Skeletal Muscle MicroRNA Patterns in Response to a Single Bout of Exercise in Females: Biomarkers for Subsequent Training Adaptation?
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bell, L.R.; Gabbett, T.J.; Davis, G.M.; Wallen, M.P.; O’Brien, B.J. Stubborn exercise responders-where to next? Sports 2022, 10, 95. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef] [PubMed]
- Sood, P.; Krek, A.; Zavolan, M.; Macino, G.; Rajewsky, N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. USA 2006, 103, 2746–2751. [Google Scholar] [CrossRef]
- Van Rooij, E.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Hill, J.; Olson, E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007, 316, 575–579. [Google Scholar] [CrossRef]
- McCarthy, J.J. MicroRNA-206: The skeletal muscle-specific myomiR. Biochim. Biophys. Acta 2008, 1779, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Quiat, D.; Johnson, B.A.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Kelm, R.J., Jr.; Olson, E.N. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 2009, 17, 662–673. [Google Scholar] [CrossRef]
- Small, E.M.; O’Rourke, J.R.; Moresi, V.; Sutherland, L.B.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. USA 2010, 107, 4218–4223. [Google Scholar] [CrossRef]
- Giagnorio, E.; Malacarne, C.; Mantegazza, R.; Bonanno, S.; Marcuzzo, S. MyomiRs and their multifaceted regulatory roles in muscle homeostasis and amyotrophic lateral sclerosis. J. Cell Sci. 2021, 134, jcs258349. [Google Scholar] [CrossRef]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Dey, B.K.; Gagan, J.; Dutta, A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell. Biol. 2011, 31, 203–214, Erratum in Mol. Cell. Biol. 2011, 31, 1329. [Google Scholar] [CrossRef]
- Goljanek-Whysall, K.; Sweetman, D.; Abu-Elmagd, M.; Chapnik, E.; Dalmay, T.; Hornstein, E.; Münsterberg, A. MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 11936–11941. [Google Scholar] [CrossRef]
- Cui, S.; Li, L.; Mubarokah, S.N.; Meech, R. Wnt/β-catenin signaling induces the myomiRs miR-133b and miR-206 to suppress Pax7 and induce the myogenic differentiation program. J. Cell. Biochem. 2019, 120, 12740–12751. [Google Scholar] [CrossRef]
- Gonçalves, T.J.M.; Boutillon, F.; Lefebvre, S.; Goffin, V.; Iwatsubo, T.; Wakabayashi, T.; Oury, F.; Armand, A.S. Collagen XXV promotes myoblast fusion during myogenic differentiation and muscle formation. Sci. Rep. 2019, 9, 5878, Erratum in Sci. Rep. 2019, 9, 17519. [Google Scholar] [CrossRef]
- Van Rooij, E.; Liu, N.; Olson, E.N. MicroRNAs flex their muscles. Trends Genet. 2008, 24, 159–166. [Google Scholar] [CrossRef]
- Callis, T.E.; Pandya, K.; Seok, H.Y.; Tang, R.H.; Tatsuguchi, M.; Huang, Z.P.; Chen, J.F.; Deng, Z.; Gunn, B.; Shumate, J.; et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Investig. 2009, 119, 2772–2786. [Google Scholar] [CrossRef]
- Gan, Z.; Rumsey, J.; Hazen, B.C.; Lai, L.; Leone, T.C.; Vega, R.B.; Xie, H.; Conley, K.E.; Auwerx, J.; Smith, S.R.; et al. Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J. Clin. Investig. 2013, 123, 2564–2575. [Google Scholar] [CrossRef]
- Alexander, M.S.; Casar, J.C.; Motohashi, N.; Myers, J.A.; Eisenberg, I.; Gonzalez, R.T.; Estrella, E.A.; Kang, P.B.; Kawahara, G.; Kunkel, L.M. Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet. Muscle 2011, 1, 27. [Google Scholar] [CrossRef]
- Alexander, M.S.; Casar, J.C.; Motohashi, N.; Vieira, N.M.; Eisenberg, I.; Marshall, J.L.; Gasperini, M.J.; Lek, A.; Myers, J.A.; Estrella, E.A.; et al. MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J. Clin. Investig. 2014, 124, 2651–2667. [Google Scholar] [CrossRef]
- Boon, H.; Sjögren, R.J.; Massart, J.; Egan, B.; Kostovski, E.; Iversen, P.O.; Hjeltnes, N.; Chibalin, A.V.; Widegren, U.; Zierath, J.R. MicroRNA-208b progressively declines after spinal cord injury in humans and is inversely related to myostatin expression. Physiol. Rep. 2015, 3, e12622. [Google Scholar] [CrossRef]
- Elia, L.; Contu, R.; Quintavalle, M.; Varrone, F.; Chimenti, C.; Russo, M.A.; Cimino, V.; De Marinis, L.; Frustaci, A.; Catalucci, D.; et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation 2009, 120, 2377–2385. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.B.; Xu, H.; Xie, S.J.; Zhou, H.; Qu, L.H. Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS ONE 2011, 6, e29173. [Google Scholar] [CrossRef] [PubMed]
- Kukreti, H.; Amuthavalli, K.; Harikumar, A.; Sathiyamoorthy, S.; Feng, P.Z.; Anantharaj, R.; Tan, S.L.; Lokireddy, S.; Bonala, S.; Sriram, S.; et al. Muscle-specific microRNA1 (miR1) targets heat shock protein 70 (HSP70) during dexamethasone-mediated atrophy. J. Biol. Chem. 2013, 288, 6663–6678. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Zhu, C.D.; Guo, J.T.; Zhao, L.H.; Zhao, J.L. miR-206 regulates the growth of the teleost tilapia (Oreochromis niloticus) through the modulation of IGF-1 gene expression. J. Exp. Biol. 2013, 216 Pt 7, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
- Baggish, A.L.; Hale, A.; Weiner, R.B.; Lewis, G.D.; Systrom, D.; Wang, F.; Wang, T.J.; Chan, S.Y. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J. Physiol. 2011, 589 Pt 16, 3983–3994. [Google Scholar] [CrossRef]
- Bye, A.; Røsjø, H.; Aspenes, S.T.; Condorelli, G.; Omland, T.; Wisløff, U. Circulating microRNAs and aerobic fitness—The HUNT-Study. PLoS ONE 2013, 8, e57496. [Google Scholar] [CrossRef]
- Cui, S.; Sun, B.; Yin, X.; Guo, X.; Chao, D.; Zhang, C.; Zhang, C.Y.; Chen, X.; Ma, J. Time-course responses of circulating microRNAs to three resistance training protocols in healthy young men. Sci. Rep. 2017, 7, 2203. [Google Scholar] [CrossRef]
- D’Souza, R.F.; Markworth, J.F.; Aasen, K.M.M.; Zeng, N.; Cameron-Smith, D.; Mitchell, C.J. Acute resistance exercise modulates microRNA expression profiles: Combined tissue and circulatory targeted analyses. PLoS ONE 2017, 12, e0181594. [Google Scholar] [CrossRef]
- Li, Y.; Yao, M.; Zhou, Q.; Cheng, Y.; Che, L.; Xu, J.; Xiao, J.; Shen, Z.; Bei, Y. Dynamic regulation of circulating microRNAs during acute exercise and long-term exercise training in basketball athletes. Front. Physiol. 2018, 9, 282. [Google Scholar] [CrossRef]
- Zhou, Q.; Shi, C.; Lv, Y.; Zhao, C.; Jiao, Z.; Wang, T. Circulating microRNAs in response to exercise training in healthy adults. Front. Genet. 2020, 11, 256. [Google Scholar] [CrossRef]
- Petek, B.J.; Wasfy, M.M. Cardiac adaption to exercise training: The female athlete. Curr. Treat. Options Cardiovasc. Med. 2018, 20, 68. [Google Scholar] [CrossRef]
- Hutchins, K.P.; Borg, D.N.; Bach, A.J.E.; Bon, J.J.; Minett, G.M.; Stewart, I.B. Female (under) representation in exercise thermoregulation research. Sports Med. Open. 2021, 7, 43. [Google Scholar] [CrossRef]
- Patel, R.; Kemp, C.L.; Hafejee, M.; Peckham, N.; Jain, V.; McCann, G.P.; Pallikadavath, S. The underrepresentation of females in studies assessing the impact of high-dose exercise on cardiovascular outcomes: A scoping review. Sports Med. Open 2021, 7, 30. [Google Scholar] [CrossRef]
- D’Lauro, C.; Jones, E.R.; Swope, L.M.; Anderson, M.N.; Broglio, S.; Schmidt, J.D. Under-representation of female athletes in research informing influential concussion consensus and position statements: An evidence review and synthesis. Br. J. Sports Med. 2022, 56, 981–987. [Google Scholar] [CrossRef]
- Smith, M.; Orchard, J.; La Gerche, A.; Gallagher, R.; Fitzpatrick, J. Fit, female or fifty-is cardiac rehabilitation “fit” for purpose for all? A Systematic Review and Meta-Analysis with Meta-Regression. Front. Cardiovasc. Med. 2022, 9, 764882. [Google Scholar] [CrossRef]
- Safdar, A.; Abadi, A.; Akhtar, M.; Hettinga, B.P.; Tarnopolsky, M.A. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS ONE 2009, 4, e5610. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.P.; Lamon, S.; Boon, H.; Wada, S.; Güller, I.; Brown, E.L.; Chibalin, A.V.; Zierath, J.R.; Snow, R.J.; Stepto, N.; et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 2013, 591, 4637–4653. [Google Scholar] [CrossRef]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef]
- Baar, K.; Wende, A.R.; Jones, T.E.; Marison, M.; Nolte, L.A.; Chen, M.; Kelly, D.P.; Holloszy, J.O. Adaptations of skeletal muscle to exercise: Rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002, 16, 1879–1886. [Google Scholar] [CrossRef]
- Geng, T.; Li, P.; Okutsu, M.; Yin, X.; Kwek, J.; Zhang, M.; Yan, Z. PGC-1alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. Am. J. Physiol. Cell Physiol. 2010, 298, C572–C579. [Google Scholar] [CrossRef]
- Yamamoto, H.; Morino, K.; Nishio, Y.; Ugi, S.; Yoshizaki, T.; Kashiwagi, A.; Maegawa, H. MicroRNA-494 regulates mitochondrial biogenesis in skeletal muscle through mitochondrial transcription factor A and Forkhead box j3. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E1419–E1427. [Google Scholar] [CrossRef] [PubMed]
- McLean, C.S.; Mielke, C.; Cordova, J.M.; Langlais, P.R.; Bowen, B.; Miranda, D.; Coletta, D.K.; Mandarino, L.J. Gene and microRNA expression responses to exercise; Relationship with insulin sensitivity. PLoS ONE 2015, 10, e0127089. [Google Scholar] [CrossRef] [PubMed]
- Eichner, L.J.; Perry, M.C.; Dufour, C.R.; Bertos, N.; Park, M.; St-Pierre, J.; Giguère, V. miR-378(∗) mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 2010, 12, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Gerin, I.; Bommer, G.T.; McCoin, C.S.; Sousa, K.M.; Krishnan, V.; MacDougald, O.A. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E198–E206. [Google Scholar] [CrossRef] [PubMed]
- Carrer, M.; Liu, N.; Grueter, C.E.; Williams, A.H.; Frisard, M.I.; Hulver, M.W.; Bassel-Duby, R.; Olson, E.N. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc. Natl. Acad. Sci. USA 2012, 109, 15330–15335. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Webb, R.; Richardson, J.A.; Olson, E.N. MyoR: A muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc. Natl. Acad. Sci. USA 1999, 96, 552–557. [Google Scholar] [CrossRef]
- Gagan, J.; Dey, B.K.; Layer, R.; Yan, Z.; Dutta, A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J. Biol. Chem. 2011, 286, 19431–19438. [Google Scholar] [CrossRef]
- Motohashi, N.; Alexander, M.S.; Shimizu-Motohashi, Y.; Myers, J.A.; Kawahara, G.; Kunkel, L.M. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J. Cell Sci. 2013, 126 Pt 12, 2678–2691. [Google Scholar] [CrossRef]
- Dalle Carbonare, L.; Dorelli, G.; Li Vigni, V.; Minoia, A.; Bertacco, J.; Cheri, S.; Deiana, M.; Innamorati, G.; Cominacini, M.; Tarperi, C.; et al. Physical activity modulates miRNAs levels and enhances MYOD expression in myoblasts. Stem Cell Rev. Rep. 2022, 18, 1865–1874. [Google Scholar] [CrossRef]
- Pham, T.T.; Ban, J.; Lee, K.; Hong, Y.; Lee, J.; Truong, A.D.; Lillehoj, H.S.; Hong, Y.H. MicroRNA gga-miR-10a-mediated transcriptional regulation of the immune genes in necrotic enteritis afflicted chickens. Dev. Comp. Immunol. 2020, 102, 103472. [Google Scholar] [CrossRef]
- Wang, X.; Ling, C.C.; Li, L.; Qin, Y.; Qi, J.; Liu, X.; You, B.; Shi, Y.; Zhang, J.; Jiang, Q.; et al. MicroRNA-10a/10b represses a novel target gene mib1 to regulate angiogenesis. Cardiovasc. Res. 2016, 110, 140–150. [Google Scholar] [CrossRef]
- Fish, J.E.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.F.; Wythe, J.D.; Ivey, K.N.; Bruneau, B.G.; Stainier, D.Y.; Srivastava, D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 2008, 15, 272–284. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Sun, C.Y.; She, X.M.; Qin, Y.; Chu, Z.B.; Chen, L.; Ai, L.S.; Zhang, L.; Hu, Y. miR-15a miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis 2013, 34, 426–435. [Google Scholar] [CrossRef]
- Fernandes, T.; Magalhães, F.C.; Roque, F.R.; Phillips, M.I.; Oliveira, E.M. Exercise training prevents the microvascular rarefaction in hypertension balancing angiogenic and apoptotic factors: Role of microRNAs-16, -21, and -126. Hypertension 2012, 59, 513–520. [Google Scholar] [CrossRef]
- Nielsen, S.; Scheele, C.; Yfanti, C.; Akerström, T.; Nielsen, A.R.; Pedersen, B.K.; Laye, M.J. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J. Physiol. 2010, 588 Pt 20, 4029–4037, Erratum in J. Physiol. 2011, 589 Pt 5, 1239; Laye, M.J. Erratum in J. Physiol. 2015, 593, 1323. [Google Scholar] [CrossRef]
- Kadi, F.; Johansson, F.; Johansson, R.; Sjöström, M.; Henriksson, J. Effects of one bout of endurance exercise on the expression of myogenin in human quadriceps muscle. Histochem. Cell Biol. 2004, 121, 329–334. [Google Scholar] [CrossRef]
- Coffey, V.G.; Shield, A.; Canny, B.J.; Carey, K.A.; Cameron-Smith, D.; Hawley, J.A. Interaction of contractile activity and training history on mRNA abundance in skeletal muscle from trained athletes. Am. J. Physiol. Endocrinol. Metab. 2006, 290, E849–E855. [Google Scholar] [CrossRef]
- Naguibneva, I.; Ameyar-Zazoua, M.; Polesskaya, A.; Ait-Si-Ali, S.; Groisman, R.; Souidi, M.; Cuvellier, S.; Harel-Bellan, A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 2006, 8, 278–284. [Google Scholar] [CrossRef]
- Sweetman, D.; Goljanek, K.; Rathjen, T.; Oustanina, S.; Braun, T.; Dalmay, T.; Münsterberg, A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev. Biol. 2008, 321, 491–499. [Google Scholar] [CrossRef]
- Keller, P.; Vollaard, N.B.; Gustafsson, T.; Gallagher, I.J.; Sundberg, C.J.; Rankinen, T.; Britton, S.L.; Bouchard, C.; Koch, L.G.; Timmons, J.A. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J. Appl. Physiol. 2011, 110, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Thiel, A.; Sudeck, G.; Gropper, H.; Maturana, F.M.; Schubert, T.; Srismith, D.; Widmann, M.; Behrens, S.; Martus, P.; Munz, B.; et al. The iReAct study—A biopsychosocial analysis of the individual response to physical activity. Contemp. Clin. Trials Commun. 2019, 17, 100508. [Google Scholar] [CrossRef] [PubMed]
- Widmann, M.; Mattioni Maturana, F.; Burgstahler, C.; Erz, G.; Schellhorn, P.; Fragasso, A.; Schmitt, A.; Nieß, A.M.; Munz, B. miRNAs as markers for the development of individualized training regimens: A pilot study. Physiol. Rep. 2022, 10, e15217. [Google Scholar] [CrossRef] [PubMed]
- Maturana, F.M.; Schellhorn, P.; Erz, G.; Burgstahler, C.; Widmann, M.; Munz, B.; Soares, R.N.; Murias, J.M.; Thiel, A.; Nieß, A.M. Individual cardiovascular responsiveness to work-matched exercise within the moderate- and severe-intensity domains. Eur. J. Appl. Physiol. 2021, 121, 2039–2059. [Google Scholar] [CrossRef] [PubMed]
- Mattioni Maturana, F.; Soares, R.N.; Murias, J.M.; Schellhorn, P.; Erz, G.; Burgstahler, C.; Widmann, M.; Munz, B.; Thiel, A.; Nieß, A.M. Responders and non-responders to aerobic exercise training: Beyond the evaluation of V̇O2max. Physiol. Rep. 2021, 9, e14951. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; L. Erlbaum associates; Academic Press: Cambridge, MA, USA, 1988. [Google Scholar]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Yin, K.J.; Olsen, K.; Hamblin, M.; Zhang, J.; Schwendeman, S.P.; Chen, Y.E. Vascular endothelial cell-specific microRNA-15a inhibits angiogenesis in hindlimb ischemia. J. Biol. Chem. 2012, 287, 27055–27064. [Google Scholar] [CrossRef]
- Suárez, Y.; Fernández-Hernando, C.; Yu, J.; Gerber, S.A.; Harrison, K.D.; Pober, J.S.; Iruela-Arispe, M.L.; Merkenschlager, M.; Sessa, W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 14082–14087. [Google Scholar] [CrossRef]
- Suárez, Y.; Sessa, W.C. MicroRNAs as novel regulators of angiogenesis. Circ. Res. 2009, 104, 442–454, Erratum in Circ. Res. 2009, 104, e55. [Google Scholar] [CrossRef]
- Anand, S.; Majeti, B.K.; Acevedo, L.M.; Murphy, E.A.; Mukthavaram, R.; Scheppke, L.; Huang, M.; Shields, D.J.; Lindquist, J.N.; Lapinski, P.E.; et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 2010, 16, 909–914. [Google Scholar] [CrossRef]
- Joris, V.; Gomez, E.L.; Menchi, L.; Lobysheva, I.; Di Mauro, V.; Esfahani, H.; Condorelli, G.; Balligand, J.L.; Catalucci, D.; Dessy, C. MicroRNA-199a-3p and microRNA-199a-5p take part to a redundant network of regulation of the NOS (NO Synthase)/NO pathway in the endothelium. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2345–2357. [Google Scholar] [CrossRef]
- Kang, T.; Lu, W.; Xu, W.; Anderson, L.; Bacanamwo, M.; Thompson, W.; Chen, Y.E.; Liu, D. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. J. Biol. Chem. 2013, 288, 34394–34402. [Google Scholar] [CrossRef]
- McCoy, C.E.; Sheedy, F.J.; Qualls, J.E.; Doyle, S.L.; Quinn, S.R.; Murray, P.J.; O’Neill, L.A. IL-10 inhibits miR-155 induction by toll-like receptors. J. Biol. Chem. 2010, 285, 20492–20498. [Google Scholar] [CrossRef]
- Onodera, Y.; Teramura, T.; Takehara, T.; Itokazu, M.; Mori, T.; Fukuda, K. Inflammation-associated miR-155 activates differentiation of muscular satellite cells. PLoS ONE 2018, 13, e0204860. [Google Scholar] [CrossRef]
- Tarnowski, M.; Kopytko, P.; Piotrowska, K. Epigenetic Regulation of inflammatory responses in the context of physical activity. Genes 2021, 12, 1313. [Google Scholar] [CrossRef]
- Chemello, F.; Grespi, F.; Zulian, A.; Cancellara, P.; Hebert-Chatelain, E.; Martini, P.; Bean, C.; Alessio, E.; Buson, L.; Bazzega, M.; et al. Transcriptomic analysis of single isolated myofibers identifies miR-27a-3p and miR-142-3p as regulators of metabolism in skeletal muscle. Cell Rep. 2019, 26, 3784–3797.e8. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, H.; Zhou, P.; Zhang, Z.; Liu, J.; Zhang, H. MicroRNA-152 promotes slow-twitch myofiber formation via targeting uncoupling protein-3 gene. Animals 2019, 9, 669. [Google Scholar] [CrossRef]
- Jia, H.; Zhao, Y.; Li, T.; Zhang, Y.; Zhu, D. miR-30e is negatively regulated by myostatin in skeletal muscle and is functionally related to fiber-type composition. Acta Biochim. Biophys. Sin. 2017, 49, 392–399. [Google Scholar] [CrossRef]
- Zhu, Y.; Long, H.T.; Zeng, L.; Tang, Y.F.; Zhao, R.B.; Lin, Z.Y.; Zhao, S.S.; Cheng, L. MiR-19b-3p regulates osteogenic differentiation of PDGFRα+ muscle cells by specifically targeting PTEN. Cell Biol. Int. 2019, 43, 565–573. [Google Scholar] [CrossRef]
- Rivas, D.A.; Peng, F.; Benard, T.; Ramos da Silva, A.S.; Fielding, R.A.; Margolis, L.M. miR-19b-3p is associated with a diametric response to resistance exercise in older adults and regulates skeletal muscle anabolism via PTEN inhibition. Am. J. Physiol. Cell Physiol. 2021, 321, C977–C991. [Google Scholar] [CrossRef]
- Singh, G.B.; Cowan, D.B.; Wang, D.Z. Tiny regulators of massive tissue: MicroRNAs in skeletal muscle development, myopathies, and cancer cachexia. Front. Oncol. 2020, 10, 598964. [Google Scholar] [CrossRef] [PubMed]
- Rivas, D.A.; Lessard, S.J.; Rice, N.P.; Lustgarten, M.S.; So, K.; Goodyear, L.J.; Parnell, L.D.; Fielding, R.A. Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF-1 signaling. FASEB J. 2014, 28, 4133–4147. [Google Scholar] [CrossRef] [PubMed]
- Hitachi, K.; Tsuchida, K. Role of microRNAs in skeletal muscle hypertrophy. Front. Physiol. 2014, 4, 408. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Li, Y.F.; Wu, G.F.; Song, Z.Y.; Lu, H.Z.; Song, C.C.; Zhang, Q.L.; Zhu, J.Y.; Yang, G.S.; Shi, X.E. MiRNA-199a-3p regulates C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal pathway. Int. J. Mol. Sci. 2013, 15, 296–308. [Google Scholar] [CrossRef]
- Meyer, S.U.; Thirion, C.; Polesskaya, A.; Bauersachs, S.; Kaiser, S.; Krause, S.; Pfaffl, M.W. TNF-α and IGF1 modify the microRNA signature in skeletal muscle cell differentiation. Cell Commun. Signal. 2015, 13, 4. [Google Scholar] [CrossRef]
- Shu, L.; Zhao, H.; Huang, W.; Hou, G.; Song, G.; Ma, H. Resveratrol upregulates mmu-miR-363-3p via the PI3K-Akt pathway to improve insulin resistance induced by a high-fat diet in mice. Diabetes Metab. Syndr. Obes. 2020, 13, 391–403. [Google Scholar] [CrossRef]
- Kong, D.; He, M.; Yang, L.; Zhou, R.; Yan, Y.Q.; Liang, Y.; Teng, C.B. MiR-17 and miR-19 cooperatively promote skeletal muscle cell differentiation. Cell. Mol. Life Sci. 2019, 76, 5041–5054. [Google Scholar] [CrossRef]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef]
- Silva, W.J.; Graça, F.A.; Cruz, A.; Silvestre, J.G.; Labeit, S.; Miyabara, E.H.; Yan, C.Y.I.; Wang, D.Z.; Moriscot, A.S. miR-29c improves skeletal muscle mass and function throughout myocyte proliferation and differentiation and by repressing atrophy-related genes. Acta Physiol. 2019, 226, e13278. [Google Scholar] [CrossRef]
- Güller, I.; Russell, A.P. MicroRNAs in skeletal muscle: Their role and regulation in development, disease and function. J. Physiol. 2010, 588 Pt 21, 4075–4087. [Google Scholar] [CrossRef]
- Zhao, Q.; Kang, Y.; Wang, H.Y.; Guan, W.J.; Li, X.C.; Jiang, L.; He, X.H.; Pu, Y.B.; Han, J.L.; Ma, Y.H.; et al. Expression profiling and functional characterization of miR-192 throughout sheep skeletal muscle development. Sci. Rep. 2016, 6, 30281. [Google Scholar] [CrossRef]
- Fochi, S.; Giuriato, G.; De Simone, T.; Gomez-Lira, M.; Tamburin, S.; Del Piccolo, L.; Schena, F.; Venturelli, M.; Romanelli, M.G. Regulation of microRNAs in satellite cell renewal, muscle function, sarcopenia and the role of exercise. Int. J. Mol. Sci. 2020, 21, 6732. [Google Scholar] [CrossRef]
- Massart, J.; Sjögren, R.J.O.; Egan, B.; Garde, C.; Lindgren, M.; Gu, W.; Ferreira, D.M.S.; Katayama, M.; Ruas, J.L.; Barrès, R.; et al. Endurance exercise training-responsive miR-19b-3p improves skeletal muscle glucose metabolism. Nat. Commun. 2021, 12, 5948. [Google Scholar] [CrossRef]
- Wang, W.; Yu, Q.; Cao, Q.; Ye, H.; Zhang, C.; Dong, Z.; Feng, D.; Zuo, J. MicroRNA-27a inhibits porcine type I muscle fibre gene expression by directly targeting peroxisome proliferator-activated receptor-γ coactivator-1α. J. Anim. Physiol. Anim. Nutr. 2023. epub ahead of print. [Google Scholar] [CrossRef]
- Chen, X.; Huang, Z.; Chen, D.; Yang, T.; Liu, G. MicroRNA-27a is induced by leucine and contributes to leucine-induced proliferation promotion in C2C12 cells. Int. J. Mol. Sci. 2013, 14, 14076–14084. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, X.; Yu, B.; He, J.; Chen, D. MicroRNA-27a promotes myoblast proliferation by targeting myostatin. Biochem. Biophys. Res. Commun. 2012, 423, 265–269. [Google Scholar] [CrossRef]
- Yokomizo, T.; Hasegawa, K.; Ishitobi, H.; Osato, M.; Ema, M.; Ito, Y.; Yamamoto, M.; Takahashi, S. Runx1 is involved in primitive erythropoiesis in the mouse. Blood 2008, 111, 4075–4080. [Google Scholar] [CrossRef]
- Kiens, B.; Richter, E.A. Utilization of skeletal muscle triacylglycerol during postexercise recovery in humans. Am. J. Physiol. 1998, 275, E332–E337. [Google Scholar] [CrossRef]
- Egan, B.; Zierath, J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef]
- Matoba, S.; Kang, J.G.; Patino, W.D.; Wragg, A.; Boehm, M.; Gavrilova, O.; Hurley, P.J.; Bunz, F.; Hwang, P.M. p53 regulates mitochondrial respiration. Science 2006, 312, 1650–1653. [Google Scholar] [CrossRef]
- Saleem, A.; Hood, D.A. Acute exercise induces tumour suppressor protein p53 translocation to the mitochondria and promotes a p53-Tfam-mitochondrial DNA complex in skeletal muscle. J. Physiol. 2013, 591, 3625–3636. [Google Scholar] [CrossRef] [PubMed]
- Achanta, G.; Sasaki, R.; Feng, L.; Carew, J.S.; Lu, W.; Pelicano, H.; Keating, M.J.; Huang, P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005, 24, 3482–3492. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.Y.; Zhuang, J.; Hwang, P.M. p53: Exercise capacity and metabolism. Curr. Opin. Oncol. 2012, 24, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Wang, P.Y.; Matsumoto, T.; Sung, H.J.; Ma, W.; Choi, J.W.; Anderson, S.A.; Leary, S.C.; Balaban, R.S.; Kang, J.G.; et al. p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circ. Res. 2009, 105, 705–712, 11p following 712. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhang, C.; Wu, R.; Sun, Y.; Levine, A.; Feng, Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. USA 2010, 107, 7455–7460. [Google Scholar] [CrossRef]
- Schild, M.; Ruhs, A.; Beiter, T.; Zügel, M.; Hudemann, J.; Reimer, A.; Krumholz-Wagner, I.; Wagner, C.; Keller, J.; Eder, K.; et al. Basal and exercise induced label-free quantitative protein profiling of m. vastus lateralis in trained and untrained individuals. J. Proteom. 2015, 122, 119–132. [Google Scholar] [CrossRef]
- Watt, K.I.; Turner, B.J.; Hagg, A.; Zhang, X.; Davey, J.R.; Qian, H.; Beyer, C.; Winbanks, C.E.; Harvey, K.F.; Gregorevic, P. The Hippo pathway effector YAP is a critical regulator of skeletal muscle fibre size. Nat. Commun. 2015, 6, 6048. [Google Scholar] [CrossRef]
- Watt, K.I.; Henstridge, D.C.; Ziemann, M.; Sim, C.B.; Montgomery, M.K.; Samocha-Bonet, D.; Parker, B.L.; Dodd, G.T.; Bond, S.T.; Salmi, T.M.; et al. Yap regulates skeletal muscle fatty acid oxidation and adiposity in metabolic disease. Nat. Commun. 2021, 12, 2887. [Google Scholar] [CrossRef]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]




| miR | Correlation Coefficient r (Spearman) | p |
|---|---|---|
| hsa-miR-15a-5p | 0.77 | 0.072 |
| hsa-miR-18a-5p | 0.6 | 0.208 |
| hsa-miR-19b-3p | 0.89 | 0.019 * |
| hsa-miR-132-3p | 0.6 | 0.208 |
| hsa-miR-155-5p | 0.66 | 0.156 |
| hsa-miR-199a-3p | 0.6 | 0.208 |
| hsa-miR-199a-5p | 0.6 | 0.208 |
| hsa-miR-199b-3p | 0.6 | 0.208 |
| hsa-miR-497-5p | 0.6 | 0.208 |
| hsa-miR-3175 | 0.77 | 0.072 |
| hsa-miR-4306 | 0.71 | 0.111 |
| hsa-miR-7151-3p | 0.77 | 0.072 |
| hsa-miR-7641 | 0.71 | 0.111 |
| hsa-miR-197-5p | −0.6 | 0.208 |
| hsa-miR-4330 | −0.6 | 0.208 |
| hsa-miR-4667-5p | −0.94 | 0.005 ** |
| hsa-miR-4743-5p | −0.77 | 0.072 |
| hsa-miR-6831-5p | −0.83 | 0.042 * |
| hsa-miR-7109-5p | −0.89 | 0.019 * |
| Signaling Pathway | p | Number of Genes | miR | ||
|---|---|---|---|---|---|
| 1 | prion diseases (hsa05020) | <1 × 10−325 | 18 | -22-5p | -27a-3p |
| -30a-3p | -30e-3p | ||||
| -106b-5p | -192-5p | ||||
| 2 | fatty acid biosynthesis (hsa00061) | <1 × 10−325 | 5 | -10a-5p | -15a-5p |
| -27a-3p | -29c-3p | ||||
| -30e-5p | -199a-3p | ||||
| -199b-3p | -1226-5p | ||||
| -2110 | |||||
| 3 | fatty acid metabolism (hsa01212) | <1 × 10−325 | 24 | -10a-5p | -15a-5p |
| -27a-3p | -29c-3p | ||||
| -30e-5p | -155-5p | ||||
| -199a-3p | -199b-3p | ||||
| -155-5p | -363-3p | ||||
| -660-5p | -1226-5p | ||||
| 4 | ECM-receptor interaction (hsa04512) | <1 × 10−325 | 43 | -22-5p | -19b-3p |
| -27a-3p | -29c-3p | ||||
| -30a-3p | -30e-3p | ||||
| -101-5p | -199a-5p | ||||
| -497-5p | |||||
| 5 | proteoglycans in cancer (hsa05205) | <1 × 10−325 | 142 | -15a-5p | -19b-3p |
| -22-5p | -27a-3p | ||||
| -29c-3p | -30a-3p | ||||
| -30e-3p | -30e-5p | ||||
| -132-3p | -155-5p | ||||
| -106b-5p | -192-5p | ||||
| -199a-3p | -199a-5p | ||||
| -497-5p | |||||
| 6 | viral carcinogenesis (hsa05203) | 9.99 × 10−16 | 123 | -15a-5p | -18a-5p |
| -19b-3p | -22-5p | ||||
| -27a-3p | -29c-3p | ||||
| -30a-3p | -30e-3p | ||||
| -30e-5p | -362-5p | ||||
| -2110 | -2115-5p | ||||
| 7 | Hippo signal transduction pathway (hsa04390) | 1.11 × 10−13 | 100 | -10a-5p | -15a-5p |
| -18a-5p | -22-5p | ||||
| -27a-3p | -29c-3p | ||||
| -30a-3p | -30e-3p | ||||
| -106b-5p | -132-3p | ||||
| -192-5p | -199a-5p | ||||
| -497-5p | |||||
| 8 | lysine degradation (hsa00310) | 2.06 × 10−11 | 33 | -15a-5p | -18a-5p |
| -22-5p | -27a-3p | ||||
| -29c-3p | -30a-3p | ||||
| -30e-3p | -30e-5p | ||||
| 106b-5p | 132-3p | ||||
| -192-5p | -197-5p | ||||
| -497-5p | -660-5p | ||||
| -1226-5p | -2110 | ||||
| -2115-5p | |||||
| 9 | cell cycle (hsa04110) | 1.65 × 10−5 | 84 | -15a-5p | -18a-5p |
| -27a-3p | -30a-3p | ||||
| -30e-3p | -30e-5p | ||||
| -101-5p | -106b-5p | ||||
| -132-3p | -192-5p | ||||
| -499-5p | |||||
| 10 | TGFβ signal transduction pathway (hsa04350) | 3.88 × 10−5 | 51 | -15a-5p | -18a-5p |
| -19b-3p | -27a-3p | ||||
| -30a-3p | -30e-3p | ||||
| -106b-5p | -132-3p | ||||
| -155-5p | -199a-5p | ||||
| -497-5p | |||||
| 11 | steroid biosynthesis (hsa00100) | 0.00016 | 12 | -18a-5p | -29c-3p |
| -30a-3p | -30e-3p | ||||
| -155-5p | -192-5p | ||||
| -199a-3p | -199b-3p | ||||
| -362-5p | -2110 | ||||
| 12 | adherens junction (hsa04520) | 0.00037 | 52 | -15a-5p | -22-5p |
| -27a-3p | -30e-5p | ||||
| -106b-5p | -199a-5p | ||||
| -362-5p | -497-5p | ||||
| 13 | p53 signal transduction pathway (hsa04115) | 0.00038 | 46 | -15a-5p | -18a-5p |
| -19b-3p | -27a-3p | ||||
| -29c-3p | -30a-3p | ||||
| -30e-3p | -30e-5p | ||||
| -106b-5p | -497-5p | ||||
| -499a-5p | -660-5p | ||||
| 14 | protein processing in the endoplasmic reticulum (hsa04141) | 0.00059 | 109 | -15a-5p | -22-5p |
| -27a-3p | -30e-5p | ||||
| -106b-5p | -197-5p | ||||
| -497-5p | |||||
| 15 | Hepatitis B (hsa05161) | 0.0029 | 88 | -15a-5p | -19b-3p |
| -27a-3p | -29c-3p | ||||
| -30a-3p | -30e-3p | ||||
| -106b-5p | -155-5p | ||||
| -497-5p | |||||
| 16 | chronic myeloic leukemia (hsa05220) | 0.0131 | 53 | -19b-3p | -27a-3p |
| -29c-3p | -30a-3p | ||||
| -30e-3p | -101-5p | ||||
| -106b-5p | -155-5p | ||||
| -497-5p | |||||
| miR | Microarray | qPCR | Concordance | ||||
|---|---|---|---|---|---|---|---|
| Fold Change | p | Fold Change | p | C | D | γ | |
| -1/-1-3p | −4.91 | 0.008 | −1.54 | 0.203 | 5 | 1 | 0.66 |
| -133a-3p | −2.34 | 0.041 | −4.07 | 0.078 | 4 | 2 | 0.33 |
| -133a-5p | −4.54 | 0.006 | +1.21 | 0.032 | 1 | 5 | −0.66 |
| -133b | −2.76 | 0.011 | −1.45 | 0.565 | 3 | 3 | 0 |
| -499a-5p | −7.61 | 0.046 | −1.33 | 0.249 | 4 | 2 | 0.33 |
| -15a-5p | −5.12 | 0.021 | −1.16 | 0.462 | 1 | 5 | −0.66 |
| -18a-5p | −7.39 | 0.028 | 1.0 | 0.576 | 1 | 5 | −0.66 |
| -19b-3p | −5.33 | 0.032 | −1.04 | 0.45 | 2 | 4 | −0.33 |
| -27a-3p | −4.64 | 0.007 | −1.78 | 0.016 | 5 | 1 | 0.66 |
| -132-3p | +6.46 | 0.028 | +1.27 | 0.614 | 4 | 2 | 0.33 |
| -155-5p | +7.32 | 0.028 | −1.16 | 0.731 | 2 | 4 | −0.33 |
| -199a-3p | −5.8 | 0.035 | −3.78 | 0.127 | 6 | 0 | 1 |
| -199a-5p | −7.69 | 0.026 | −1.49 | 0.023 | 6 | 0 | 1 |
| -378a-5p | −1.99 | 0.018 | −1.29 | 0.01 | 5 | 1 | 0.66 |
| -497-5p | −6.46 | 0.043 | −1.29 | 0.206 | 3 | 3 | 0 |
| miR | Fold Change | p | |
|---|---|---|---|
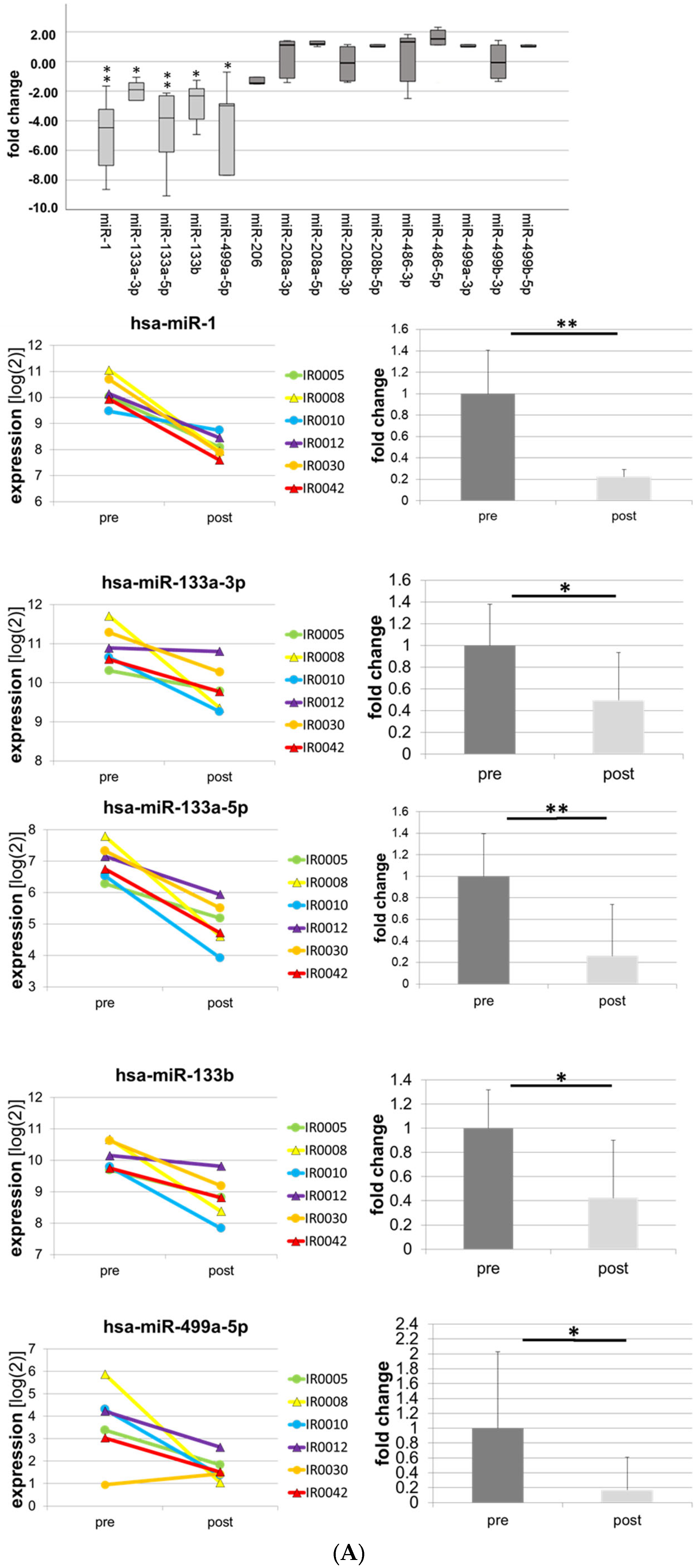
| -1-3p | −8.64 | 0.000 | ** |
| -133a-3p | −7.56 | 0.000 | ** |
| -133a-5p | −3.64 | 0.034 | * |
| -133b | −7.28 | 0.002 | ** |
| -499a-5p | −5.24 | 0.000 | ** |
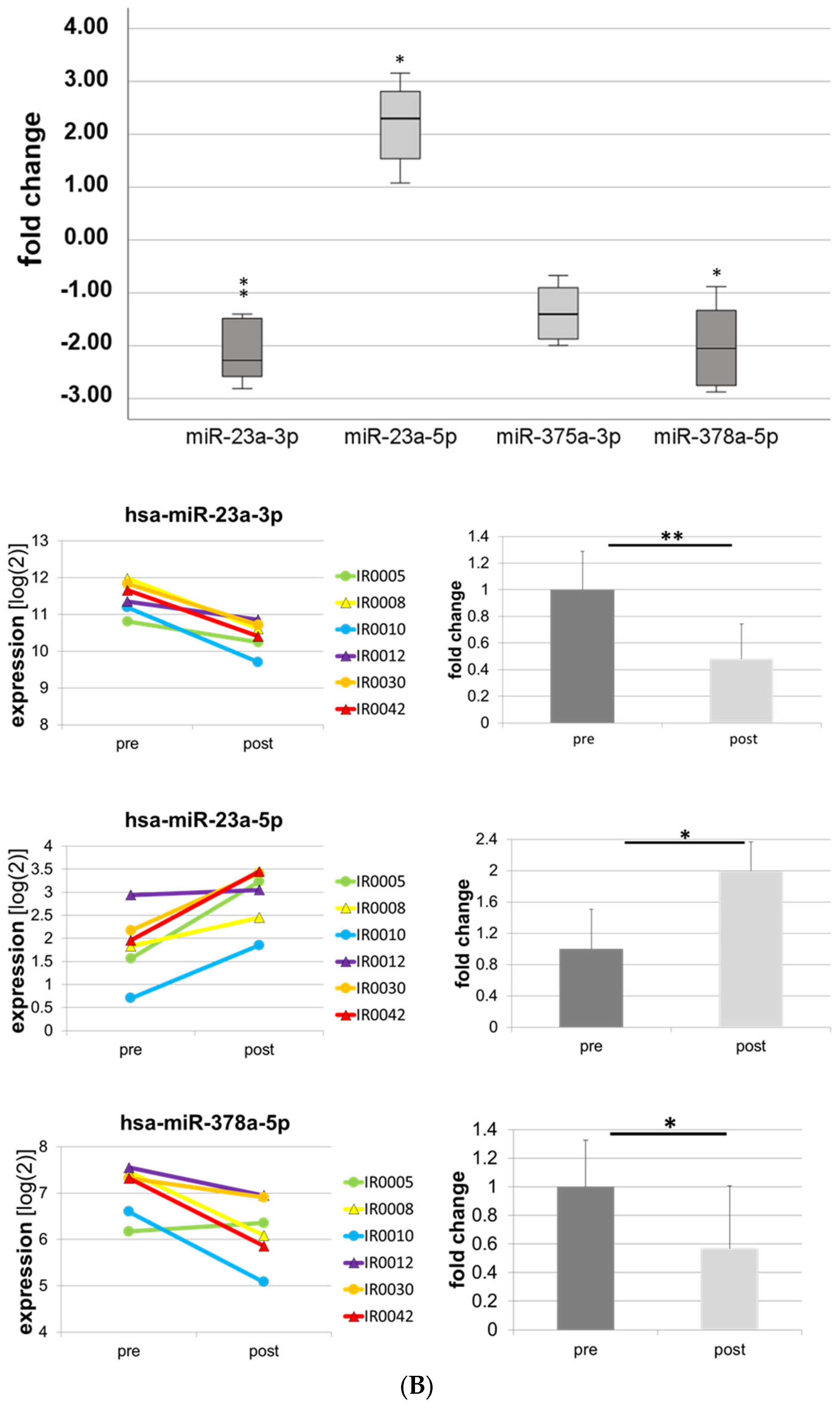
| -27a-3p | −2.12 | 0.000 | ** |
| -378a-5p | −3.33 | 0.000 | ** |
| -199a-3p | −2.41 | 0.015 | * |
| -199a-5p | −1.77 | 0.000 | ** |
| -497-5p | −1.78 | 0.000 | ** |
| -15a-5p | −1.48 | 0.09 | n.s. |
| -18a-5p | −1.59 | 0.222 | n.s. |
| -19b-3p | −1.7 | 0.015 | * |
| -132-3p | +1.3 | 0.275 | n.s. |
| -155-5p | +2.92 | 0.001 | ** |
| miR | 1-3p | 133a-3p | 133a-5p | 133b | 499a-5p | 378a-5p | 497-5p | 199a-5p | 27a-3p | 15a-5p | 18a-5p | 19b-3p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-3p | - | 0.921 | 0.965 | 0.942 | 0.819 | 0.838 | ||||||
| 133a-3p | 0.921 | - | 0.947 | 0.956 | 0.924 | 0.878 | ||||||
| 133a-5p | 0.965 | 0.947 | - | 0.958 | 0.981 | 0.946 | ||||||
| 133b | 0.942 | 0.956 | 0.958 | - | 0.847 | 0.840 | ||||||
| 499a-5p | 0.819 | 0.924 | 0.981 | 0.847 | - | |||||||
| 378a-5p | 0.838 | 0.878 | 0.946 | 0.840 | - | |||||||
| 497-5p | - | 0.683 | 0.511 | |||||||||
| 199a-5p | 0.683 | - | ||||||||||
| 27a-3p | 0.511 | - | ||||||||||
| 15a-5p | - | 0.683 | 0.511 | |||||||||
| 18a-5p | 0.683 | - | 0.767 | |||||||||
| 19b-3p | 0.511 | 0.767 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grieb, A.; Schmitt, A.; Fragasso, A.; Widmann, M.; Mattioni Maturana, F.; Burgstahler, C.; Erz, G.; Schellhorn, P.; Nieß, A.M.; Munz, B. Skeletal Muscle MicroRNA Patterns in Response to a Single Bout of Exercise in Females: Biomarkers for Subsequent Training Adaptation? Biomolecules 2023, 13, 884. https://doi.org/10.3390/biom13060884
Grieb A, Schmitt A, Fragasso A, Widmann M, Mattioni Maturana F, Burgstahler C, Erz G, Schellhorn P, Nieß AM, Munz B. Skeletal Muscle MicroRNA Patterns in Response to a Single Bout of Exercise in Females: Biomarkers for Subsequent Training Adaptation? Biomolecules. 2023; 13(6):884. https://doi.org/10.3390/biom13060884
Chicago/Turabian StyleGrieb, Alexandra, Angelika Schmitt, Annunziata Fragasso, Manuel Widmann, Felipe Mattioni Maturana, Christof Burgstahler, Gunnar Erz, Philipp Schellhorn, Andreas M. Nieß, and Barbara Munz. 2023. "Skeletal Muscle MicroRNA Patterns in Response to a Single Bout of Exercise in Females: Biomarkers for Subsequent Training Adaptation?" Biomolecules 13, no. 6: 884. https://doi.org/10.3390/biom13060884






