ATP13A4 Upregulation Drives the Elevated Polyamine Transport System in the Breast Cancer Cell Line MCF7
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Polyamines and Inhibitors
2.3. Cell Culture and Lentiviral Transduction
2.4. RT-qPCR
2.5. BODIPY-Polyamine Uptake
2.6. Measurement of Cell Viability and Cell Death
2.7. Colony Formation Assay
2.8. Western Blotting
2.9. Statistical Analysis
3. Results
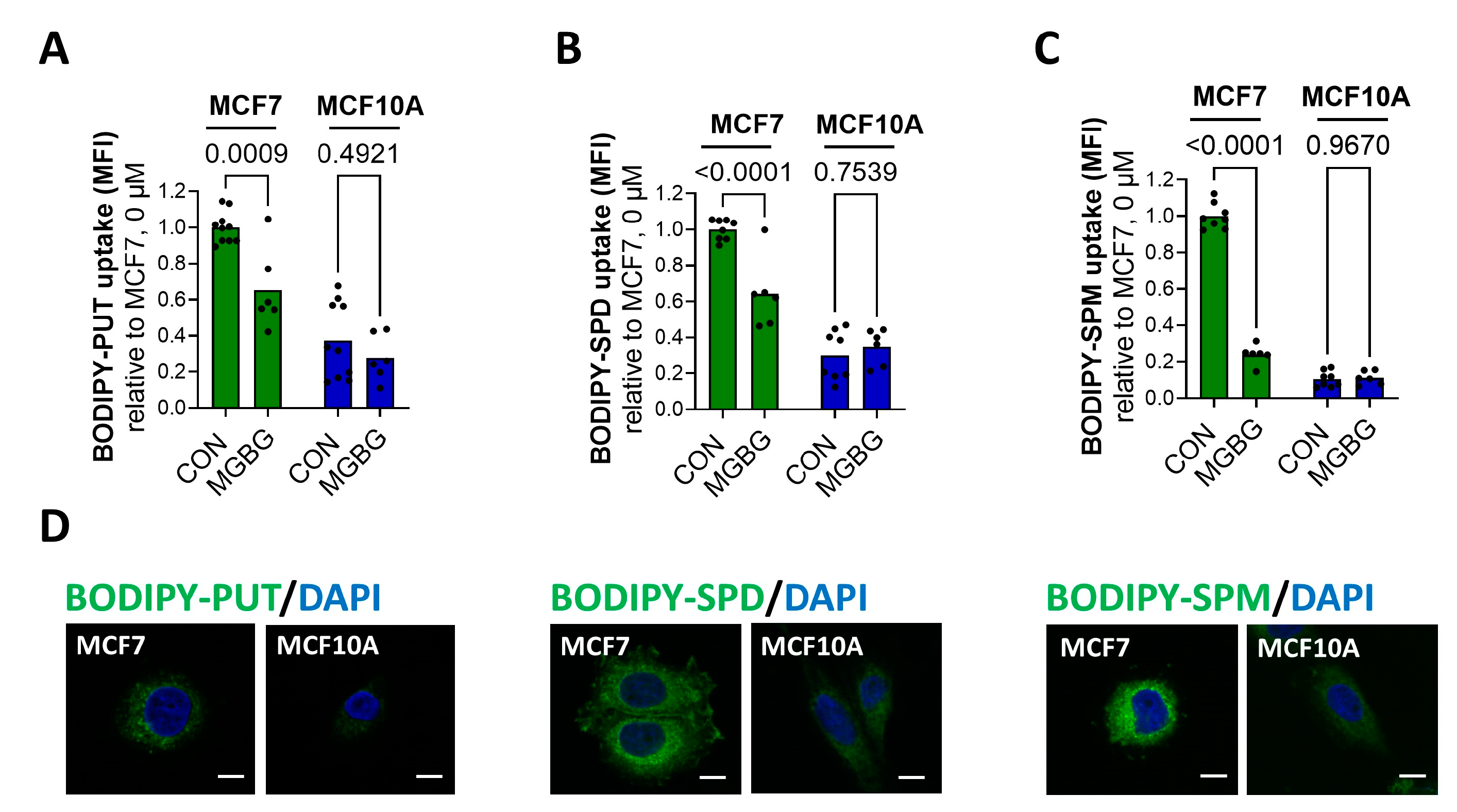
3.1. MCF7 Cells Show Enhanced PTS Activity Compared to MCF10A Cells
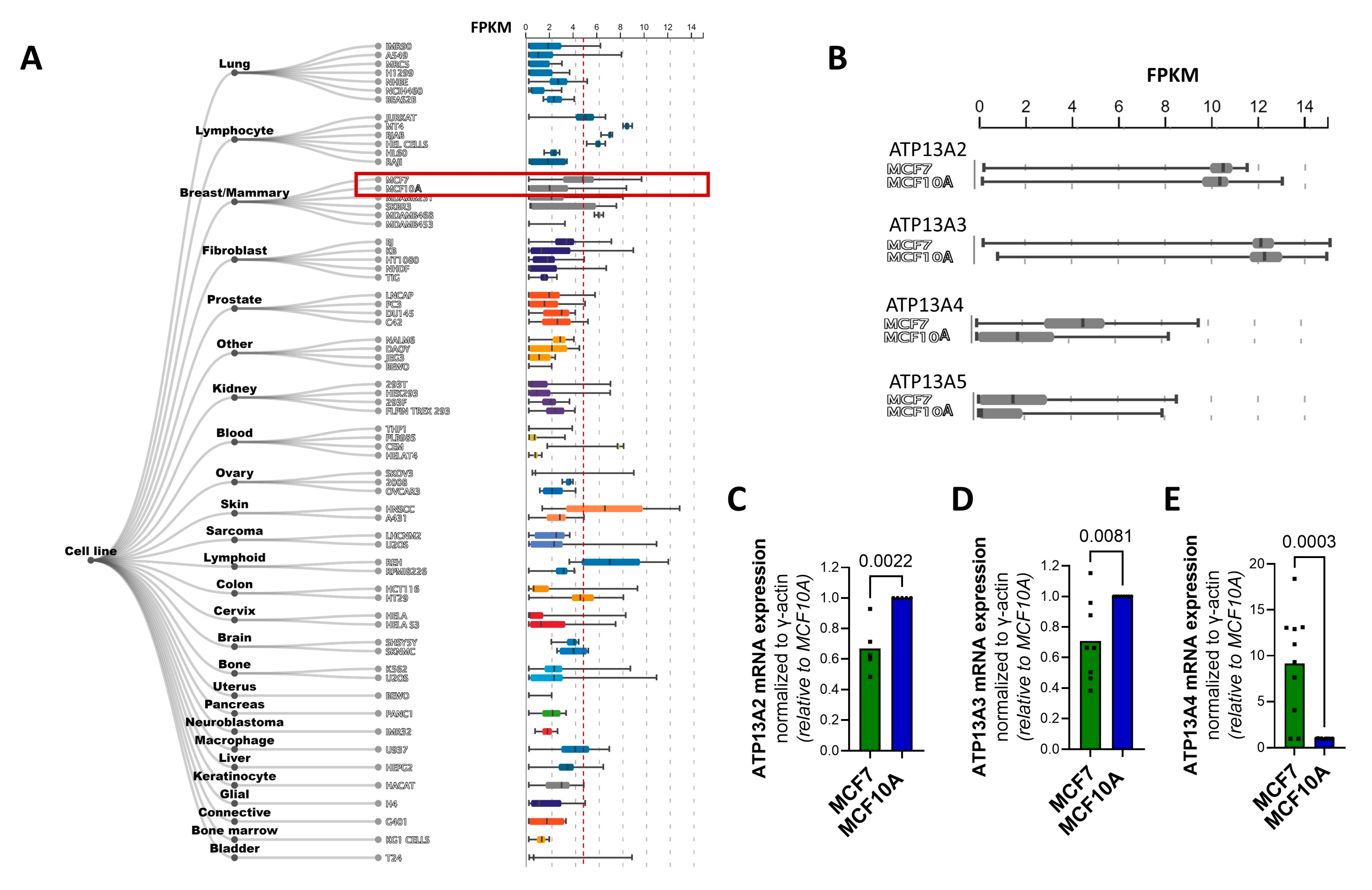
3.2. The P5B ATPase ATP13A4 Is Upregulated in MCF7 versus MCF10A Cells
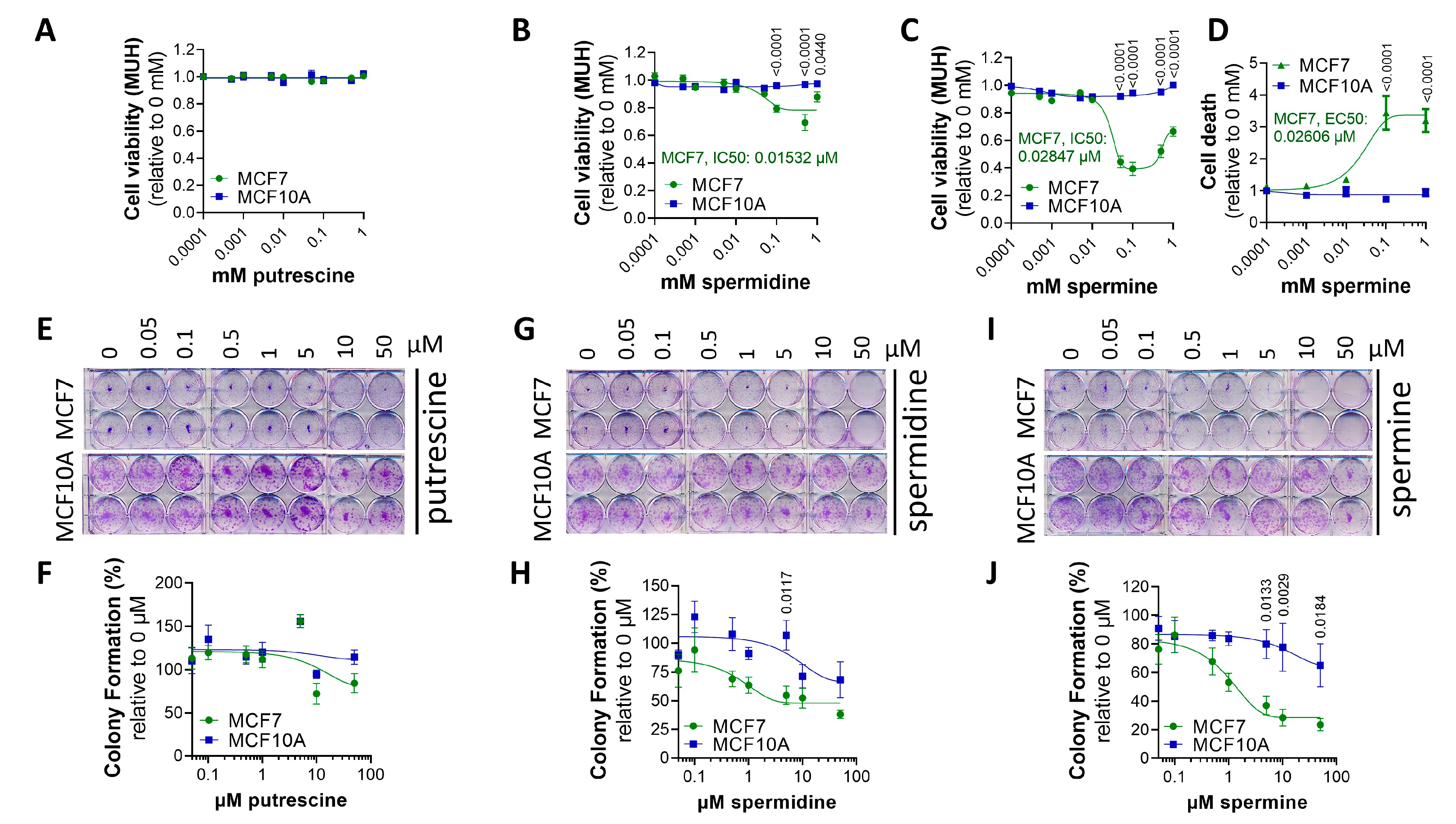
3.3. MCF7 Cells Are More Sensitive to Polyamine Cytotoxicity
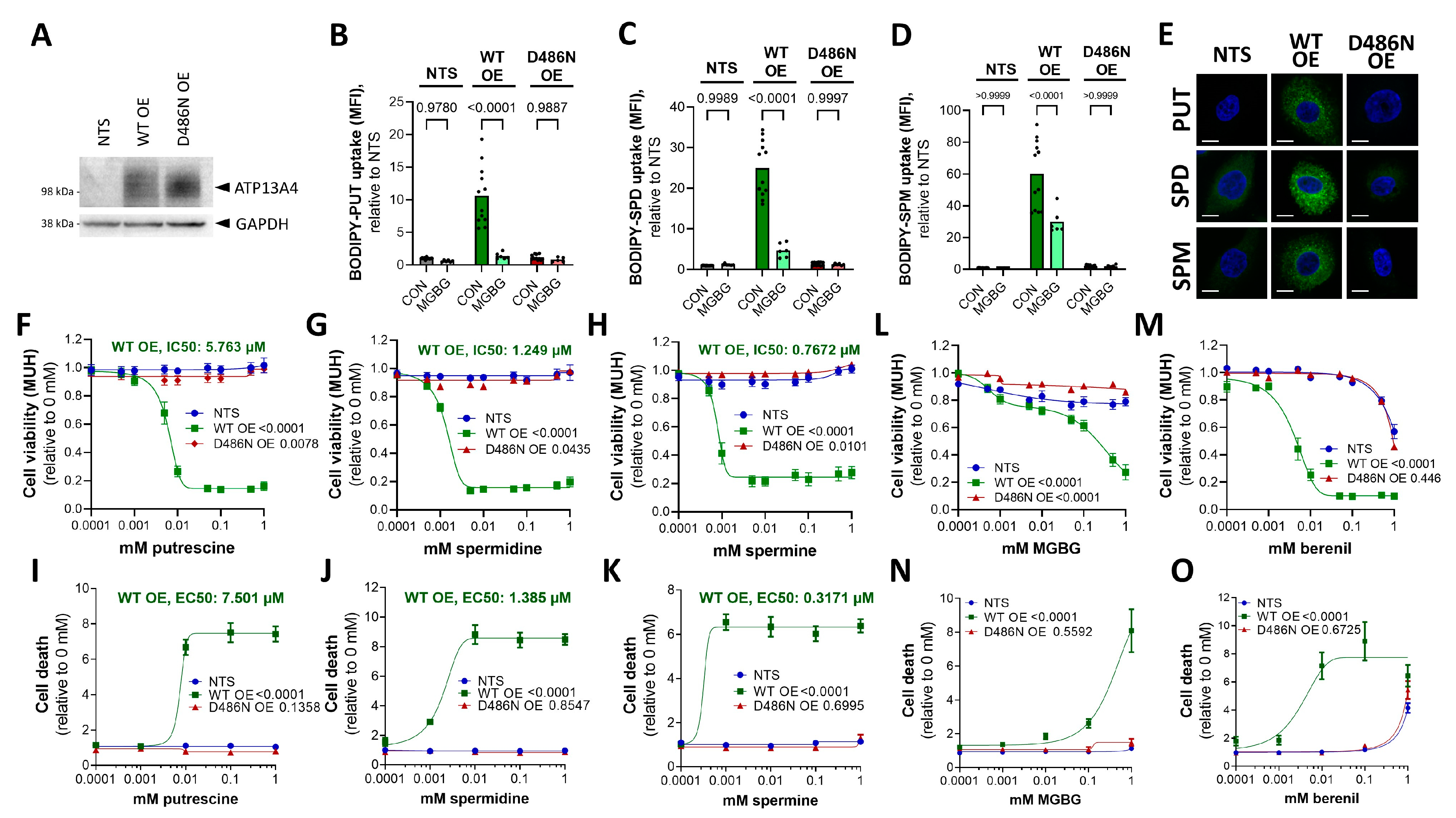
3.4. Overexpression of ATP13A4 in MCF10A Cells Induces an MCF7 Polyamine Phenotype
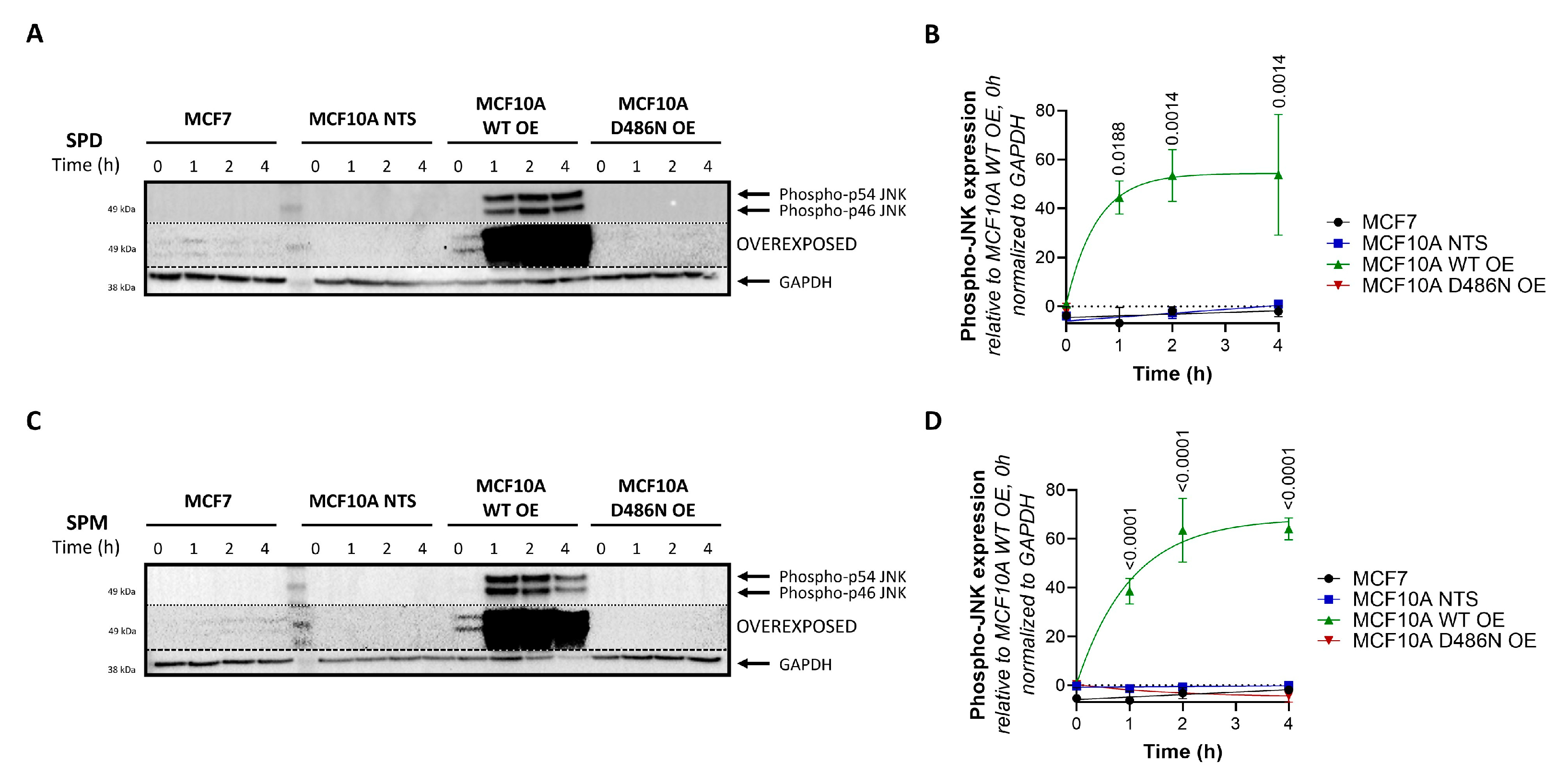
3.5. ATP13A4 Overexpression Triggers Activation of the JNK Signaling Pathway in MCF10A Cells
4. Discussion
4.1. ATP13A4 Emerges as a Novel Polyamine Transporter in the Mammalian PTS
4.2. Is ATP13A4 Implicated in the Upregulated PTS of Other Cancer Types?
4.3. Interplay of ATP13A4, Polyamine Toxicity and JNK Signaling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Azfar, M.; van Veen, S.; Houdou, M.; Hamouda, N.N.; Eggermont, J.; Vangheluwe, P. P5B-ATPases in the mammalian polyamine transport system and their role in disease. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119354. [Google Scholar] [CrossRef] [PubMed]
- van Veen, S.; Martin, S.; Van den Haute, C.; Benoy, V.; Lyons, J.; Vanhoutte, R.; Kahler, J.P.; Decuypere, J.P.; Gelders, G.; Lambie, E.; et al. ATP13A2 deficiency disrupts lysosomal polyamine export. Nature 2020, 578, 419–424. [Google Scholar] [CrossRef]
- Hamouda, N.N.; Van den Haute, C.; Vanhoutte, R.; Sannerud, R.; Azfar, M.; Mayer, R.; Cortés Calabuig, Á.; Swinnen, J.V.; Agostinis, P.; Baekelandt, V.; et al. ATP13A3 is a major component of the enigmatic mammalian polyamine transport system. J. Biol. Chem. 2021, 296, 100182. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Meng, Y.; Wu, X.; Sun, Y. Polyamines and related signaling pathways in cancer. Cancer Cell Int. 2020, 20, 539. [Google Scholar] [CrossRef]
- Leveque, J.; Foucher, F.; Bansard, J.Y.; Havouis, R.; Grall, J.Y.; Moulinoux, J.P. Polyamine profiles in tumor, normal tissue of the homologous breast, blood, and urine of breast cancer sufferers. Breast Cancer Res. Treat. 2000, 60, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Novita Sari, I.; Setiawan, T.; Seock Kim, K.; Toni Wijaya, Y.; Won Cho, K.; Young Kwon, H. Metabolism and function of polyamines in cancer progression. Cancer Lett. 2021, 519, 91–104. [Google Scholar] [CrossRef]
- Corral, M.; Wallace, H.M. Upregulation of Polyamine Transport in Human Colorectal Cancer Cells. Biomolecules 2020, 10, 499. [Google Scholar] [CrossRef]
- Palmer, A.J.; Wallace, H.M. The polyamine transport system as a target for anticancer drug development. Amino Acids 2010, 38, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Wada, M.; Canals, D.; Adada, M.; Coant, N.; Salama, M.F.; Helke, K.L.; Arthur, J.S.; Shroyer, K.R.; Kitatani, K.; Obeid, L.M.; et al. P38 delta MAPK promotes breast cancer progression and lung metastasis by enhancing cell proliferation and cell detachment. Oncogene 2017, 36, 6649–6657. [Google Scholar] [CrossRef]
- Whyte, J.; Bergin, O.; Bianchi, A.; McNally, S.; Martin, F. Key signalling nodes in mammary gland development and cancer. Mitogen-activated protein kinase signalling in experimental models of breast cancer progression and in mammary gland development. Breast Cancer Res. 2009, 11, 209. [Google Scholar] [CrossRef]
- Akinyele, O.; Wallace, H.M. Understanding the Polyamine and mTOR Pathway Interaction in Breast Cancer Cell Growth. Med. Sci. 2022, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.J.; Thomas, T. Cellular and Animal Model Studies on the Growth Inhibitory Effects of Polyamine Analogues on Breast Cancer. Med. Sci. 2018, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Fahrmann, J.F.; Vykoukal, J.; Fleury, A.; Tripathi, S.; Dennison, J.B.; Murage, E.; Wang, P.; Yu, C.Y.; Capello, M.; Creighton, C.J.; et al. Association Between Plasma Diacetylspermine and Tumor Spermine Synthase With Outcome in Triple-Negative Breast Cancer. J. Natl. Cancer Inst. 2020, 112, 607–616. [Google Scholar] [CrossRef]
- Rajeeve, V.; Pearce, W.; Cascante, M.; Vanhaesebroeck, B.; Cutillas, P.R. Polyamine production is downstream and upstream of oncogenic PI3K signalling and contributes to tumour cell growth. Biochem. J. 2013, 450, 619–628. [Google Scholar] [CrossRef]
- Khan, A.; Gamble, L.D.; Upton, D.H.; Ung, C.; Yu, D.M.T.; Ehteda, A.; Pandher, R.; Mayoh, C.; Hebert, S.; Jabado, N.; et al. Dual targeting of polyamine synthesis and uptake in diffuse intrinsic pontine gliomas. Nat. Commun. 2021, 12, 971. [Google Scholar] [CrossRef]
- Soule, H.D.; Maloney, T.M.; Wolman, S.R.; Peterson, W.D., Jr.; Brenz, R.; McGrath, C.M.; Russo, J.; Pauley, R.J.; Jones, R.F.; Brooks, S.C. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990, 50, 6075–6086. [Google Scholar] [PubMed]
- Soule, H.D.; Vazguez, J.; Long, A.; Albert, S.; Brennan, M. A human cell line from a pleural effusion derived from a breast carcinoma. Natl. Cancer Inst. 1973, 51, 1409–1416. [Google Scholar] [CrossRef]
- Comsa, S.; Cimpean, A.M.; Raica, M. The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research. Anticancer Res. 2015, 35, 3147–3154. [Google Scholar]
- Houdou, M.; Jacobs, N.; Coene, J.; Azfar, M.; Vanhoutte, R.; Van den Haute, C.; Eggermont, J.; Daniels, V.; Verhelst, S.H.L.; Vangheluwe, P. Novel Green Fluorescent Polyamines to Analyze ATP13A2 and ATP13A3 Activity in the Mammalian Polyamine Transport System. Biomolecules 2023, 13, 337. [Google Scholar] [CrossRef]
- Jianmongkol, S.; Vuletich, J.L.; Bender, A.T.; Demady, D.R.; Osawa, Y. Aminoguanidine-mediated inactivation and alteration of neuronal nitric-oxide synthase. J. Biol. Chem. 2000, 275, 13370–13376. [Google Scholar] [CrossRef]
- Ding, K.; Sandgren, S.; Mani, K.; Belting, M.; Fransson, L.A. Modulations of glypican-1 heparan sulfate structure by inhibition of endogenous polyamine synthesis. Mapping of spermine-binding sites and heparanase, heparin lyase, and nitric oxide/nitrite cleavage sites. J. Biol. Chem. 2001, 276, 46779–46791. [Google Scholar] [CrossRef] [PubMed]
- Belting, M.; Mani, K.; Jonsson, M.; Cheng, F.; Sandgren, S.; Jonsson, S.; Ding, K.; Delcros, J.G.; Fransson, L.A. Glypican-1 is a vehicle for polyamine uptake in mammalian cells: A pivital role for nitrosothiol-derived nitric oxide. J. Biol. Chem. 2003, 278, 47181–47189. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Osipyan, A.; Song, S.; Chen, D.; Schut, R.A.; van Merkerk, R.; van der Wouden, P.E.; Cool, R.H.; Quax, W.J.; Melgert, B.N.; et al. Thieno[2,3-d]pyrimidine-2,4(1H,3H)-dione Derivative Inhibits d-Dopachrome Tautomerase Activity and Suppresses the Proliferation of Non-Small Cell Lung Cancer Cells. J. Med. Chem. 2022, 65, 2059–2077. [Google Scholar] [CrossRef] [PubMed]
- Vanhoutte, R.; Kahler, J.P.; Martin, S.; van Veen, S.; Verhelst, S.H.L. Clickable Polyamine Derivatives as Chemical Probes for the Polyamine Transport System. Chembiochem 2018, 19, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Konig, S.G.; Oz, S.; Kramer, R. A polyamine-modified near-infrared fluorescent probe for selective staining of live cancer cells. Chem. Commun. 2015, 51, 7360–7363. [Google Scholar] [CrossRef]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef]
- Blaschko, H.; Friedman, P.J.; Hawes, R.; Nilsson, K. The amine oxidases of mammalian plasma. J. Physiol. 1959, 145, 384–404. [Google Scholar] [CrossRef]
- Kunimoto, S.; Nosaka, C.; Xu, C.Z.; Takeuchi, T. Serum effect on cellular uptake of spermidine, spergualin, 15-deoxyspergualin, and their metabolites by L5178Y cells. J. Antibiot. 1989, 42, 116–122. [Google Scholar] [CrossRef]
- Li, H.; Prever, L.; Hirsch, E.; Gulluni, F. Targeting PI3K/AKT/mTOR Signaling Pathway in Breast Cancer. Cancers 2021, 13, 3517. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Ray, R.M.; Viar, M.J.; Johnson, L.R. Polyamines are required for activation of c-Jun NH2-terminal kinase and apoptosis in response to TNF-alpha in IEC-6 cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 285, G980–G991. [Google Scholar] [CrossRef]
- Sorensen, D.M.; Holemans, T.; van Veen, S.; Martin, S.; Arslan, T.; Haagendahl, I.W.; Holen, H.W.; Hamouda, N.N.; Eggermont, J.; Palmgren, M.; et al. Parkinson disease related ATP13A2 evolved early in animal evolution. PLoS ONE 2018, 13, e0193228. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, M.; Zhang, S.; Yin, J.; Zhang, P.; Xuan, X.; Wang, P.; Liu, Z.; Zhou, B.; Yang, M. Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2. Cell Discov. 2021, 7, 106. [Google Scholar] [CrossRef]
- Sim, S.I.; von Bulow, S.; Hummer, G.; Park, E. Structural basis of polyamine transport by human ATP13A2 (PARK9). Mol. Cell 2021, 81, 4635–4649.e8. [Google Scholar] [CrossRef]
- Tillinghast, J.; Drury, S.; Bowser, D.; Benn, A.; Lee, K.P.K. Structural mechanisms for gating and ion selectivity of the human polyamine transporter ATP13A2. Mol. Cell 2021, 81, 4650–4662.e4. [Google Scholar] [CrossRef]
- Tomita, A.; Daiho, T.; Kusakizako, T.; Yamashita, K.; Ogasawara, S.; Murata, T.; Nishizawa, T.; Nureki, O. Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2. Mol. Cell 2021, 81, 4799–4809.e5. [Google Scholar] [CrossRef]
- Li, P.; Wang, K.; Salustros, N.; Gronberg, C.; Gourdon, P. Structure and transport mechanism of P5B-ATPases. Nat. Commun. 2021, 12, 3973. [Google Scholar] [CrossRef]
- Zheng, K.; Li, T. Prediction of ATPase cation transporting 13A2 molecule in Petromyzon marinus and pan-cancer analysis into human tumors from an evolutionary perspective. Immunogenetics 2021, 73, 277–289. [Google Scholar] [CrossRef]
- Chen, Q.; Zhong, L.; Zhou, C.; Feng, Y.; Liu, Q.X.; Zhou, D.; Lu, X.; Du, G.S.; Jian, D.; Luo, H.; et al. Knockdown of Parkinson’s disease-related gene ATP13A2 reduces tumorigenesis via blocking autophagic flux in colon cancer. Cell Biosci. 2020, 10, 144. [Google Scholar] [CrossRef]
- Huang, J.; Xu, S.; Yu, Z.; Zheng, Y.; Yang, B.; Ou, Q. ATP13A2 is a Prognostic Biomarker and Correlates with Immune Infiltrates in Hepatocellular Carcinoma. J. Gastrointest. Surg. 2023, 27, 56–66. [Google Scholar] [CrossRef]
- Li, G.; Gao, Y.; Li, K.; Lin, A.; Jiang, Z. Genomic analysis of biomarkers related to the prognosis of acute myeloid leukemia. Oncol. Lett. 2020, 20, 1824–1834. [Google Scholar] [CrossRef]
- Liu, Q.X.; Zheng, H.; Deng, X.F.; Zhou, D.; Dai, J.G. Status of the Parkinson’s disease gene family expression in non-small-cell lung cancer. World J. Surg. Oncol. 2015, 13, 238. [Google Scholar] [CrossRef]
- Li, S.; Xu, S.; Chen, Y.; Zhou, J.; Ben, S.; Guo, M.; Chu, H.; Gu, D.; Zhang, Z.; Wang, M. Metal Exposure Promotes Colorectal Tumorigenesis via the Aberrant N(6)-Methyladenosine Modification of ATP13A3. Environ. Sci. Technol. 2023, 57, 2864–2876. [Google Scholar] [CrossRef]
- Madan, M.; Patel, A.; Skruber, K.; Geerts, D.; Altomare, D.A.; Iv, O.P. ATP13A3 and caveolin-1 as potential biomarkers for difluoromethylornithine-based therapies in pancreatic cancers. Am. J. Cancer Res. 2016, 6, 1231–1252. [Google Scholar]
- Sekhar, V.; Andl, T.; Phanstiel, O.T. ATP13A3 facilitates polyamine transport in human pancreatic cancer cells. Sci. Rep. 2022, 12, 4045. [Google Scholar] [CrossRef]
- Davis, S.J.; Sheppard, K.E.; Anglesio, M.S.; George, J.; Traficante, N.; Fereday, S.; Intermaggio, M.P.; Menon, U.; Gentry-Maharaj, A.; Lubinski, J.; et al. Enhanced GAB2 Expression Is Associated with Improved Survival in High-Grade Serous Ovarian Cancer and Sensitivity to PI3K Inhibition. Mol. Cancer Ther. 2015, 14, 1495–1503. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Long, Y.; Li, Y.; Huang, M. Coexisting of COX7A2L-ALK, LINC01210-ALK, ATP13A4-ALK and Acquired SLCO2A1-ALK in a Lung Adenocarcinoma with Rearrangements Loss During the Treatment of Crizotinib and Ceritinib: A Case Report. OncoTargets Ther. 2020, 13, 8313–8316. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Chen, Y.; Zhuang, H.; Chen, X.; Shi, Z.; Wang, X. Spermidine-induced growth inhibition and apoptosis via autophagic activation in cervical cancer. Oncol. Rep. 2018, 39, 2845–2854. [Google Scholar] [CrossRef]
- Poulin, R.; Pelletier, G.; Pegg, A.E. Induction of apoptosis by excessive polyamine accumulation in ornithine decarboxylase-overproducing L1210 cells. Biochem. J. 1995, 311 Pt 3, 723–727. [Google Scholar] [CrossRef]
- Sakamoto, A.; Sahara, J.; Kawai, G.; Yamamoto, K.; Ishihama, A.; Uemura, T.; Igarashi, K.; Kashiwagi, K.; Terui, Y. Cytotoxic Mechanism of Excess Polyamines Functions through Translational Repression of Specific Proteins Encoded by Polyamine Modulon. Int. J. Mol. Sci. 2020, 21, 2406. [Google Scholar] [CrossRef]
- Murray Stewart, T.; Dunston, T.T.; Woster, P.M.; Casero, R.A., Jr. Polyamine catabolism and oxidative damage. J. Biol. Chem. 2018, 293, 18736–18745. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wu, W.; Jacevic, V.; Franca, T.C.C.; Wang, X.; Kuca, K. Selective inhibitors for JNK signalling: A potential targeted therapy in cancer. J. Enzyme Inhib. Med. Chem. 2020, 35, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Girnius, N.; Edwards, Y.J.; Garlick, D.S.; Davis, R.J. The cJUN NH(2)-terminal kinase (JNK) signaling pathway promotes genome stability and prevents tumor initiation. eLife 2018, 7, e36389. [Google Scholar] [CrossRef]
- Itah, Z.; Chaudhry, S.; Raju Ponny, S.; Aydemir, O.; Lee, A.; Cavanagh-Kyros, J.; Tournier, C.; Muller, W.J.; Davis, R.J. HER2-driven breast cancer suppression by the JNK signaling pathway. Proc. Natl. Acad. Sci. USA 2023, 120, e2218373120. [Google Scholar] [CrossRef]
- Insua-Rodriguez, J.; Pein, M.; Hongu, T.; Meier, J.; Descot, A.; Lowy, C.M.; De Braekeleer, E.; Sinn, H.P.; Spaich, S.; Sutterlin, M.; et al. Stress signaling in breast cancer cells induces matrix components that promote chemoresistant metastasis. EMBO Mol. Med. 2018, 10, e9003. [Google Scholar] [CrossRef]
- Pein, M.; Insua-Rodriguez, J.; Hongu, T.; Riedel, A.; Meier, J.; Wiedmann, L.; Decker, K.; Essers, M.A.G.; Sinn, H.P.; Spaich, S.; et al. Metastasis-initiating cells induce and exploit a fibroblast niche to fuel malignant colonization of the lungs. Nat. Commun. 2020, 11, 1494. [Google Scholar] [CrossRef]
- Semba, T.; Wang, X.; Xie, X.; Cohen, E.N.; Reuben, J.M.; Dalby, K.N.; Long, J.P.; Phi, L.T.H.; Tripathy, D.; Ueno, N.T. Identification of the JNK-Active Triple-Negative Breast Cancer Cluster Associated With an Immunosuppressive Tumor Microenvironment. J. Natl. Cancer Inst. 2022, 114, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Tabor, C.W.; Rosenthal, S.M. Pharmacology of spermine and spermidine; some effects on animals and bacteria. J. Pharmacol. Exp. Ther. 1956, 116, 139–155. [Google Scholar] [PubMed]

| Gene | NCBI Accession Number | Primer Sequences |
|---|---|---|
| γ-actin | NM_001199954.3 | F: CACTGAGCGAGGCTACAGCTT R: TTGATGTCGCGCACGATTT |
| ATP13A2 | XM_005245810 | F: CATGGCTCTGTACAGCCTGA R: CTCATGAGCACTGCCACTGT |
| ATP13A3 | XM_047448904 | F: TACTGTGGAGCACTGATG R: GAGTTGCCACCATGTCATGC |
| ATP13A4 | XM_047449063 | F: CCAGCACGCTCTGCTCAATG R: GAAGATGGATCCGGCAAGGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Veen, S.; Kourti, A.; Ausloos, E.; Van Asselberghs, J.; Van den Haute, C.; Baekelandt, V.; Eggermont, J.; Vangheluwe, P. ATP13A4 Upregulation Drives the Elevated Polyamine Transport System in the Breast Cancer Cell Line MCF7. Biomolecules 2023, 13, 918. https://doi.org/10.3390/biom13060918
van Veen S, Kourti A, Ausloos E, Van Asselberghs J, Van den Haute C, Baekelandt V, Eggermont J, Vangheluwe P. ATP13A4 Upregulation Drives the Elevated Polyamine Transport System in the Breast Cancer Cell Line MCF7. Biomolecules. 2023; 13(6):918. https://doi.org/10.3390/biom13060918
Chicago/Turabian Stylevan Veen, Sarah, Antria Kourti, Elke Ausloos, Joris Van Asselberghs, Chris Van den Haute, Veerle Baekelandt, Jan Eggermont, and Peter Vangheluwe. 2023. "ATP13A4 Upregulation Drives the Elevated Polyamine Transport System in the Breast Cancer Cell Line MCF7" Biomolecules 13, no. 6: 918. https://doi.org/10.3390/biom13060918






