Sperm and Oocyte Chromosomal Abnormalities
Abstract
:1. Introduction
2. Sperm
2.1. History of Sperm Chromosome Analysis
2.2. Results of Sperm Chromosome Analysis Using Zona-Free Hamster Eggs and Mouse Eggs
2.3. Sperm Chromosome Analysis Using Sperm-FISH
2.4. Results of Chromosome Analysis in Normal Males
2.5. Results of Chromosome Analysis in Men with Abnormal Semen Findings
2.6. Male Age and Sperm Chromosome Aberration Rate
2.7. Sperm Chromosome Aberration Rates in Men with Chromosome Aberrations
2.7.1. 47, XYY Males
2.7.2. 47, XXY Males
2.8. Male with Robertsonian Translocation
2.9. Males Carrying Reciprocal Translocations
2.10. Males with Chromosomal Inversions
2.11. Sperm Chromosome Testing and Clinical Application/Genetic Counseling
3. Oocyte
3.1. Meiosis of the Oocyte
3.2. The Mechanism Underlying Abnormal Development by Meiosis in Oocytes
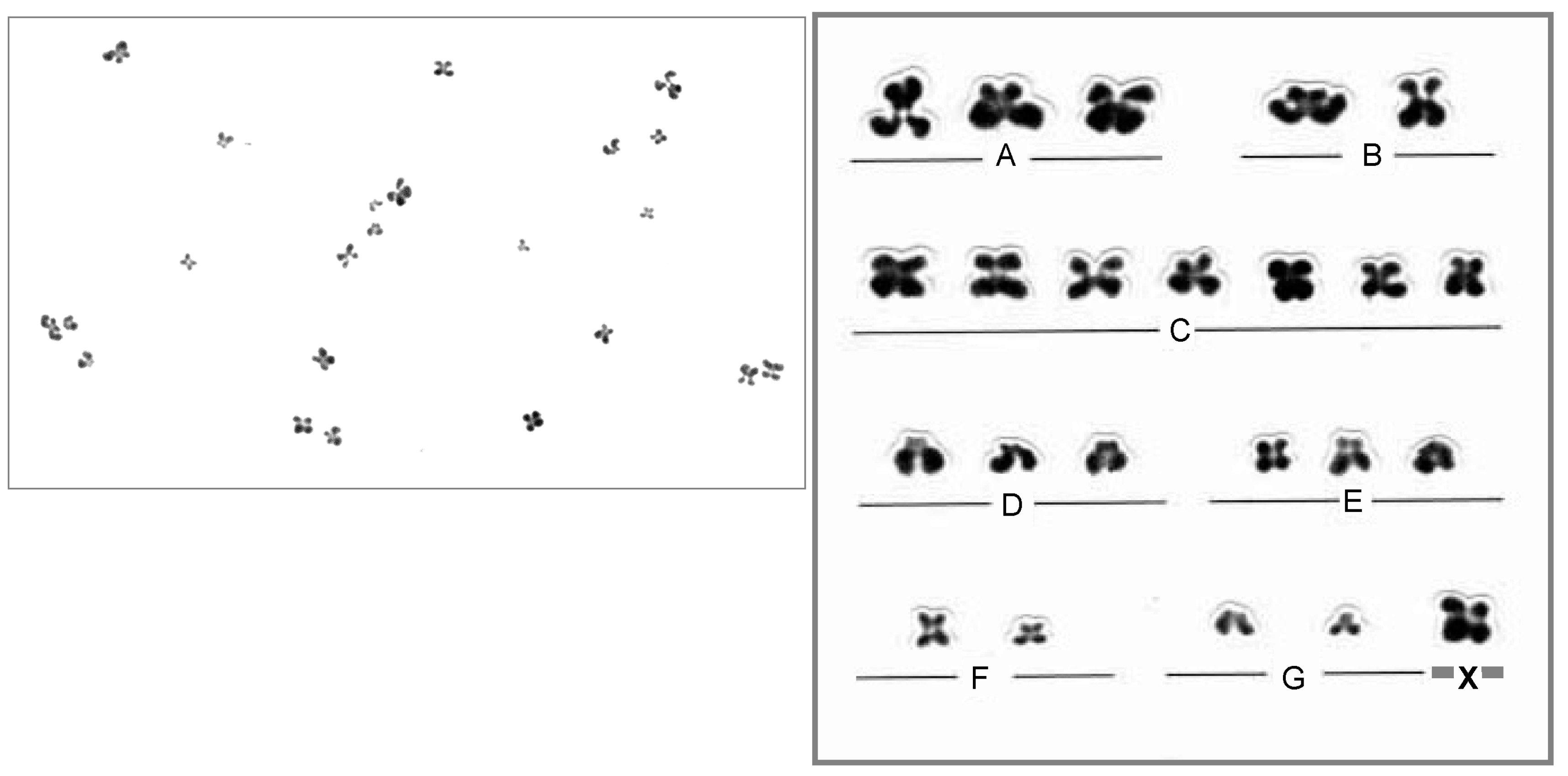
3.3. Oocyte Chromosome Analysis Methods
3.4. Chromosome Abnormalities in Oocytes
4. Embryo
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, S.; Shang, Y.; Liu, Y.; Zhai, B.; Yang, X.; Zhang, L. Crossover patterns under meiotic chromosome program. Asian J. Androl. 2021, 23, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Charalambous, C.; Webster, A.; Schuh, M. Aneuploidy in mammalian oocytes and the impact of maternal ageing. Nat. Rev. Mol. Cell Biol. 2023, 24, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Rudak, E.; Jacobs, P.A.; Yanagimachi, R. Direct analysis of the chromosome constitution of human spermatozoa. Nature 1978, 274, 911–913. [Google Scholar] [CrossRef] [PubMed]
- Kamiguchi, Y.; Mikamo, K. An improved, efficient method for analyzing human sperm chromosomes using zona-free hamster ova. Am. J. Hum. Genet. 1986, 38, 724–740. [Google Scholar] [CrossRef]
- Yanagimachi, R. Intracytoplasmic sperm injection experiments using the mouse as a model. Hum. Reprod. 1998, 13, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Brandriff, B.; Gordon, L.; Ashworth, L.; Watchmaker, G.; Moore, D., 2nd; Wyrobek, A.J.; Carrano, A.V. Chromosomes of human sperm: Variability among normal individuals. Hum. Genet. 1985, 70, 18–24. [Google Scholar] [CrossRef]
- Martin, R.H.; Ko, E.; Rademaker, A. Distribution of aneuploidy in human gametes: Comparison between human sperm and oocytes. Am. J. Med. Genet. 1991, 39, 321–331. [Google Scholar] [CrossRef]
- Watanabe, S. A detailed cytogenetic analysis of large numbers of fresh and frozen-thawed human sperm after ICSI into mouse oocytes. Hum. Reprod. 2003, 18, 1150–1157. [Google Scholar] [CrossRef] [Green Version]
- Carrell, D.T.; Emery, B.R. Use of automated imaging and analysis technology for the detection of aneuploidy in human sperm. Fertil. Steril. 2008, 90, 434–437. [Google Scholar] [CrossRef]
- Shi, Q.; Martin, R.H. Aneuploidy in human spermatozoa: FISH analysis in men with constitutional chromosomal abnormalities, and in infertile men. Reproduction 2001, 121, 655–666. [Google Scholar] [CrossRef]
- Zhu, S.; Zhu, Y.; Zhang, F.; Wu, J.; Lei, C.; Jiang, F. Comprehensive chromosome FISH assessment of sperm aneuploidy in normozoospermic males. J. Assist. Reprod. Genet. 2022, 39, 1887–1900. [Google Scholar] [CrossRef]
- Miharu, N. Chromosome abnormalities in sperm from infertile men with normal somatic karyotypes: Oligozoospermia. Cytogenet. Genome Res. 2005, 111, 347–351. [Google Scholar] [CrossRef]
- Piomboni, P.; Stendardi, A.; Gambera, L. Chromosomal aberrations and aneuploidies of spermatozoa. Adv. Exp. Med. Biol. 2014, 791, 27–52. [Google Scholar]
- Vidal, F.; Blanco, J.; Egozcue, J. Chromosomal abnormalities in sperm. Mol. Cell Endocrinol. 2001, 183 (Suppl. S1), S51–S54. [Google Scholar] [CrossRef] [PubMed]
- Egozcue, J.; Blanco, J.; Anton, E.; Egozcue, S.; Sarrate, Z.; Vidal, F. Genetic analysis of sperm and implications of severe male infertility—A review. Placenta 2003, 24 (Suppl. B), S62–S65. [Google Scholar] [CrossRef]
- Collodel, G.; Capitani, S.; Baccetti, B.; Pammolli, A.; Moretti, E. Sperm aneuploidies and low progressive motility. Hum. Reprod. 2007, 22, 1893–1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrell, D.T.; Emery, B.R.; Wilcox, A.L.; Campbell, B.; Erickson, L.; Hatasaka, H.H.; Jones, K.P.; Peterson, C.M. Sperm chromosome aneuploidy as related to male factor infertility and some ultrastructure defects. Arch. Androl. 2004, 50, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, L.; Meseguer, M.; Mateu, E.; Mercader, A.; Peinado, V.; Bori, L.; Campos-Galindo, I.; Milán, M.; García-Herrero, S.; Simón, C.; et al. Sperm chromosomal abnormalities and their contribution to human embryo aneuploidy. Biol. Reprod. 2019, 101, 1091–1101. [Google Scholar] [CrossRef]
- Sarrate, Z.; Vidal, F.; Blanco, J. Role of sperm fluorescent in situ hybridization studies in infertile patients: Indications, study approach, and clinical relevance. Fertil. Steril. 2010, 93, 1892–1902. [Google Scholar] [CrossRef]
- Buwe, A.; Guttenbach, M.; Schmid, M. Effect of paternal age on the frequency of cytogenetic abnormalities in human spermatozoa. Cytogenet. Genome Res. 2005, 111, 213–228. [Google Scholar] [CrossRef]
- Fonseka, K.G.; Griffin, D.K. Is there a paternal age effect for aneuploidy? Cytogenet. Genome Res. 2011, 133, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Templado, C.; Donate, A.; Giraldo, J.; Bosch, M.; Estop, A. Advanced age increases chromosome structural abnormalities in human spermatozoa. Eur. J. Hum. Genet. 2011, 19, 145–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, R.H.; Spriggs, E.; Ko, E.; Rademaker, A.W. The relationship between paternal age, sex ratios, and aneuploidy frequencies in human sperm, as assessed by multicolor FISH. Am. J. Hum. Genet. 1995, 57, 1395–1399. [Google Scholar]
- Bosch, M.; Rajmil, O.; Egozcue, J.; Templado, C. Linear increase of structural and numerical chromosome 9 abnormalities in human sperm regarding age. Eur. J. Hum. Genet. 2003, 11, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Rousseaux, S.; Hazzouri, M.; Pelletier, R.; Monteil, M.; Usson, Y.; Sèle, B. Disomy rates for chromosomes 14 and 21 studied by fluorescent in-situ hybridization in spermatozoa from three men over 60 years of age. Mol. Hum. Reprod. 1998, 4, 695–699. [Google Scholar] [CrossRef]
- Donate, A.; Estop, A.M.; Giraldo, J.; Templado, C. Paternal Age and Numerical Chromosome Abnormalities in Human Spermatozoa. Cytogenet. Genome Res. 2016, 148, 241–248. [Google Scholar] [CrossRef]
- Iqbal, F. Meiotic Behavior of Extra Sex Chromosomes in Patients with the 47,XXY and 47,XYY Karyotype and Its Ultimate Consequences for Spermatogenesis. Crit. Rev. Eukaryot. Gene Expr. 2020, 30, 19–37. [Google Scholar] [CrossRef]
- Gonzalez-Merino, E.; Hans, C.; Abramowicz, M.; Englert, Y.; Emiliani, S. Aneuploidy study in sperm and preimplantation embryos from nonmosaic 47,XYY men. Fertil. Steril. 2007, 88, 600–606. [Google Scholar] [CrossRef]
- Sa, R.; Ferraz, L.; Barros, A.; Sousa, M. The Klinefelter Syndrome and Testicular Sperm Retrieval Outcomes. Genes 2023, 14, 647. [Google Scholar] [CrossRef]
- Forabosco, A.; Percesepe, A.; Santucci, S. Incidence of non-age-dependent chromosomal abnormalities: A population-based study on 88965 amniocenteses. Eur. J. Hum. Genet. 2009, 17, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Harton, G.L.; Tempest, H.G. Chromosomal disorders and male infertility. Asian J. Androl. 2012, 14, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Baccetti, B.; Collodel, G.; Marzella, R.; Moretti, E.; Piomboni, P.; Scapigliati, G.; Serafini, F. Ultrastructural studies of spermatozoa from infertile males with Robertsonian translocations and 18, X, Y aneuploidies. Hum. Reprod. 2005, 20, 2295–2300. [Google Scholar] [CrossRef] [Green Version]
- Anton, E.; Vidal, F.; Blanco, J. Interchromosomal effect analyses by sperm FISH: Incidence and distribution among reorganization carriers. Syst. Biol. Reprod. Med. 2011, 57, 268–278. [Google Scholar] [CrossRef]
- Yapan, C.; Beyazyurek, C.; Ekmekci, C.; Kahraman, S. The Largest Paracentric Inversion, the Highest Rate of Recombinant Spermatozoa. Case Report: 46,XY, inv(2)(q21.2q37.3) and Literature Review. Balkan J. Med. Genet. 2014, 17, 55–62. [Google Scholar]
- Morel, F.; Laudier, B.; Guérif, F.; Couet, M.L.; Royère, D.; Roux, C.; Bresson, J.L.; Amice, V.; De Braekeleer, M.; Douet-Guilbert, N. Meiotic segregation analysis in spermatozoa of pericentric inversion carriers using fluorescence in-situ hybridization. Hum. Reprod. 2007, 22, 136–141. [Google Scholar] [CrossRef] [Green Version]
- Ramasamy, R.; Besada, S.; Lamb, D.J. Fluorescent in situ hybridization of human sperm: Diagnostics, indications, and therapeutic implications. Fertil. Steril. 2014, 102, 1534–1539. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Peng, X.; Gong, F.; Huang, C.; Peng, Y.; Long, X.; Lin, G.; Zhu, W. The role of total chromosomal disomy in human spermatozoa as a predictor of the outcome of pre-implantation genetic screening. Fertil. Steril. 2020, 113, 1196–1204.e1191. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, S.; Findikli, N.; Biricik, A.; Oncu, N.; Ogur, C.; Sertyel, S.; Karlikaya, G.; Karagozoglu, H.; Saglam, Y. Preliminary FISH studies on spermatozoa and embryos in patients with variable degrees of teratozoospermia and a history of poor prognosis. Reprod. Biomed. Online 2006, 12, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, S.I.; Hassold, T.J.; Hunt, P.A. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 2012, 13, 493–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikwar, M.; MacFarlane, A.J.; Marchetti, F. Mechanisms of oocyte aneuploidy associated with advanced maternal age. Mutat. Res. Rev. Mutat. Res. 2020, 785, 108320. [Google Scholar] [CrossRef] [PubMed]
- Angell, R.R. Predivision in human oocytes at meiosis I: A mechanism for trisomy formation in man. Hum. Genet. 1991, 86, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, M.; Fujiwara, R.; Nishizawa, H.; Ito, M.; Kogo, H.; Inagaki, H.; Ohye, T.; Kato, T.; Fujii, T.; Kurahashi, H. Age-related decrease of meiotic cohesins in human oocytes. PLoS ONE 2014, 9, e96710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennabi, I.; Terret, M.E.; Verlhac, M.H. Meiotic spindle assembly and chromosome segregation in oocytes. J. Cell Biol. 2016, 215, 611–619. [Google Scholar] [CrossRef] [Green Version]
- Mikamo, K.; Kamiguchi, Y. Primary incidences of spontaneous chromosomal anomalies and their origins and causal mechanisms in the Chinese hamster. Mutat. Res. 1983, 108, 265–278. [Google Scholar] [CrossRef]
- Rubio, C.; Rodrigo, L.; Garcia-Pascual, C.; Peinado, V.; Campos-Galindo, I.; Garcia-Herrero, S.; Simón, C. Clinical application of embryo aneuploidy testing by next-generation sequencing. Biol. Reprod. 2019, 101, 1083–1090. [Google Scholar] [CrossRef]
- Tejada, M.I.; Mendoza, R.; Corcóstegui, B.; Benito, J.A. Chromosome studies in human unfertilized oocytes and uncleaved zygotes after treatment with gonadotropin-releasing hormone analogs. Fertil. Steril. 1991, 56, 874–880. [Google Scholar] [CrossRef]
- Bongso, A.; Chye, N.S.; Ratnam, S.; Sathananthan, H.; Wong, P.C. Chromosome anomalies in human oocytes failing to fertilize after insemination in vitro. Hum. Reprod. 1988, 3, 645–649. [Google Scholar] [CrossRef]
- De Sutter, P.; Dhont, M.; Vandekerckhove, D. Hormonal stimulation for in vitro fertilization: A comparison of fertilization rates and cytogenetic findings in unfertilized oocytes. J. Assist. Reprod. Genet. 1992, 9, 254–258. [Google Scholar] [CrossRef]
- Ma, S.; Kalousek, D.K.; Yuen, B.H.; Gomel, V.; Katagiri, S.; Moon, Y.S. Chromosome investigation in in vitro fertilization failure. J. Assist. Reprod. Genet. 1994, 11, 445–451. [Google Scholar] [CrossRef]
- Zenzes, M.T.; Wang, P.; Casper, R.F. Cigarette smoking may affect meiotic maturation of human oocytes. Hum. Reprod. 1995, 10, 3213–3217. [Google Scholar] [PubMed]
- Pellestor, F.; Andréo, B.; Arnal, F.; Humeau, C.; Demaille, J. Mechanisms of non-disjunction in human female meiosis: The co-existence of two modes of malsegregation evidenced by the karyotyping of 1397 In Vitro unfertilized oocytes. Hum. Reprod. 2002, 17, 2134–2145. [Google Scholar] [CrossRef] [Green Version]
- Lim, A.S.; Ho, A.T.; Tsakok, M.F. Chromosomes of oocytes failing In Vitro fertilization. Hum. Reprod. 1995, 10, 2570–2575. [Google Scholar] [CrossRef] [PubMed]
- Sengoku, K.; Tamate, K.; Takuma, N.; Yoshida, T.; Goishi, K.; Ishikawa, M. The chromosomal normality of unfertilized oocytes from patients with polycystic ovarian syndrome. Hum. Reprod. 1997, 12, 474–477. [Google Scholar] [CrossRef] [Green Version]
- Nakaoka, Y.; Okamoto, E.; Miharu, N.; Ohama, K. Chromosome analysis in human oocytes remaining unfertilized after in-vitro insemination: Effect of maternal age and fertilization rate. Hum. Reprod. 1998, 13, 419–424. [Google Scholar] [CrossRef] [Green Version]
- Kamiguchi, Y.; Rosenbusch, B.; Sterzik, K.; Mikamo, K. Chromosomal analysis of unfertilized human oocytes prepared by a gradual fixation-air drying method. Hum. Genet. 1993, 90, 533–541. [Google Scholar] [CrossRef]
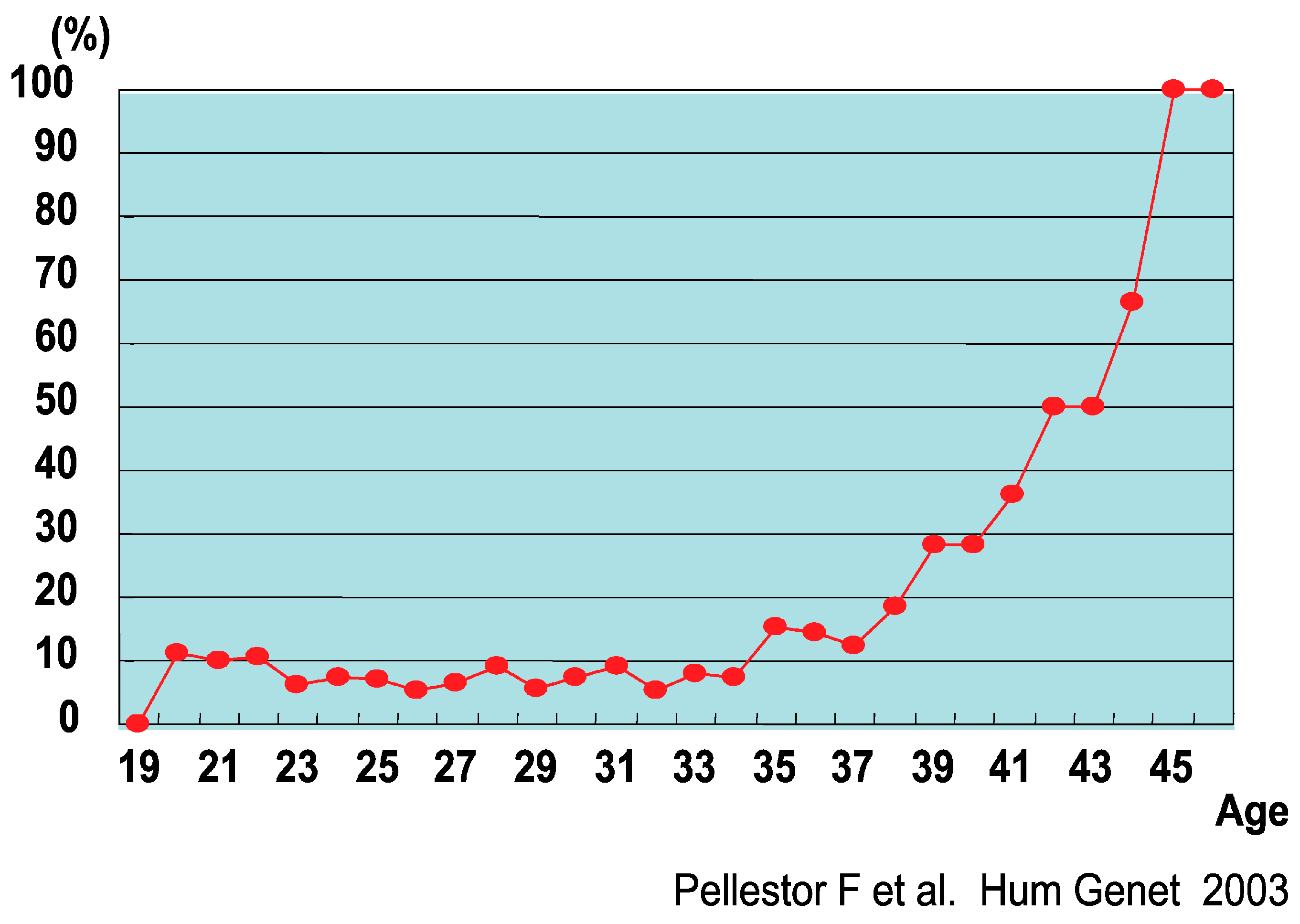
- Pellestor, F.; Andréo, B.; Arnal, F.; Humeau, C.; Demaille, J. Maternal aging and chromosomal abnormalities: New data drawn from in vitro unfertilized human oocytes. Hum. Genet. 2003, 112, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Dyban, A.; Freidine, M.; Severova, E.; Cieslak, J.; Ivakhnenko, V.; Verlinsky, Y. Detection of aneuploidy in human oocytes and corresponding first polar bodies by fluorescent in situ hybridization. J. Assist. Reprod. Genet. 1996, 13, 73–78. [Google Scholar] [CrossRef]
- Anahory, T.; Andréo, B.; Régnier-Vigouroux, G.; Soulie, J.P.; Baudouin, M.; Demaille, J.; Pellestor, F. Sequential multiple probe fluorescence in-situ hybridization analysis of human oocytes and polar bodies by combining centromeric labelling and whole chromosome painting. Mol. Hum. Reprod. 2003, 9, 577–585. [Google Scholar] [CrossRef] [Green Version]
- Honda, N.; Miharu, N.; Hara, T.; Samura, O.; Honda, H.; Ohama, K. Chromosomal FISH analysis of unfertilized human oocytes and polar bodies. J. Hum. Genet. 2002, 47, 488–491. [Google Scholar] [CrossRef] [PubMed]
- Cupisti, S.; Conn, C.M.; Fragouli, E.; Whalley, K.; Mills, J.A.; Faed, M.J.; Delhanty, J.D. Sequential FISH analysis of oocytes and polar bodies reveals aneuploidy mechanisms. Prenat. Diagn. 2003, 23, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Verlinsky, Y.; Cieslak, J.; Ivakhnenko, V.; Evsikov, S.; Wolf, G.; White, M.; Lifchez, A.; Kaplan, B.; Moise, J.; Valle, J.; et al. Prevention of age-related aneuploidies by polar body testing of oocytes. J. Assist. Reprod. Genet. 1999, 16, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Kuliev, A.; Cieslak, J.; Ilkevitch, Y.; Verlinsky, Y. Chromosomal abnormalities in a series of 6,733 human oocytes in preimplantation diagnosis for age-related aneuploidies. Reprod. Biomed. Online 2003, 6, 54–59. [Google Scholar] [CrossRef]
- Gabriel, A.S.; Thornhill, A.R.; Ottolini, C.S.; Gordon, A.; Brown, A.P.; Taylor, J.; Bennett, K.; Handyside, A.; Griffin, D.K. Array comparative genomic hybridisation on first polar bodies suggests that non-disjunction is not the predominant mechanism leading to aneuploidy in humans. J. Med. Genet. 2011, 48, 433–437. [Google Scholar] [CrossRef]
- Fragouli, E.; Alfarawati, S.; Spath, K.; Jaroudi, S.; Sarasa, J.; Enciso, M.; Wells, D. The origin and impact of embryonic aneuploidy. Hum. Genet. 2013, 132, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Verpoest, W.; Staessen, C.; Bossuyt, P.M.; Goossens, V.; Altarescu, G.; Bonduelle, M.; Devesa, M.; Eldar-Geva, T.; Gianaroli, L.; Griesinger, G.; et al. Preimplantation genetic testing for aneuploidy by microarray analysis of polar bodies in advanced maternal age: A randomized clinical trial. Hum. Reprod. 2018, 33, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Capalbo, A.; Bono, S.; Spizzichino, L.; Biricik, A.; Baldi, M.; Colamaria, S.; Ubaldi, F.M.; Rienzi, L.; Fiorentino, F. Sequential comprehensive chromosome analysis on polar bodies, blastomeres and trophoblast: Insights into female meiotic errors and chromosomal segregation in the preimplantation window of embryo development. Hum. Reprod. 2013, 28, 509–518. [Google Scholar] [CrossRef]
- Gruhn, J.R.; Zielinska, A.P.; Shukla, V.; Blanshard, R.; Capalbo, A.; Cimadomo, D.; Nikiforov, D.; Chan, A.C.; Newnham, L.J.; Vogel, I.; et al. Chromosome errors in human eggs shape natural fertility over reproductive life span. Science 2019, 365, 1466–1469. [Google Scholar] [CrossRef] [Green Version]
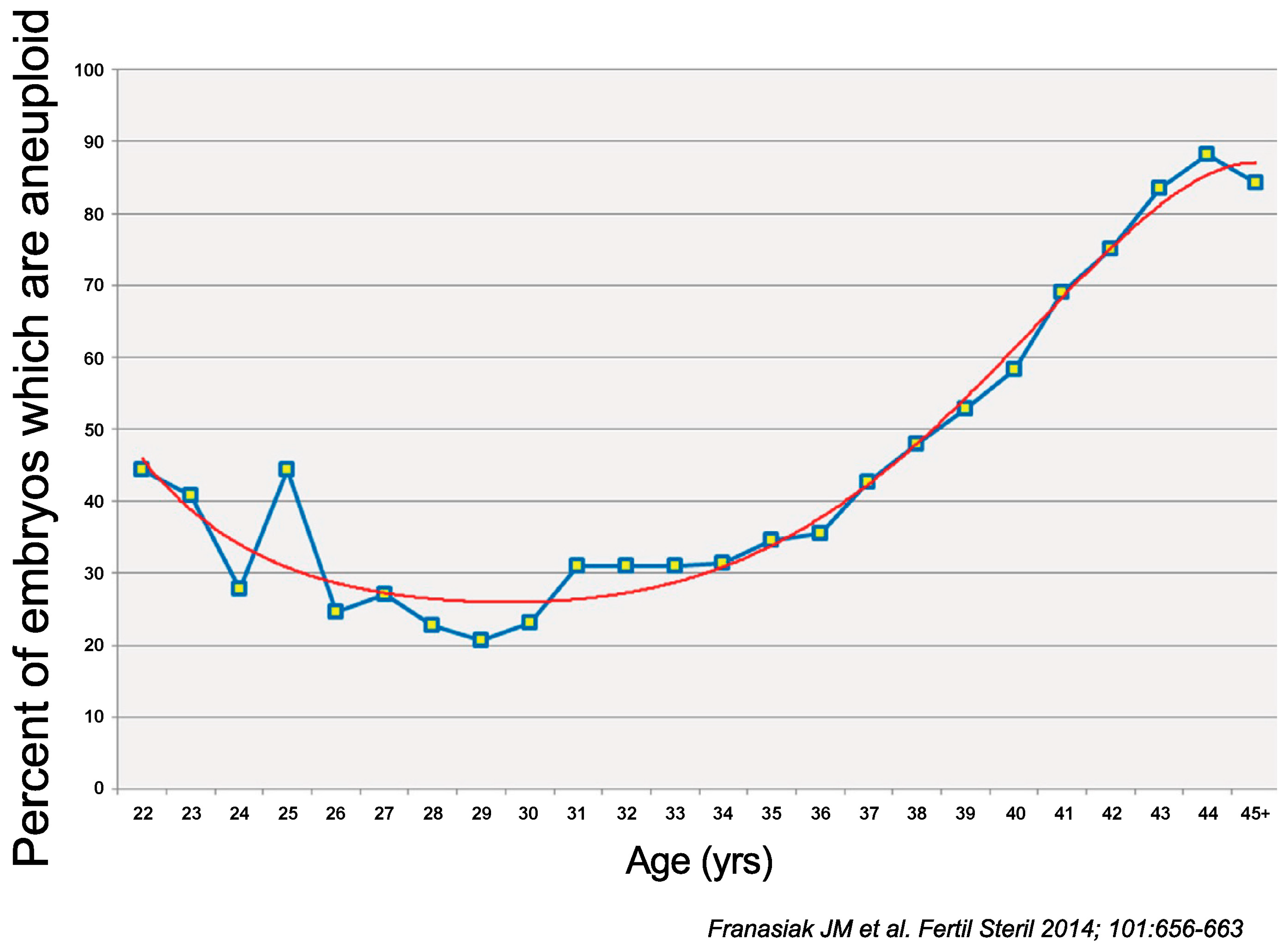
- Franasiak, J.M.; Forman, E.J.; Hong, K.H.; Werner, M.D.; Upham, K.M.; Treff, N.R.; Scott, R.T., Jr. The nature of aneuploidy with increasing age of the female partner: A review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil. Steril. 2014, 101, 656–663.e651. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, J.; Collins, G.S.; Salem, S.A.; Liu, X.; Lyle, S.S.; Peck, A.C.; Sills, E.S.; Salem, R.D. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: Results from a randomized pilot study. Mol. Cytogenet. 2012, 5, 24. [Google Scholar] [CrossRef] [Green Version]
- Munné, S.; Wells, D. Detection of mosaicism at blastocyst stage with the use of high-resolution next-generation sequencing. Fertil. Steril. 2017, 107, 1085–1091. [Google Scholar] [CrossRef] [Green Version]
- Rana, B.; Lambrese, K.; Mendola, R.; Xu, J.; Garrisi, J.; Miller, K.; Marin, D.; Treff, N.R. Identifying parental and cell-division origins of aneuploidy in the human blastocyst. Am. J. Hum. Genet. 2023, 110, 565–574. [Google Scholar] [CrossRef] [PubMed]
| Method | Pros | Cons |
|---|---|---|
| Zona-free hamster eggs | Can be analyzed by karyotype | Technically difficult Only swim-up sperm can be analyzed Difficult to analyze multiple sperm |
| ICSI of mouse egg | Can be analyzed by karyotype | Difficult to analyze multiple sperm |
| Sperm-FISH | Capable of analyzing multiple sperm | Difficult to analyze structural anomalies Signal analysis standards and accuracy vary between laboratories |
| Karyotype | XX Disomy (%) | YY Disomy (%) | XY Disomy (%) |
|---|---|---|---|
| 47, XYY | 1.65 ± 2.31 | 1.54 ± 1.53 | 4.43 ± 6.03 |
| 47, XYY/46, XY | 0.17 ± 0.16 | 0.50 ± 0.45 | 0.48 ± 0.31 |
| 47, XXY | 4.64 ± 2.56 | 0.30 ± 0.46 | 11.1 ± 6.89 |
| 47, XXY/46, XY | 0.40 ± 0.31 | 0.42 ± 0.49 | 1.22 ± 0.71 |
| Seminal Parameters | Altered FISH Results | % |
|---|---|---|
| Astheteratozoospermic | 6/71 | 8.5 |
| Asthenozoospermic | 4/67 | 6.0 |
| Normozoospermic | 4/34 | 11.8 |
| Oligoasthenoteratozoospermic | 13/62 | 21.0 |
| Oligoasthenozoospermic | 17/51 | 33.3 |
| Oligoteratozoospermic | 2/13 | 15.4 |
| Oligozoospermic | 2/4 | 50.0 |
| Teratozoospermic | 1/17 | 5.9 |
| Total | 49/319 | 15.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samura, O.; Nakaoka, Y.; Miharu, N. Sperm and Oocyte Chromosomal Abnormalities. Biomolecules 2023, 13, 1010. https://doi.org/10.3390/biom13061010
Samura O, Nakaoka Y, Miharu N. Sperm and Oocyte Chromosomal Abnormalities. Biomolecules. 2023; 13(6):1010. https://doi.org/10.3390/biom13061010
Chicago/Turabian StyleSamura, Osamu, Yoshiharu Nakaoka, and Norio Miharu. 2023. "Sperm and Oocyte Chromosomal Abnormalities" Biomolecules 13, no. 6: 1010. https://doi.org/10.3390/biom13061010
APA StyleSamura, O., Nakaoka, Y., & Miharu, N. (2023). Sperm and Oocyte Chromosomal Abnormalities. Biomolecules, 13(6), 1010. https://doi.org/10.3390/biom13061010





