OTUD7B Activates Wnt Signaling Pathway through the Interaction with LEF1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmids
2.3. Reagents and Antibodies
2.4. Shor Interfering RNAs (siRNA)
2.5. Site-Directed Mutagenesis
2.6. Immunoprecipitation and Pulldown Assays
2.7. Deubiquitination Assay
2.8. Cycloheximide Chase Assay
2.9. Western Blotting
2.10. Luciferase Assay
2.11. Cytoplasmic and Nuclear Fractionation
2.12. Pulldown Assay with Nuclear and Cytoplasmic Fractions
2.13. Quantitation of Immunoblot Images
2.14. Quantitative PCR Array
2.15. Statistical Analysis
3. Results
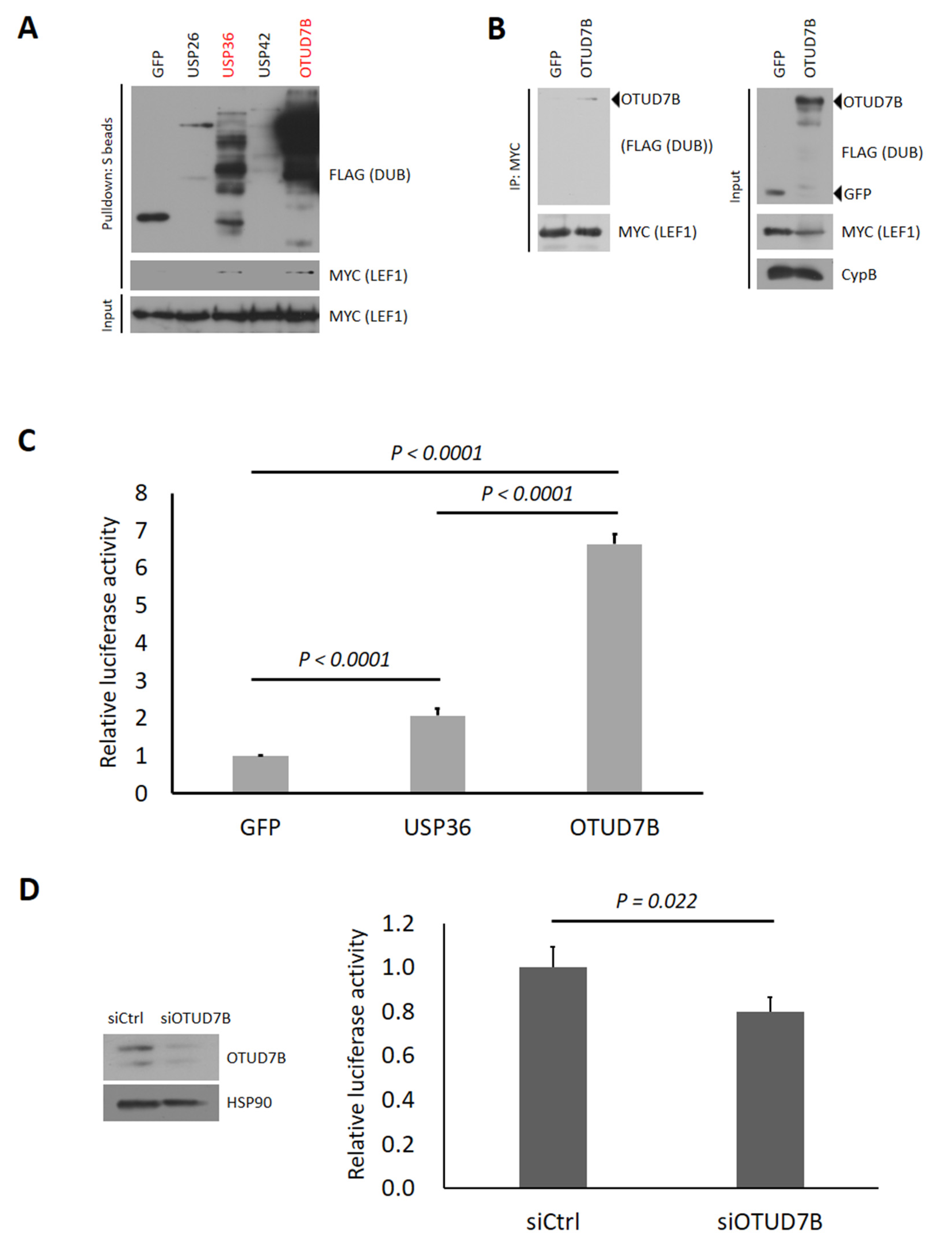
3.1. Identification of Human Deubiquitinases (DUBs) Interacting with LEF1
3.2. OTUD7B Promotes and Its Depletion Suppresses the Transcriptional Activity of LEF1
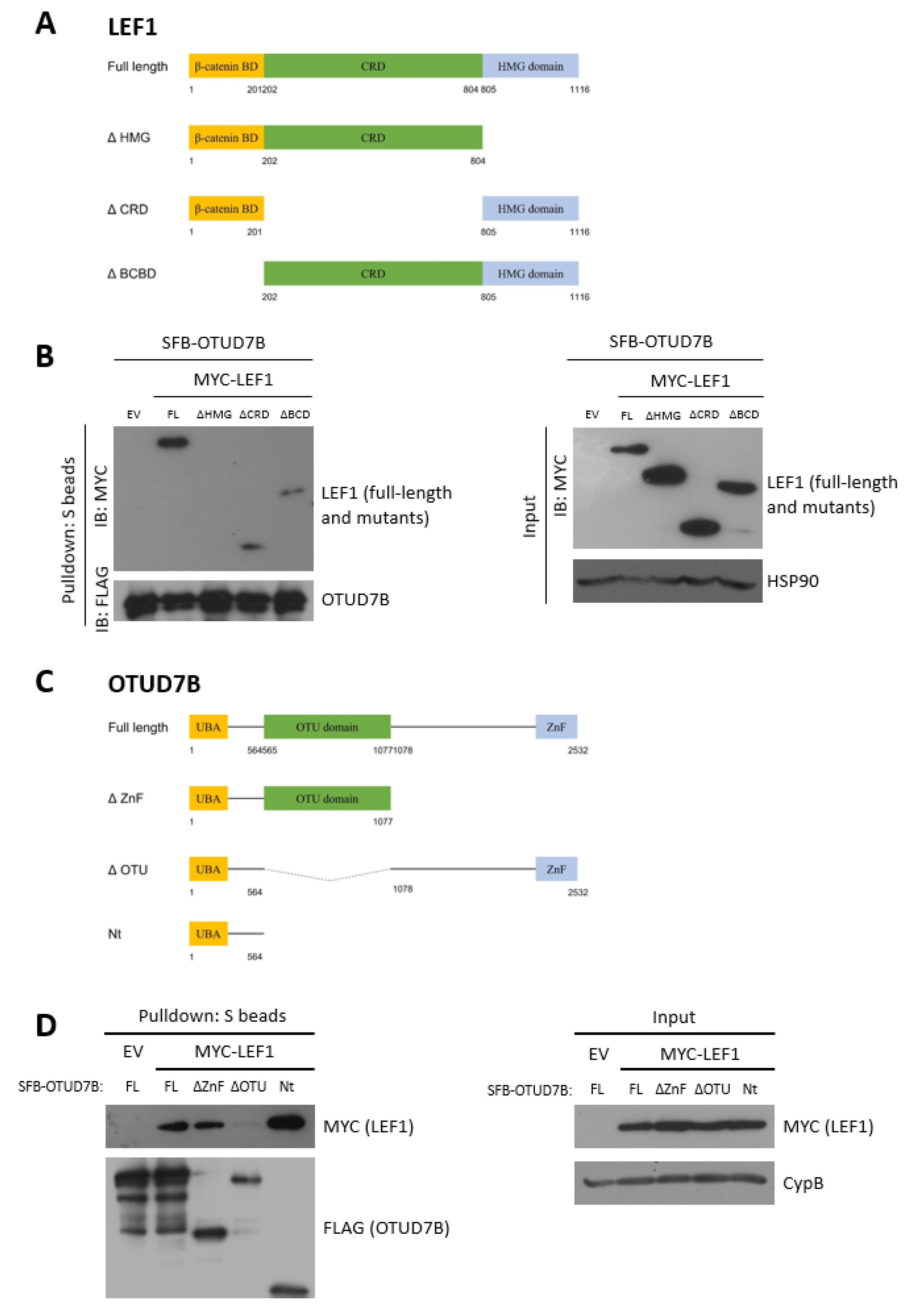
3.3. LEF1 Interacts with OTUD7B through the HMG and UBA Domains, Respectively
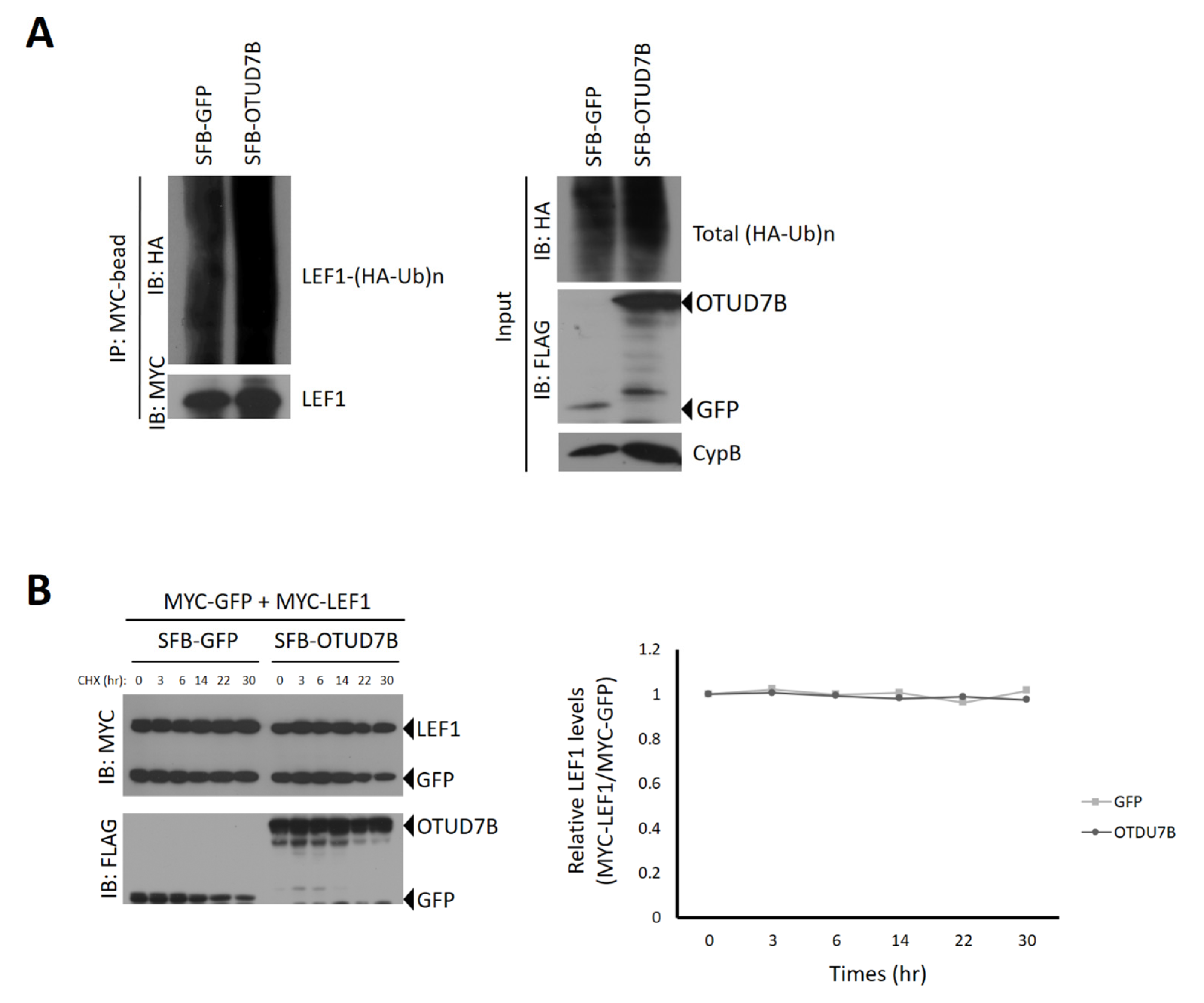
3.4. OTUD7B Does Not Deubiquitinate nor Stabilize LEF1
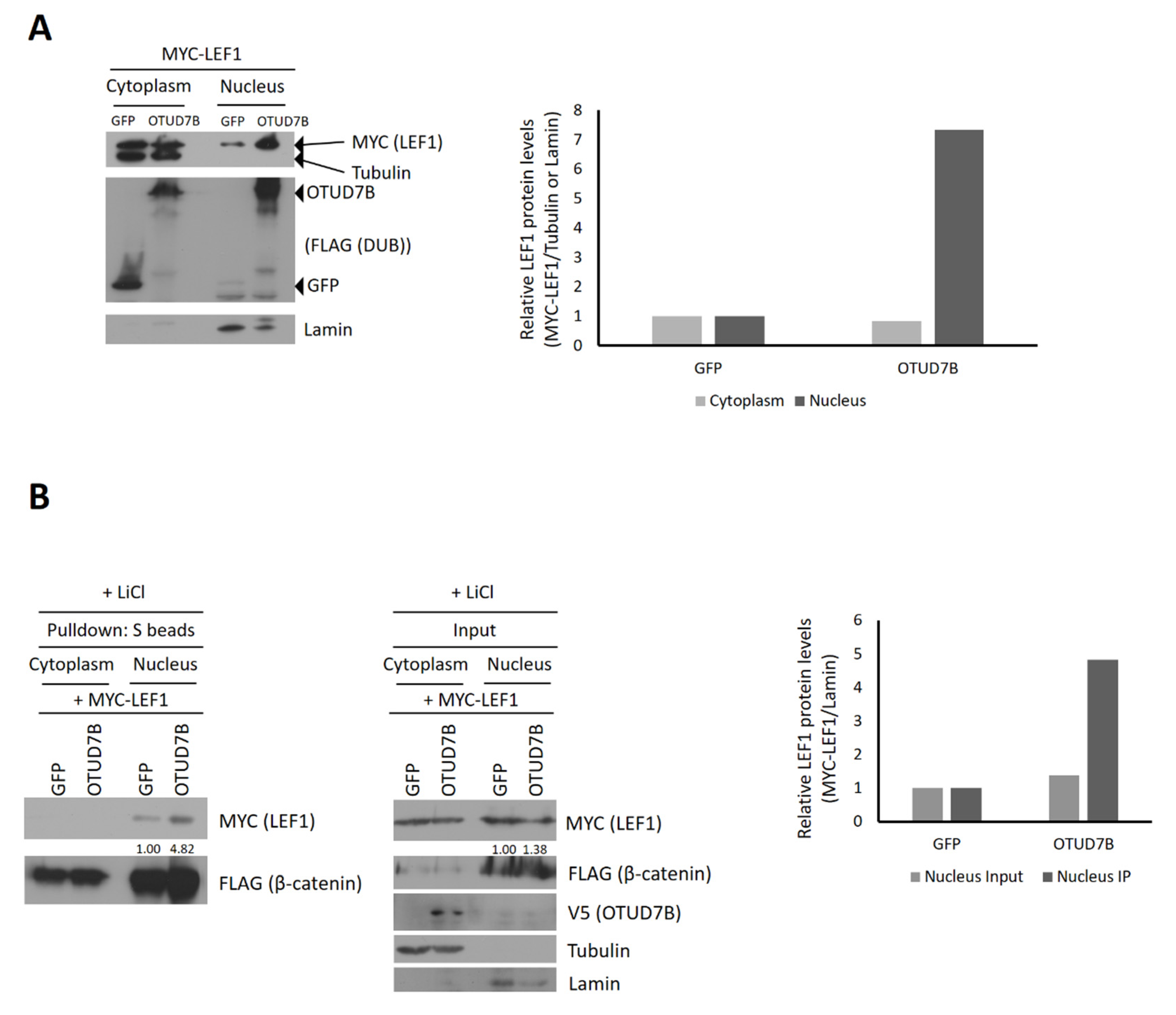
3.5. OTUD7B Promotes the Nuclear Translocation of LEF1
3.6. OTUD7B Promotes the Interaction between LEF1 and β-Catenin
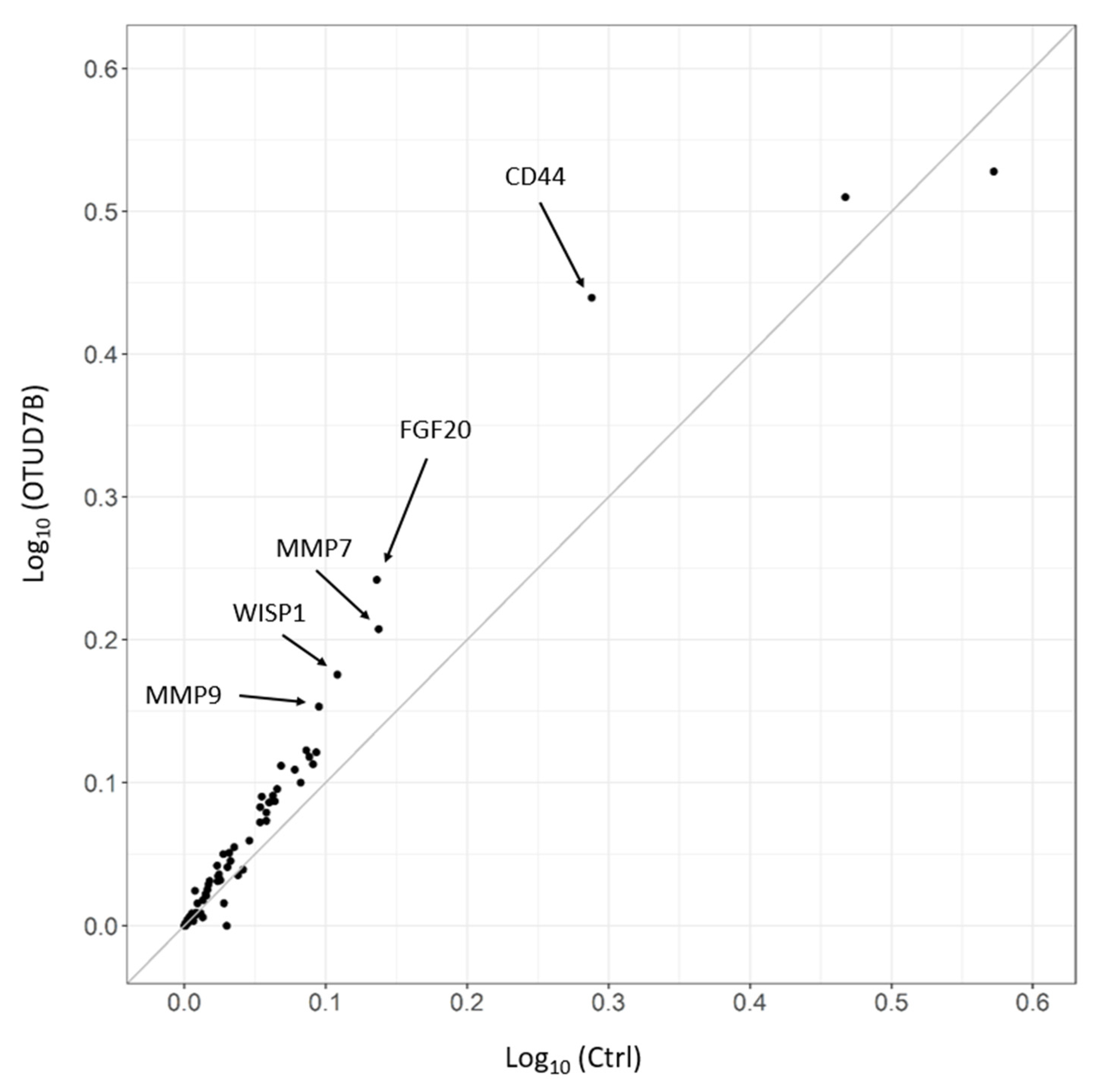
3.7. OTUD7B Upregulates the Expression of Wnt Target Genes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xing, S.; Gai, K.; Li, X.; Shao, P.; Zeng, Z.; Zhao, X.; Zhao, X.; Chen, X.; Paradee, W.J.; Meyerholz, D.K.; et al. Tcf1 and Lef1 are required for the immunosuppressive function of regulatory T cells. J. Exp. Med. 2019, 216, 847–866. [Google Scholar] [CrossRef]
- Sun, H.; He, Z.; Xi, Q.; Zhao, F.; Hu, J.; Wang, J.; Liu, X.; Zhao, Z.; Li, M.; Luo, Y.; et al. Lef1 and Dlx3 May Facilitate the Maturation of Secondary Hair Follicles in the Skin of Gansu Alpine Merino. Genes 2022, 13, 1326. [Google Scholar] [CrossRef]
- Phan, Q.M.; Fine, G.M.; Salz, L.; Herrera, G.G.; Wildman, B.; Driskell, I.M.; Driskell, R.R. Lef1 expression in fibroblasts maintains developmental potential in adult skin to regenerate wounds. Elife 2020, 9, e60066. [Google Scholar] [CrossRef]
- Steinke, F.C.; Xue, H.H. From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells. Immunol. Res. 2014, 59, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; O’Riordan, M.; Okamura, R.; Devaney, E.; Willert, K.; Nusse, R.; Grosschedl, R. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity 2000, 13, 15–24. [Google Scholar] [CrossRef] [Green Version]
- Held, W.; Clevers, H.; Grosschedl, R. Redundant functions of TCF-1 and LEF-1 during T and NK cell development, but unique role of TCF-1 for Ly49 NK cell receptor acquisition. Eur. J. Immunol. 2003, 33, 1393–1398. [Google Scholar] [CrossRef]
- Yu, S.; Zhou, X.; Steinke, F.C.; Liu, C.; Chen, S.C.; Zagorodna, O.; Jing, X.; Yokota, Y.; Meyerholz, D.K.; Mullighan, C.G.; et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 2012, 37, 813–826. [Google Scholar] [CrossRef] [Green Version]
- Petropoulos, K.; Arseni, N.; Schessl, C.; Stadler, C.R.; Rawat, V.P.; Deshpande, A.J.; Heilmeier, B.; Hiddemann, W.; Quintanilla-Martinez, L.; Bohlander, S.K.; et al. A novel role for Lef-1, a central transcription mediator of Wnt signaling, in leukemogenesis. J. Exp. Med. 2008, 205, 515–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutterbach, B.; Westendorf, J.J.; Linggi, B.; Isaac, S.; Seto, E.; Hiebert, S.W. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 2000, 275, 651–656. [Google Scholar] [CrossRef] [Green Version]
- Hecht, A.; Stemmler, M.P. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J. Biol. Chem. 2003, 278, 3776–3785. [Google Scholar] [CrossRef] [Green Version]
- Boras, K.; Hamel, P.A. Alx4 binding to LEF-1 regulates N-CAM promoter activity. J. Biol. Chem. 2002, 277, 1120–1127. [Google Scholar] [CrossRef] [Green Version]
- Behrens, J.; von Kries, J.P.; Kuhl, M.; Bruhn, L.; Wedlich, D.; Grosschedl, R.; Birchmeier, W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 1996, 382, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, S.; Bruhn, L.; Sieber, H.; Pichler, A.; Melchior, F.; Grosschedl, R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001, 15, 3088–3103. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/beta-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target Ther. 2022, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Santiago, L.; Daniels, G.; Wang, D.; Deng, F.M.; Lee, P. Wnt signaling pathway protein LEF1 in cancer, as a biomarker for prognosis and a target for treatment. Am. J. Cancer Res. 2017, 7, 1389–1406. [Google Scholar]
- Ota, S.; Ishitani, S.; Shimizu, N.; Matsumoto, K.; Itoh, M.; Ishitani, T. NLK positively regulates Wnt/beta-catenin signalling by phosphorylating LEF1 in neural progenitor cells. EMBO J. 2012, 31, 1904–1915. [Google Scholar] [CrossRef] [Green Version]
- Ishitani, T.; Ninomiya-Tsuji, J.; Matsumoto, K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol. Cell Biol. 2003, 23, 1379–1389. [Google Scholar] [CrossRef] [Green Version]
- Yamada, M.; Ohnishi, J.; Ohkawara, B.; Iemura, S.; Satoh, K.; Hyodo-Miura, J.; Kawachi, K.; Natsume, T.; Shibuya, H. NARF, an nemo-like kinase (NLK)-associated ring finger protein regulates the ubiquitylation and degradation of T cell factor/lymphoid enhancer factor (TCF/LEF). J. Biol. Chem. 2006, 281, 20749–20760. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Lee, S.; Kim, J.R.; Jho, E.H. Pja2 Inhibits Wnt/beta-catenin Signaling by Reducing the Level of TCF/LEF1. Int. J. Stem Cells 2018, 11, 242–247. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Kim, J. OTUD6A Is an Aurora Kinase A-Specific Deubiquitinase. Int. J. Mol. Sci. 2021, 22, 1936. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Alavi Naini, F.; Sun, Y.; Ma, L. Ubiquitin-specific peptidase 2a (USP2a) deubiquitinates and stabilizes beta-catenin. Am. J. Cancer Res. 2018, 8, 1823–1836. [Google Scholar] [PubMed]
- Grosschedl, R.; Giese, K.; Pagel, J. HMG domain proteins: Architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994, 10, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Stros, M.; Launholt, D.; Grasser, K.D. The HMG-box: A versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 2007, 64, 2590–2606. [Google Scholar] [CrossRef]
- Dikic, I.; Wakatsuki, S.; Walters, K.J. Ubiquitin-binding domains—From structures to functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 659–671. [Google Scholar] [CrossRef]
- Hu, H.; Brittain, G.C.; Chang, J.H.; Puebla-Osorio, N.; Jin, J.; Zal, A.; Xiao, Y.; Cheng, X.; Chang, M.; Fu, Y.X.; et al. OTUD7B controls non-canonical NF-kappaB activation through deubiquitination of TRAF3. Nature 2013, 494, 371–374. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Wang, H.; Xiao, Y.; Jin, J.; Chang, J.H.; Zou, Q.; Xie, X.; Cheng, X.; Sun, S.C. Otud7b facilitates T cell activation and inflammatory responses by regulating Zap70 ubiquitination. J. Exp. Med. 2016, 213, 399–414. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Jie, Z.; Joo, D.; Ordureau, A.; Liu, P.; Gan, W.; Guo, J.; Zhang, J.; North, B.J.; Dai, X.; et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature 2017, 545, 365–369. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Liu, S.; Sagum, C.; Chen, J.; Singh, R.; Chaturvedi, A.; Horton, J.R.; Kashyap, T.R.; Fushman, D.; Cheng, X.; et al. Crosstalk between Lys63- and Lys11-polyubiquitin signaling at DNA damage sites is driven by Cezanne. Genes Dev. 2019, 33, 1702–1717. [Google Scholar] [CrossRef]
- Xie, W.; Tian, S.; Yang, J.; Cai, S.; Jin, S.; Zhou, T.; Wu, Y.; Chen, Z.; Ji, Y.; Cui, J. OTUD7B deubiquitinates SQSTM1/p62 and promotes IRF3 degradation to regulate antiviral immunity. Autophagy 2022, 18, 2288–2302. [Google Scholar] [CrossRef]
- Wang, J.H.; Wei, W.; Guo, Z.X.; Shi, M.; Guo, R.P. Decreased Cezanne expression is associated with the progression and poor prognosis in hepatocellular carcinoma. J. Transl. Med. 2015, 13, 41. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Cui, L.; Ding, N.; Zhu, L.; Qu, X.; Dong, W.; Du, J.; Liu, Q. Expressions of insulin-like growth factor receptor-1 and cezanne-1 in lung adenocarcinoma. Med. Oncol. 2017, 34, 78. [Google Scholar] [CrossRef]
- Chiu, H.W.; Lin, H.Y.; Tseng, I.J.; Lin, Y.F. OTUD7B upregulation predicts a poor response to paclitaxel in patients with triple-negative breast cancer. Oncotarget 2018, 9, 553–565. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Yang, C.; Wang, R.; Wu, J.; Zhang, Y.; Liu, D.; Sun, X.; Li, X.; Ren, H.; Qin, S. OTUD7B suppresses Smac mimetic-induced lung cancer cell invasion and migration via deubiquitinating TRAF3. J. Exp. Clin. Cancer Res. 2020, 39, 244. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wu, Z.; Tian, Z.; Chen, W.; Wu, G. OTUD7B stabilizes estrogen receptor alpha and promotes breast cancer cell proliferation. Cell Death Dis. 2021, 12, 534. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Cai, K.; Zheng, D.; Liu, Y.; Li, L.; He, Z.; Sun, C.; Yu, C. RHBDL2 promotes the proliferation, migration, and invasion of pancreatic cancer by stabilizing the N1ICD via the OTUD7B and activating the Notch signaling pathway. Cell Death Dis. 2022, 13, 945. [Google Scholar] [CrossRef] [PubMed]
- Bonacci, T.; Suzuki, A.; Grant, G.D.; Stanley, N.; Cook, J.G.; Brown, N.G.; Emanuele, M.J. Cezanne/OTUD7B is a cell cycle-regulated deubiquitinase that antagonizes the degradation of APC/C substrates. EMBO J. 2018, 37, e98701. [Google Scholar] [CrossRef]
- Wang, Y.N.; Yamaguchi, H.; Hsu, J.M.; Hung, M.C. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene 2010, 29, 3997–4006. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Cao, X.; Ali-Osman, F.; Keir, S.; Lo, H.W. EGFR and EGFRvIII interact with PUMA to inhibit mitochondrial translocalization of PUMA and PUMA-mediated apoptosis independent of EGFR kinase activity. Cancer Lett. 2010, 294, 101–110. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, E.; Rushworth, L.; Baccarini, M.; Kolch, W. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science 2004, 306, 2267–2270. [Google Scholar] [CrossRef]
- Niault, T.; Sobczak, I.; Meissl, K.; Weitsman, G.; Piazzolla, D.; Maurer, G.; Kern, F.; Ehrenreiter, K.; Hamerl, M.; Moarefi, I.; et al. From autoinhibition to inhibition in trans: The Raf-1 regulatory domain inhibits Rok-alpha kinase activity. J. Cell Biol. 2009, 187, 335–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiener, R.; Zhang, X.; Wang, T.; Wolberger, C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 2012, 483, 618–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakada, S.; Tai, I.; Panier, S.; Al-Hakim, A.; Iemura, S.; Juang, Y.C.; O’Donnell, L.; Kumakubo, A.; Munro, M.; Sicheri, F.; et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 2010, 466, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Juang, Y.C.; Landry, M.C.; Sanches, M.; Vittal, V.; Leung, C.C.; Ceccarelli, D.F.; Mateo, A.R.; Pruneda, J.N.; Mao, D.Y.; Szilard, R.K.; et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 2012, 45, 384–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinelt, N.; van Wijk, S.J.L. Ubiquitin-dependent and -independent functions of OTULIN in cell fate control and beyond. Cell Death Differ. 2021, 28, 493–504. [Google Scholar] [CrossRef]
- Prieve, M.G.; Guttridge, K.L.; Munguia, J.; Waterman, M.L. Differential importin-alpha recognition and nuclear transport by nuclear localization signals within the high-mobility-group DNA binding domains of lymphoid enhancer factor 1 and T-cell factor 1. Mol. Cell Biol. 1998, 18, 4819–4832. [Google Scholar] [CrossRef] [Green Version]
- An, W.; Luong, L.A.; Bowden, N.P.; Yang, M.; Wu, W.; Zhou, X.; Liu, C.; Niu, K.; Luo, J.; Zhang, C.; et al. Cezanne is a critical regulator of pathological arterial remodelling by targeting beta-catenin signalling. Cardiovasc. Res. 2022, 118, 638–653. [Google Scholar] [CrossRef]
- Pareja, F.; Ferraro, D.A.; Rubin, C.; Cohen-Dvashi, H.; Zhang, F.; Aulmann, S.; Ben-Chetrit, N.; Pines, G.; Navon, R.; Crosetto, N.; et al. Deubiquitination of EGFR by Cezanne-1 contributes to cancer progression. Oncogene 2012, 31, 4599–4608. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.D.; Shen, Y.; Qiao, S.; Liu, W.W.; Zheng, L.; Wang, Y.N.; Cui, N.; Wang, Y.F.; Zhao, S.; Shi, J.H. Upregulation of OTUD7B (Cezanne) Promotes Tumor Progression via AKT/VEGF Pathway in Lung Squamous Carcinoma and Adenocarcinoma. Front. Oncol. 2019, 9, 862. [Google Scholar] [CrossRef] [Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.; Piao, H.-l.; Kim, J. OTUD7B Activates Wnt Signaling Pathway through the Interaction with LEF1. Biomolecules 2023, 13, 1001. https://doi.org/10.3390/biom13061001
Lee Y, Piao H-l, Kim J. OTUD7B Activates Wnt Signaling Pathway through the Interaction with LEF1. Biomolecules. 2023; 13(6):1001. https://doi.org/10.3390/biom13061001
Chicago/Turabian StyleLee, Yuri, Hai-long Piao, and Jongchan Kim. 2023. "OTUD7B Activates Wnt Signaling Pathway through the Interaction with LEF1" Biomolecules 13, no. 6: 1001. https://doi.org/10.3390/biom13061001
APA StyleLee, Y., Piao, H.-l., & Kim, J. (2023). OTUD7B Activates Wnt Signaling Pathway through the Interaction with LEF1. Biomolecules, 13(6), 1001. https://doi.org/10.3390/biom13061001




