 2007, 43, 585–599. [Google Scholar] [CrossRef]
- Müller, A.; Florek, M. 5-Azacytidine/Azacitidine. Recent Results Cancer Res. 2010, 184, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Gertz, M.; Steegborn, C. Using Mitochondrial Sirtuins as Drug Targets: Disease Implications and Available Compounds. Cell Mol. Life Sci. 2016, 73, 2871–2896. [Google Scholar] [CrossRef] [PubMed]
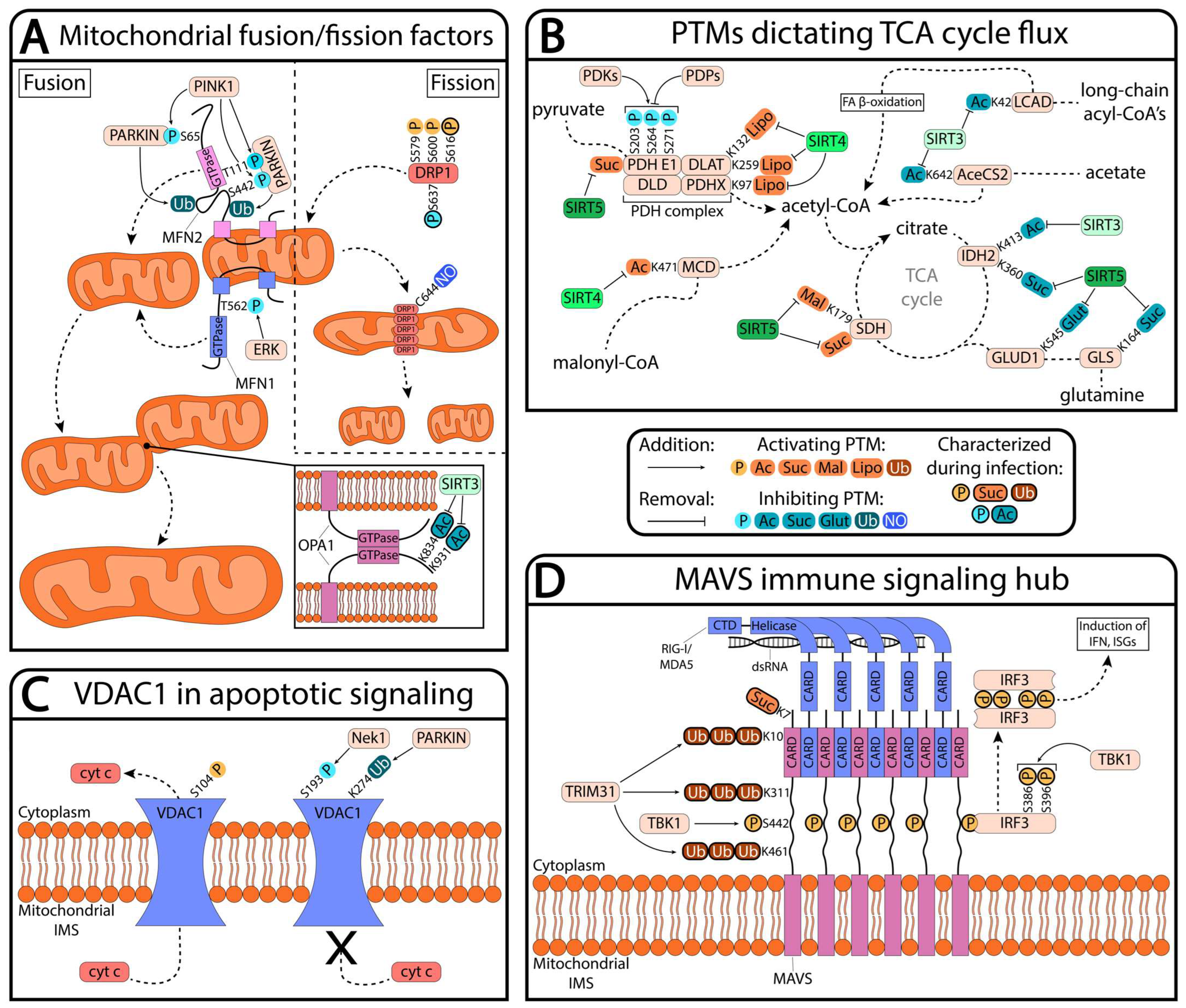
| Substrate | Modification | Site | Function | References |
|---|---|---|---|---|
| AceCS2 | Acetylation | Lys642 | Inhibits enzymatic activity | [54] |
| DRP1 | Phosphorylation | Ser579, Ser600, Ser616; Ser637 | Activates DRP1 binding to mitochondria; inhibits DRP1 binding | [54,55,56,57,58,59,60,61] |
| S-nitrosylation | Cys644 | Inhibits DRP1 oligomerization | [62] | |
| GLS | Succinylation | Lys164 | Inhibits enzymatic activity | [63] |
| GLUD1 | Glutarylation | Lys545 | Inhibits enzymatic activity | [64] |
| IDH2 | Acetylation | Lys413 | Inhibits enzymatic activity | [65] |
| Succinylation | Lys360 | Inhibits enzymatic activity | [66] | |
| LCAD | Acetylation | Lys42 | Inhibits enzymatic activity | [67] |
| MAVS | Polyubiquitination | Lys10, Lys311, Lys461 | Promote MAVS aggregation | [68] |
| Succinylation | Lys7 | Promotes MAVS aggregation | [69] | |
| Phosphorylation | Ser442 | Activates IRF3, innate immune signaling | [33,70] | |
| MCD | Acetylation | Lys471 | Activates enzymatic activity | [71] |
| MFN1 | Phosphorylation | Thr562 | Inhibits mitochondrial fusion | [72] |
| MFN2 | Phosphorylation | Ser65, Thr111, Ser442 | Inhibit mitochondrial fusion | [73,74] |
| OPA1 | Acetylation | Lys834, Lys931 | Inhibit mitochondrial fusion | [49] |
| PDHA1 | Phosphorylation | Ser203, Ser264, Ser271 | Inhibit enzymatic activity | [75,76,77,78] |
| Lipoylation | Lys97, Lys132, Lys259 | Activate enzymatic activity | [79,80] | |
| SDH | Malonylation | Lys179 | Activates enzymatic activity | [81] |
| VDAC1 | Phosphorylation | Ser104; Ser193 | Promotes cytochrome c release; inhibits cytochrome c release | [82,83,84] |
| Ubiquitination | Lys274 | Inhibits apoptotic signaling | [85] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.W.; Tyl, M.D.; Cristea, I.M. Orchestration of Mitochondrial Function and Remodeling by Post-Translational Modifications Provide Insight into Mechanisms of Viral Infection. Biomolecules 2023, 13, 869. https://doi.org/10.3390/biom13050869
Park JW, Tyl MD, Cristea IM. Orchestration of Mitochondrial Function and Remodeling by Post-Translational Modifications Provide Insight into Mechanisms of Viral Infection. Biomolecules. 2023; 13(5):869. https://doi.org/10.3390/biom13050869
Chicago/Turabian StylePark, Ji Woo, Matthew D. Tyl, and Ileana M. Cristea. 2023. "Orchestration of Mitochondrial Function and Remodeling by Post-Translational Modifications Provide Insight into Mechanisms of Viral Infection" Biomolecules 13, no. 5: 869. https://doi.org/10.3390/biom13050869
APA StylePark, J. W., Tyl, M. D., & Cristea, I. M. (2023). Orchestration of Mitochondrial Function and Remodeling by Post-Translational Modifications Provide Insight into Mechanisms of Viral Infection. Biomolecules, 13(5), 869. https://doi.org/10.3390/biom13050869






