Uncovering the Underlying Mechanisms Blocking Replication of Bluetongue Virus Serotype 26 (BTV-26) in Culicoides Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Viruses
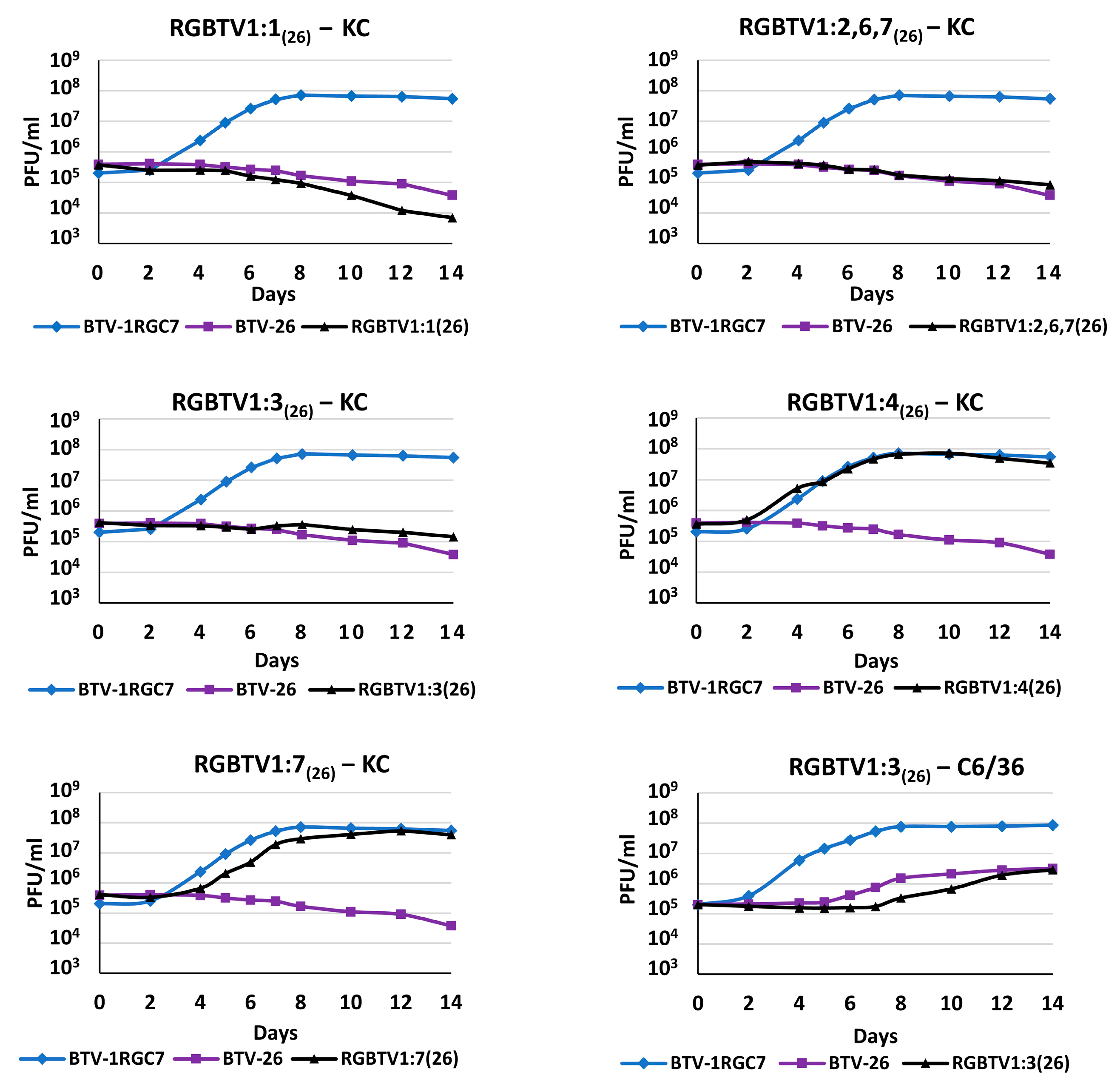
2.2. Infection of BSR, C6/36 or KC Cells with BTV-1RGC7, BTV-26, RGBTV1:1(26), RGBTV1:2,6,7(26), RGBTV1:3(26), RGBTV1:4(26) or RGBTV1:7(26)
2.3. Purification of BTV-1RGC7 and BTV-26 Grown in BSR Cells and Assessment of Binding of Virus Particles to Cell Membranes of KC Cells
2.4. Construction of Expression Plasmids and Assessment of Their Functionality on Insect Cells
2.5. Golden Gate Cloning
2.6. Expression of VP1, VP2, VP3 and VP7 of BTV-1RGC7 or BTV-26 in BSR or KC Cells Using pCI-G or pKCi
2.7. Expression of VP1, VP2, VP3 and VP7 of BTV-1RGC7 and/or BTV-26 in C6/36 Cells Using the pAE Plasmid
2.8. Western Blot
2.9. Quantification of Protein Expression
2.10. Design of Real-Time PCR Primers
2.11. Assessing Stability of Viral mRNA Derived from Transcription of pKCi Plasmids within KC Cells
2.12. Bioinformatic Analyses
3. Results
3.1. Infection of BSR, C6/36 or KC Cells with BTV-1RGC7, BTV-26, RGBTV1:1(26), RGBTV1:2,6,7(26), RGBTV1:3(26), RGBTV1:4(26) or RGBTV1:7(26)
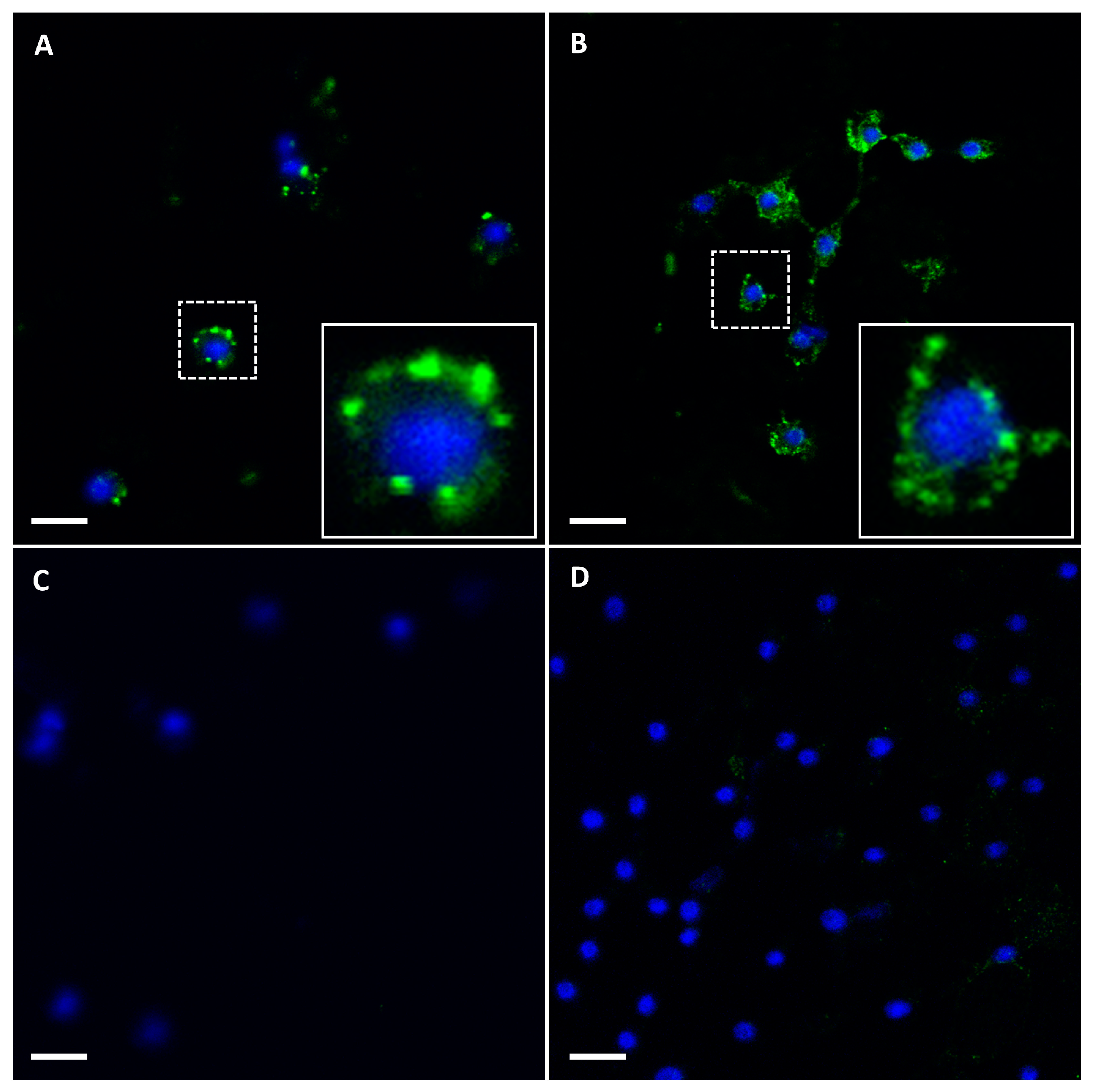
3.2. Binding of Purified Virus Particles of BTV-1RGC7 and Wild-Type BTV-26 to Cell Membranes of KC Cells
3.3. Assessing Stability of Viral mRNA Derived from Transcription of pKCi Plasmids within KC Cells
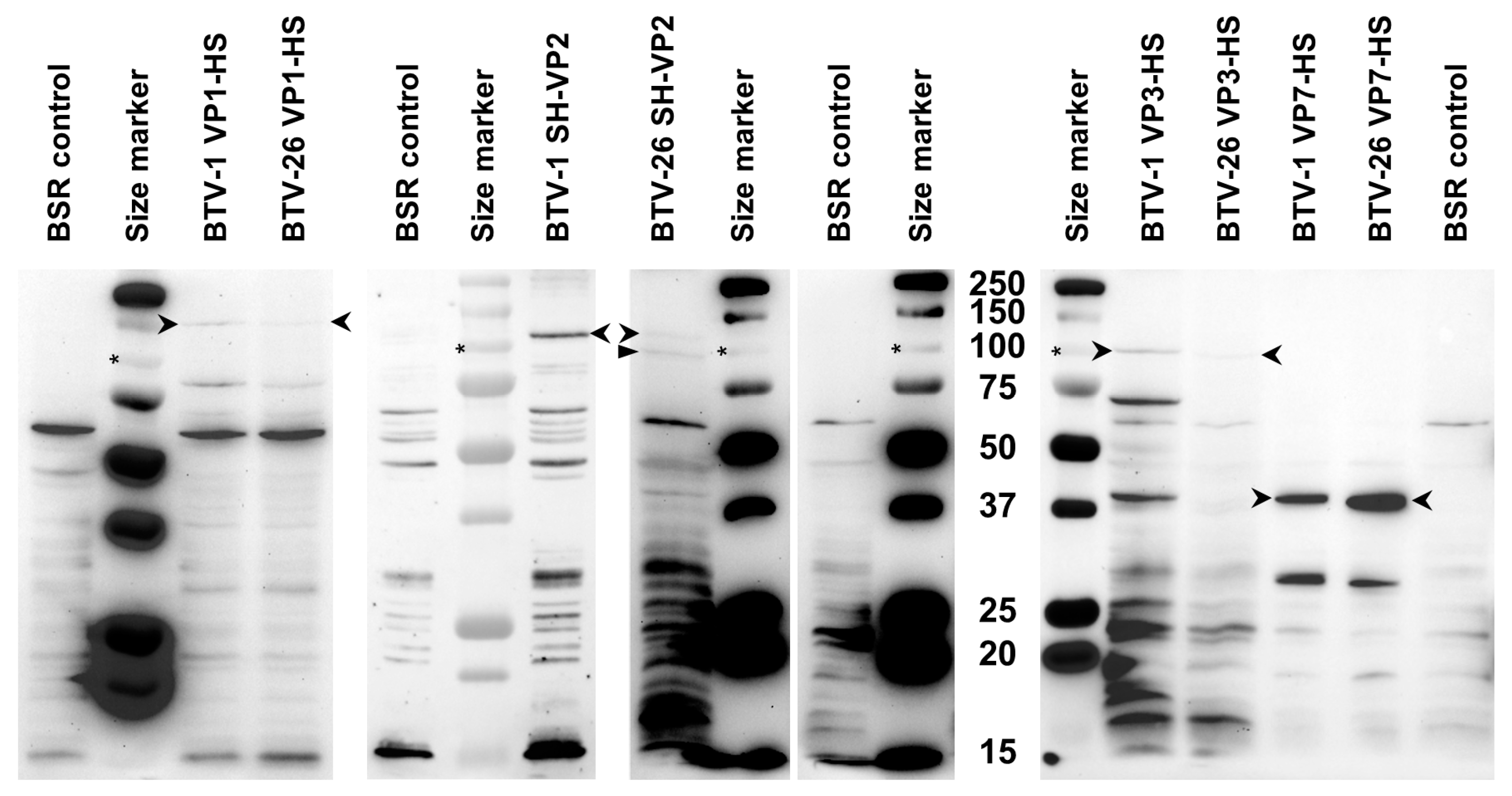
3.4. Expression of VP1, VP2, VP3 and VP7 of BTV-1RGC7 or BTV-26 in BSR Cells Using the pCI-G Plasmid
3.5. Expression of VP1, VP2, VP3 and VP7 of BTV-1RGC7 and/or BTV-26 in C6/36 Cells Using pAE Plasmids
3.6. Expression of VP1, VP2, VP3 and VP7 of BTV-1RGC7 or BTV-26 in KC Cells Using pKCi Plasmids
3.6.1. Expression of VP1
3.6.2. Expression of VP2 and Analysis of the RNA Sequence Context
3.6.3. Expression of VP3
3.6.4. Expression of VP7
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Attoui, H.; Mohd Jaafar, F.; Mertens, P.P.C. In the Reovirales a new taxonomic order: Families Sedoreoviridae and Spinareoviridae. In Proceedings of the 13th International dsRNA Virus Symposium, Houffalize, Belgium, 24–28 September 2018; Leuven University Press: Leuven, Belgium, 2018. [Google Scholar]
- Matthijnssens, J.; Attoui, H.; Banyai, K.; Brussaard, C.P.D.; Danthi, P.; Del Vas, M.; Dermody, T.S.; Duncan, R.; Fang, Q.; Johne, R.; et al. ICTV Virus Taxonomy Profile: Sedoreoviridae 2022. J. Gen. Virol. 2022, 103, 001782. [Google Scholar] [CrossRef] [PubMed]
- Mohd Jaafar, F.; Belhouchet, M.; Belaganahalli, M.; Tesh, R.B.; Mertens, P.P.; Attoui, H. Full-genome characterisation of Orungo, Lebombo and Changuinola viruses provides evidence for co-evolution of orbiviruses with their arthropod vectors. PLoS ONE 2014, 9, e86392. [Google Scholar] [CrossRef] [PubMed]
- Belaganahalli, M.N.; Maan, S.; Maan, N.S.; Brownlie, J.; Tesh, R.; Attoui, H.; Mertens, P.P. Genetic characterization of the tick-borne orbiviruses. Viruses 2015, 7, 2185–2209. [Google Scholar] [CrossRef] [PubMed]
- Attoui, H.; Billoir, F.; Cantaloube, J.F.; Biagini, P.; de Micco, P.; de Lamballerie, X. Strategies for the sequence determination of viral dsRNA genomes. J. Virol. Methods 2000, 89, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.A.; Renzullo, S.; Mader, M.; Chaignat, V.; Worwa, G.; Thuer, B. Genetic characterization of toggenburg orbivirus, a new bluetongue virus, from goats, Switzerland. Emerg. Infect. Dis. 2008, 14, 1855–1861. [Google Scholar] [CrossRef]
- Ries, C.; Vogtlin, A.; Hussy, D.; Jandt, T.; Gobet, H.; Hilbe, M.; Burgener, C.; Schweizer, L.; Hafliger-Speiser, S.; Beer, M.; et al. Putative Novel Atypical BTV Serotype ‘36’ Identified in Small Ruminants in Switzerland. Viruses 2021, 13, 721. [Google Scholar] [CrossRef]
- Batten, C.; Darpel, K.; Henstock, M.; Fay, P.; Veronesi, E.; Gubbins, S.; Graves, S.; Frost, L.; Oura, C. Evidence for transmission of bluetongue virus serotype 26 through direct contact. PLoS ONE 2014, 9, e96049. [Google Scholar] [CrossRef]
- Batten, C.A.; Henstock, M.R.; Steedman, H.M.; Waddington, S.; Edwards, L.; Oura, C.A. Bluetongue virus serotype 26: Infection kinetics, pathogenesis and possible contact transmission in goats. Vet. Microbiol. 2013, 162, 62–67. [Google Scholar] [CrossRef]
- Breard, E.; Schulz, C.; Sailleau, C.; Bernelin-Cottet, C.; Viarouge, C.; Vitour, D.; Guillaume, B.; Caignard, G.; Gorlier, A.; Attoui, H.; et al. Bluetongue virus serotype 27: Experimental infection of goats, sheep and cattle with three BTV-27 variants reveal atypical characteristics and likely direct contact transmission BTV-27 between goats. Transbound. Emerg. Dis. 2018, 65, e251–e263. [Google Scholar] [CrossRef]
- Bumbarov, V.; Golender, N.; Jenckel, M.; Wernike, K.; Beer, M.; Khinich, E.; Zalesky, O.; Erster, O. Characterization of bluetongue virus serotype 28. Transbound. Emerg. Dis. 2020, 67, 171–182. [Google Scholar] [CrossRef]
- Belhouchet, M.; Mohd Jaafar, F.; Firth, A.E.; Grimes, J.M.; Mertens, P.P.; Attoui, H. Detection of a fourth orbivirus non-structural protein. PLoS ONE 2011, 6, e25697. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.; Hardy, A.; Barry, G.; Pinto, R.M.; Caporale, M.; Melzi, E.; Hughes, J.; Taggart, A.; Janowicz, A.; Varela, M.; et al. Characterization of a second open reading frame in genome segment 10 of bluetongue virus. J. Gen. Virol. 2015, 96, 3280–3293. [Google Scholar] [CrossRef]
- Mohd Jaafar, F.; Monsion, B.; Mertens, P.P.C.; Attoui, H. Identification of Orbivirus Non-Structural Protein 5 (NS5), Its Role and Interaction with RNA/DNA in Infected Cells. Int. J. Mol. Sci. 2023, 24, 6845. [Google Scholar] [CrossRef] [PubMed]
- Ratinier, M.; Caporale, M.; Golder, M.; Franzoni, G.; Allan, K.; Nunes, S.F.; Armezzani, A.; Bayoumy, A.; Rixon, F.; Shaw, A.; et al. Identification and characterization of a novel non-structural protein of bluetongue virus. PLoS Pathog. 2011, 7, e1002477. [Google Scholar] [CrossRef] [PubMed]
- Pullinger, G.D.; Guimera Busquets, M.; Nomikou, K.; Boyce, M.; Attoui, H.; Mertens, P.P. Identification of the Genome Segments of Bluetongue Virus Serotype 26 (Isolate KUW2010/02) that Restrict Replication in a Culicoides sonorensis Cell Line (KC Cells). PLoS ONE 2016, 11, e0149709. [Google Scholar] [CrossRef]
- Chaignat, V.; Worwa, G.; Scherrer, N.; Hilbe, M.; Ehrensperger, F.; Batten, C.; Cortyen, M.; Hofmann, M.; Thuer, B. Toggenburg Orbivirus, a new bluetongue virus: Initial detection, first observations in field and experimental infection of goats and sheep. Vet. Microbiol. 2009, 138, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Skehel, J.J.; Joklik, W.K. Studies on the in vitro transcription of reovirus RNA catalyzed by reovirus cores. Virology 1969, 39, 822–831. [Google Scholar] [CrossRef]
- Huismans, H.; Verwoerd, D.W. Control of transcription during the expression of the bluetongue virus genome. Virology 1973, 52, 81–88. [Google Scholar] [CrossRef]
- Sato, M.; Maeda, N.; Yoshida, H.; Urade, M.; Saito, S. Plaque formation of herpes virus hominis type 2 and rubella virus in variants isolated from the colonies of BHK21/WI-2 cells formed in soft agar. Arch. Virol. 1977, 53, 269–273. [Google Scholar] [CrossRef]
- Wechsler, S.J.; McHolland, L.E.; Wilson, W.C. A RNA virus in cells from Culicoides variipennis. J. Invertebr. Pathol. 1991, 57, 200–205. [Google Scholar] [CrossRef]
- Attoui, H.; Monsion, B.; Klonjkowski, B.; Zientara, S.; Mertens, P.P.C.; Mohd Jaafar, F. Identification of the Genome Segments of Bluetongue Virus Type 26/Type 1 Reassortants Influencing Horizontal Transmission in a Mouse Model. Viruses 2021, 13, 2208. [Google Scholar] [CrossRef] [PubMed]
- Mohd Jaafar, F.; Belhouchet, M.; Vitour, D.; Adam, M.; Breard, E.; Zientara, S.; Mertens, P.P.; Attoui, H. Immunisation with bacterial expressed VP2 and VP5 of bluetongue virus (BTV) protect alpha/beta interferon-receptor knock-out (IFNAR−/−) mice from homologous lethal challenge. Vaccine 2014, 32, 4059–4067. [Google Scholar] [CrossRef] [PubMed]
- Mertens, P.P.; Burroughs, J.N.; Anderson, J. Purification and properties of virus particles, infectious subviral particles, and cores of bluetongue virus serotypes 1 and 4. Virology 1987, 157, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.A.; Gross, T.L.; Myles, K.M.; Adelman, Z.N. Validation of novel promoter sequences derived from two endogenous ubiquitin genes in transgenic Aedes aegypti. Insect. Mol. Biol. 2010, 19, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Mohd Jaafar, F.; Attoui, H.; Gallian, P.; Isahak, I.; Wong, K.T.; Cheong, S.K.; Nadarajah, V.S.; Cantaloube, J.F.; Biagini, P.; De Micco, P.; et al. Recombinant VP9-based enzyme-linked immunosorbent assay for detection of immunoglobulin G antibodies to Banna virus (genus Seadornavirus). J. Virol. Methods 2004, 116, 55–61. [Google Scholar] [CrossRef]
- Marshall, O.J. PerlPrimer: Cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 2004, 20, 2471–2472. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Honer Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Harrison, R.L.; Jarvis, D.L. Protein N-glycosylation in the baculovirus-insect cell expression system and engineering of insect cells to produce “mammalianized” recombinant glycoproteins. Adv. Virus Res. 2006, 68, 159–191. [Google Scholar]
- Shi, X.; Jarvis, D.L. Protein N-glycosylation in the baculovirus-insect cell system. Curr. Drug Targets 2007, 8, 1116–1125. [Google Scholar] [CrossRef]
- Walski, T.; De Schutter, K.; Van Damme, E.J.M.; Smagghe, G. Diversity and functions of protein glycosylation in insects. Insect. Biochem. Mol. Biol. 2017, 83, 21–34. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Shivakoti, S.; Ding, K.; Cui, Y.; Roy, P.; Zhou, Z.H. In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating. Proc. Natl. Acad. Sci. USA 2019, 116, 16535–16540. [Google Scholar] [CrossRef] [PubMed]
- Manole, V.; Laurinmaki, P.; Van Wyngaardt, W.; Potgieter, C.A.; Wright, I.M.; Venter, G.J.; van Dijk, A.A.; Sewell, B.T.; Butcher, S.J. Structural insight into African horsesickness virus infection. J. Virol. 2012, 86, 7858–7866. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Nomikou, K.; Veronesi, E.; Bachanek-Bankowska, K.; Belaganahalli, M.N.; Attoui, H.; Mertens, P.P. Complete genome characterisation of a novel 26th bluetongue virus serotype from Kuwait. PLoS ONE 2011, 6, e26147. [Google Scholar] [CrossRef] [PubMed]
- Maan, S.; Maan, N.S.; Nomikou, K.; Batten, C.; Antony, F.; Belaganahalli, M.N.; Samy, A.M.; Reda, A.A.; Al-Rashid, S.A.; El Batel, M.; et al. Novel bluetongue virus serotype from Kuwait. Emerg. Infect. Dis. 2011, 17, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, P. Key Mechanistic Principles and Considerations Concerning RNA Interference. Front. Plant Sci. 2020, 11, 1237. [Google Scholar] [CrossRef]
- Schnettler, E.; Ratinier, M.; Watson, M.; Shaw, A.E.; McFarlane, M.; Varela, M.; Elliott, R.M.; Palmarini, M.; Kohl, A. RNA interference targets arbovirus replication in Culicoides cells. J. Virol. 2013, 87, 2441–2454. [Google Scholar] [CrossRef] [PubMed]
- Guimera Busquets, M.; Pullinger, G.D.; Darpel, K.E.; Cooke, L.; Armstrong, S.; Simpson, J.; Palmarini, M.; Fragkoudis, R.; Mertens, P.P.C. An Early Block in the Replication of the Atypical Bluetongue Virus Serotype 26 in Culicoides Cells Is Determined by Its Capsid Proteins. Viruses 2021, 13, 919. [Google Scholar] [CrossRef]
- Matsuo, E.; Roy, P. Bluetongue virus VP1 polymerase activity in vitro: Template dependency, dinucleotide priming and cap dependency. PLoS ONE 2011, 6, e27702. [Google Scholar] [CrossRef]
- Grimes, J.M.; Burroughs, J.N.; Gouet, P.; Diprose, J.M.; Malby, R.; Zientara, S.; Mertens, P.P.; Stuart, D.I. The atomic structure of the bluetongue virus core. Nature 1998, 395, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Boyce, M.; Bhattacharya, B.; Zhang, X.; Schein, S.; Roy, P.; Zhou, Z.H. Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 6292–6297. [Google Scholar] [CrossRef] [PubMed]
- Pooggin, M.M.; Ryabova, L.A. Ribosome Shunting, Polycistronic Translation, and Evasion of Antiviral Defenses in Plant Pararetroviruses and Beyond. Front. Microbiol. 2018, 9, 644. [Google Scholar] [CrossRef] [PubMed]
- Yueh, A.; Schneider, R.J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996, 10, 1557–1567. [Google Scholar] [CrossRef]
- Racine, T.; Duncan, R. Facilitated leaky scanning and atypical ribosome shunting direct downstream translation initiation on the tricistronic S1 mRNA of avian reovirus. Nucleic. Acids Res. 2010, 38, 7260–7272. [Google Scholar] [CrossRef]
- Luttermann, C.; Meyers, G. The importance of inter- and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA. Genes Dev. 2009, 23, 331–344. [Google Scholar] [CrossRef]
- Yueh, A.; Schneider, R.J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000, 14, 414–421. [Google Scholar] [CrossRef]
- Topilina, N.I.; Mills, K.V. Recent advances in in vivo applications of intein-mediated protein splicing. Mob. DNA 2014, 5, 5. [Google Scholar] [CrossRef]
- Ogata, H.; Raoult, D.; Claverie, J.M. A new example of viral intein in Mimivirus. Virol. J. 2005, 2, 8. [Google Scholar] [CrossRef]
- Nanda, A.; Nasker, S.S.; Mehra, A.; Panda, S.; Nayak, S. Inteins in Science: Evolution to Application. Microorganisms 2020, 8, 2004. [Google Scholar] [CrossRef]
- Diprose, J.M.; Burroughs, J.N.; Sutton, G.C.; Goldsmith, A.; Gouet, P.; Malby, R.; Overton, I.; Zientara, S.; Mertens, P.P.; Stuart, D.I.; et al. Translocation portals for the substrates and products of a viral transcription complex: The bluetongue virus core. EMBO J. 2001, 20, 7229–7239. [Google Scholar] [CrossRef] [PubMed]
- Gouet, P.; Diprose, J.M.; Grimes, J.M.; Malby, R.; Burroughs, J.N.; Zientara, S.; Stuart, D.I.; Mertens, P.P. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 1999, 97, 481–490. [Google Scholar] [CrossRef] [PubMed]




| Viral Protein (VP)/Virus | pKCi Derived Plasmids | pCI-G Derived Plasmids |
|---|---|---|
| VP1/BTV-26 | pKCi-BTV26-SH-VP1 pKCi-BTV26-VP1-HS | pCI-BTV26-VP1-HS |
| VP2/BTV-26 | pKCi-BTV26-SH-VP2 pKCi-BTV26-VP2-HS | pCI-BTV26-SH-VP2 |
| VP3/BTV-26 | pKCi-BTV26-SH-VP3 | pCI-BTV26-VP3-HS |
| VP7/BTV-26 | pKCi-BTV26-VP7-HS | pCI-BTV26-VP7-HS |
| VP1/BTV-1 | pKCi-BTV1-SH-VP1 pKCi-BTV1-VP1-HS | pCI-BTV1-VP1-HS |
| VP2/BTV-1 | pKCi-BTV1-SH-VP2 | pCI-BTV1-SH-VP2 |
| VP3/BTV-1 | pKCi-BTV1-SH-VP3 | pCI-BTV1-VP3-HS |
| VP7/BTV-1 | pKCi-BTV1-VP7-HS | pCI-BTV1-VP7-HS |
| Virus | Protein Theoretical Molecular Weight in kDa (Native/Tagged) | |||
|---|---|---|---|---|
| VP1 | VP2 | VP3 | VP7 | |
| BTV-1 | 149.7/153.5 | 111.9/115.9 | 103.3/107.1 | 38.5/42.4 |
| BTV-26 | 150.0/153.8 | 110.9/114.9 | 102.9/106.7 | 38.5/42.4 |
| Virus | Protein Estimated Molecular Weight in kDa | |||
|---|---|---|---|---|
| VP1 | VP2 | VP3 | VP7 | |
| BTV-1 | 141.7 | 116.2 | 97.9 | 38.2 |
| BTV-26 | 147.9 | 103.0 | 97.8 | 37.4 |
| 9.9 kDa Deletion | 12.4 kDa Deletion | 13.2 kDa Deletion | GC Content of Seg-2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Region I | MFE (kcal/mol) | GC% | Region II | MFE (kcal/mol) | GC% | Region III | MFE (kcal/mol) | GC% | |
| BTV-1_914-1172 | 61.8 | 43.24 | BTV-1_1455-1792 | 81.3 | 42.07 | BTV-1_2290-2635 | 98.4 | 44.51 | 42.65 |
| BTV-4_911-1169 | 66.6 | 45.56 | BTV-4_1446-1770 | 79.5 | 40.92 | BTV-4_2275-2620 | 88.7 | 44.22 | 42.84 |
| BTV-6_902-1160 | 65.8 | 41.31 | BTV-6_1440-1767 | 78.6 | 42.07 | BTV-6_2272-2617 | 93.1 | 42.49 | 42.43 |
| BTV-9_911-1169 | 66.8 | 44.40 | BTV-9_1437-1764 | 78.8 | 43.60 | BTV-9_2272-2617 | 81.6 | 38.44 | 41.70 |
| BTV-26_911-1169 | 84.7 | 51.74 | BTV-26_1446-1770 | 90.7 | 43.08 | BTV-26_2278-2623 | 116.3 | 45.35 | 44.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monsion, B.; Mohd Jaafar, F.; Mertens, P.P.C.; Attoui, H. Uncovering the Underlying Mechanisms Blocking Replication of Bluetongue Virus Serotype 26 (BTV-26) in Culicoides Cells. Biomolecules 2023, 13, 878. https://doi.org/10.3390/biom13060878
Monsion B, Mohd Jaafar F, Mertens PPC, Attoui H. Uncovering the Underlying Mechanisms Blocking Replication of Bluetongue Virus Serotype 26 (BTV-26) in Culicoides Cells. Biomolecules. 2023; 13(6):878. https://doi.org/10.3390/biom13060878
Chicago/Turabian StyleMonsion, Baptiste, Fauziah Mohd Jaafar, Peter P. C. Mertens, and Houssam Attoui. 2023. "Uncovering the Underlying Mechanisms Blocking Replication of Bluetongue Virus Serotype 26 (BTV-26) in Culicoides Cells" Biomolecules 13, no. 6: 878. https://doi.org/10.3390/biom13060878
APA StyleMonsion, B., Mohd Jaafar, F., Mertens, P. P. C., & Attoui, H. (2023). Uncovering the Underlying Mechanisms Blocking Replication of Bluetongue Virus Serotype 26 (BTV-26) in Culicoides Cells. Biomolecules, 13(6), 878. https://doi.org/10.3390/biom13060878










