Central Nervous System Targeted Protein Degraders
Abstract
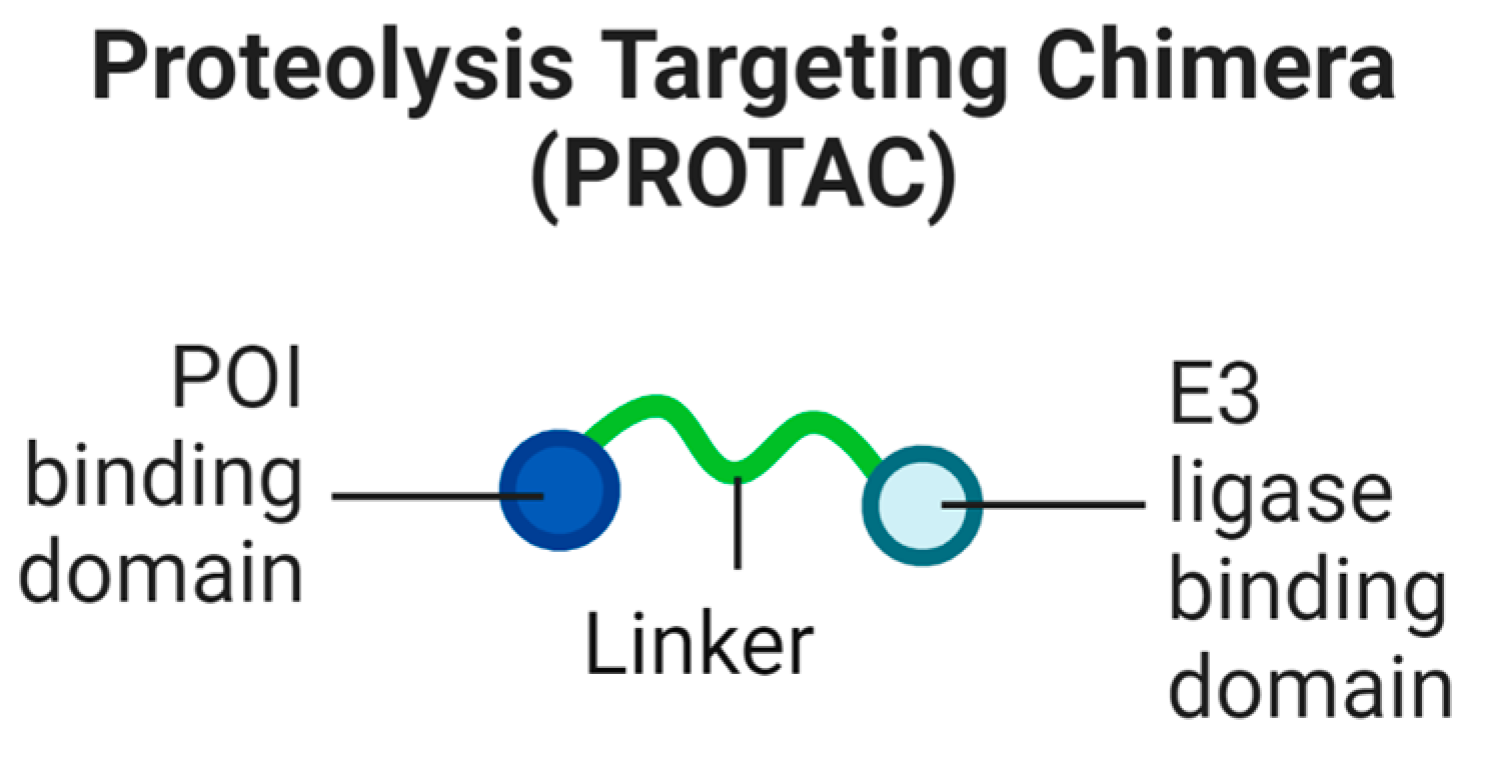
1. Proteolysis-Targeting Chimeras (PROTACs)
1.1. Necessary Steps for Targeted Protein Degradation
- Cellular uptake of the PROTAC to the appropriate intracellular compartment containing the ubiquitination machinery and the POI;
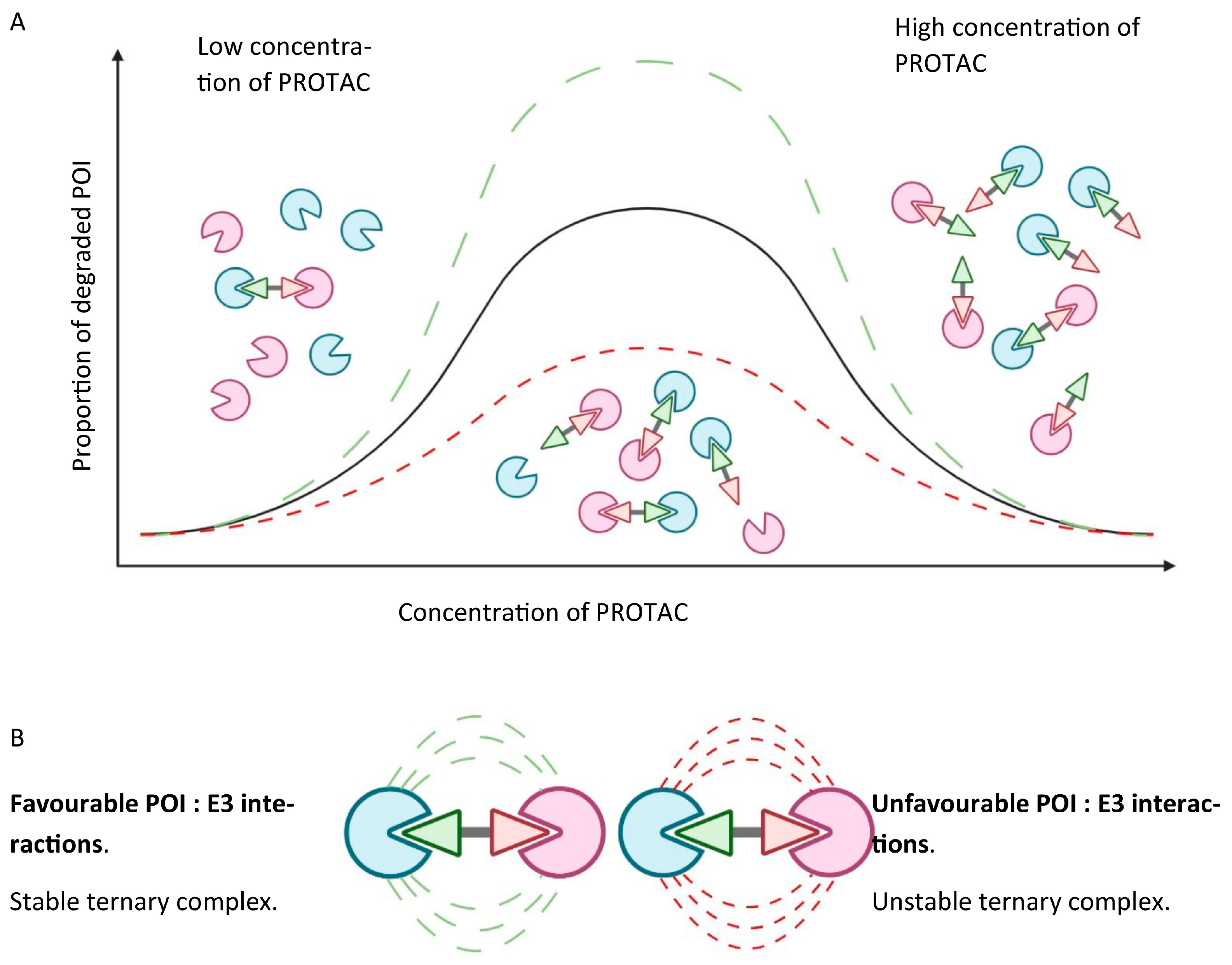
- Ternary complex formation to enable ubiquitin transfer once the PROTAC is inside the cell. This relies on simultaneously binding to the POI and E3 ligase, which in turn is reliant on the binding affinity of the PROTAC to both proteins. Albeit well described, the thermodynamics of ternary complex formation are less intuitively simple compared to a two-body system. Specifically, a “hook effect”, characterised by a bell-shaped dose–response curve, which occurs when the PROTAC forms 1:1 ligand-bound complexes with either the POI or the E3 ligase, leads to ineffectual complex formation at higher concentrations of the PROTAC, and may give rise to possible issues around in vivo dose selection. Secondary protein–protein interactions (PPIs) may also favour or hinder ternary complex formation through cooperativity or steric clashes, respectively. Figure 3 graphically depicts the concentration-dependent “hook effect”;
- Once formed, the ternary complex must accommodate the two bound proteins to occupy a favourable conformation, such that ubiquitin transfer may take place to a suitable acceptor site, commonly a surface lysine. Ubiquitin transfer must occur quickly, at a rate faster than dissociation of the ternary complex;
- Targeted induced polyubiquitination should also kinetically outcompete deubiquitinases, which belong to a large family encompassing wide-ranging substrate specificities;
- Furthermore, the motif of the transferred ubiquitin residues should facilitate facile recognition through the proteasome to bring about actual degradation;
- Even if all the previous steps are successful, and the POI is degraded by the proteasome, this does not guarantee a decreased steady state level of protein. The de novo resynthesis rate of the POI, which may vary significantly between cell types, must be markedly slower than the rate of induced degradation. Likewise, the initial reduction of equilibrium protein levels may not persist over time if the loss of mature protein triggers the induction of feedback mechanisms that upregulate either the translation or transcription of the new protein.
The Hook Effect
1.2. Blood–Brain Barrier Permeability
1.3. Solubility of PROTACs
1.4. Choice of E3 Ligase
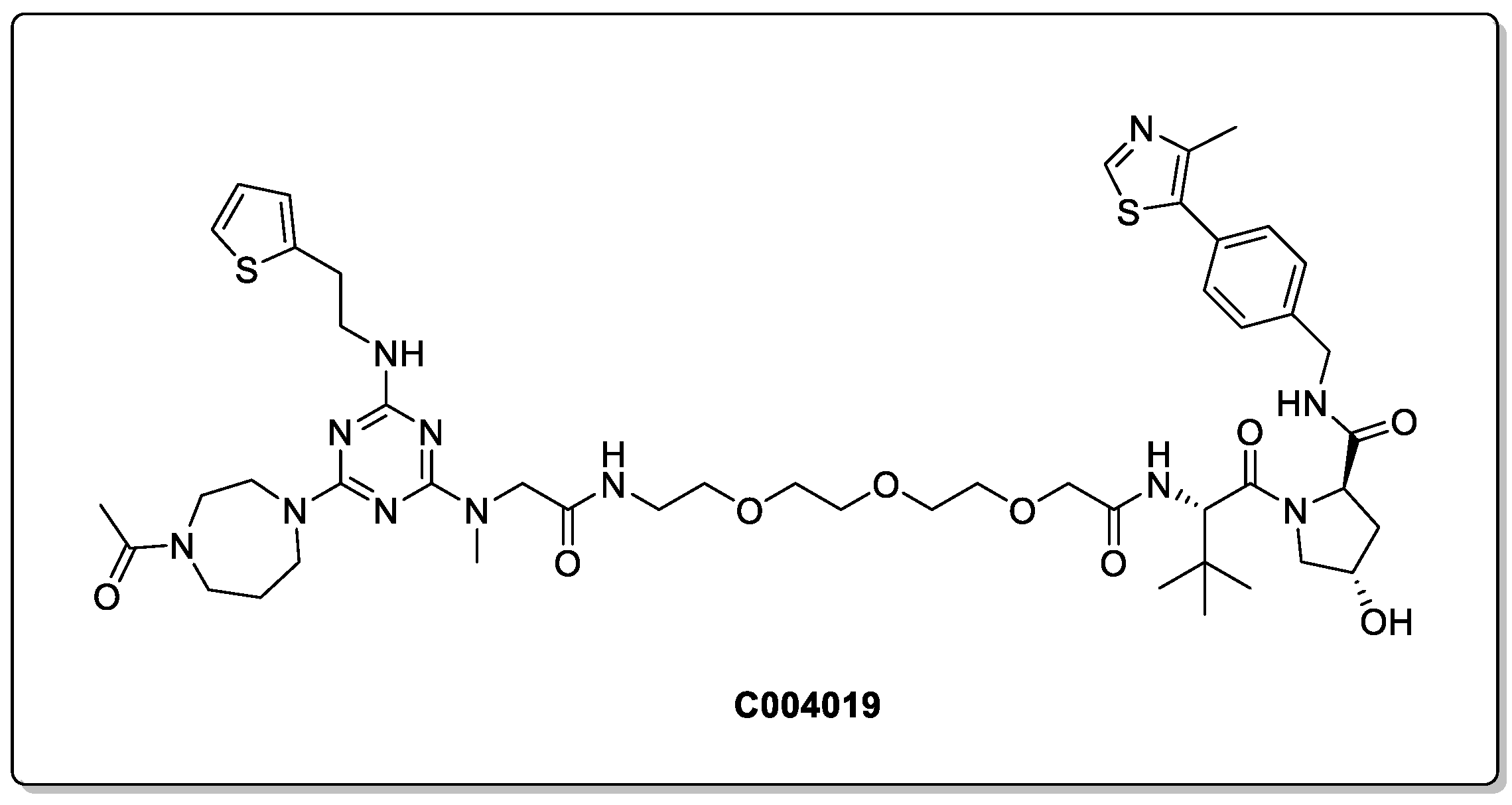
1.5. Role of the Linker
2. Treating Neurodegenerative Diseases Using PROTACs
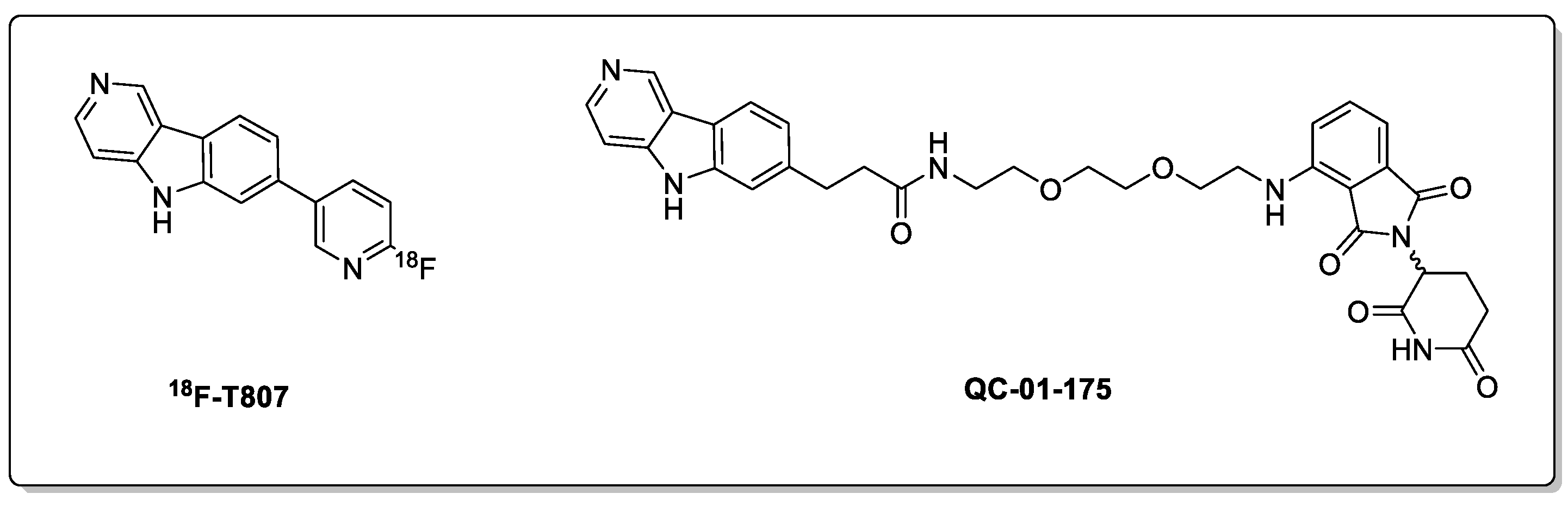
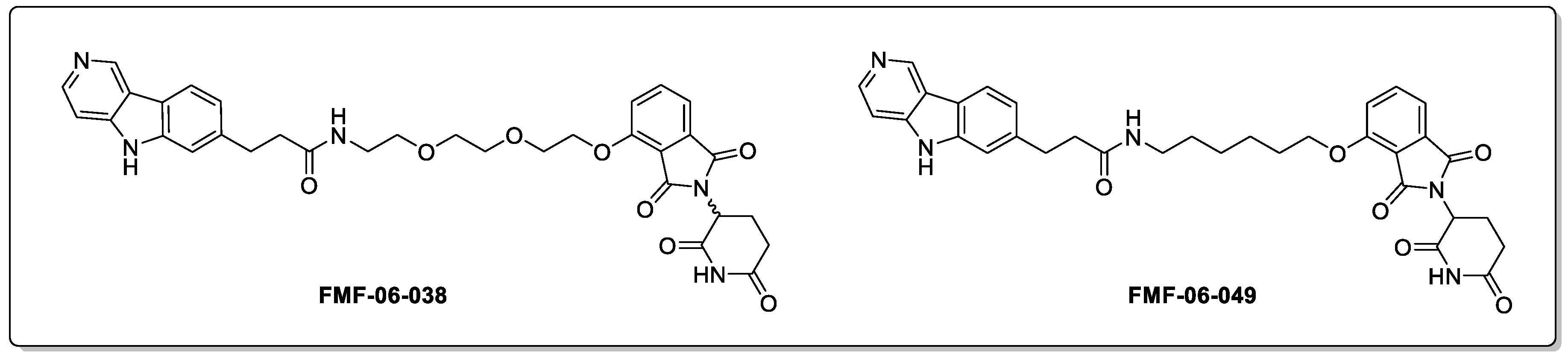
2.1. Tau and Alzheimer’s Disease (AD)
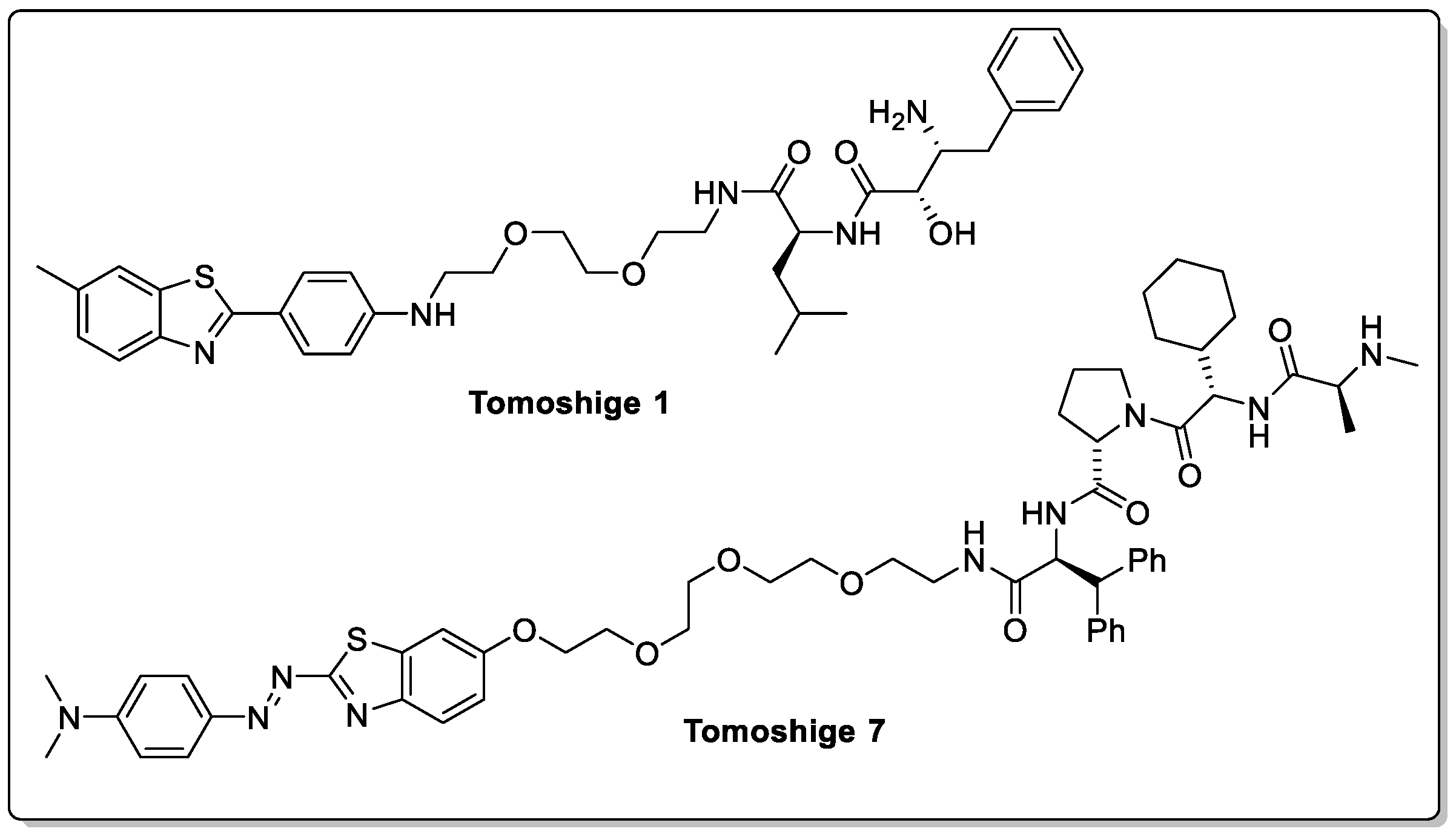
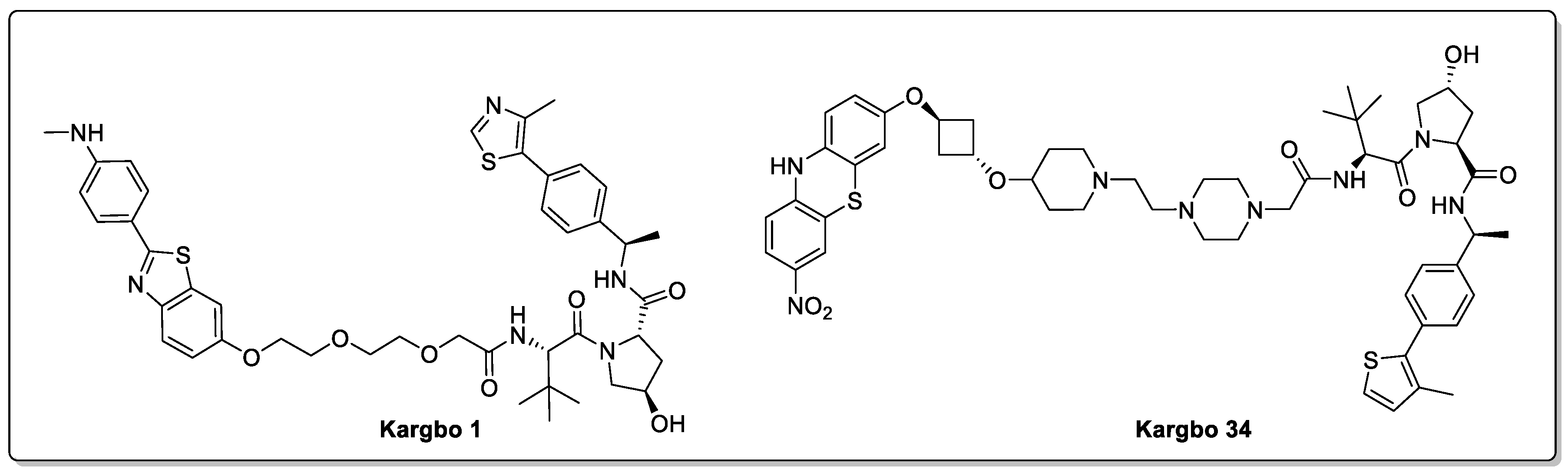
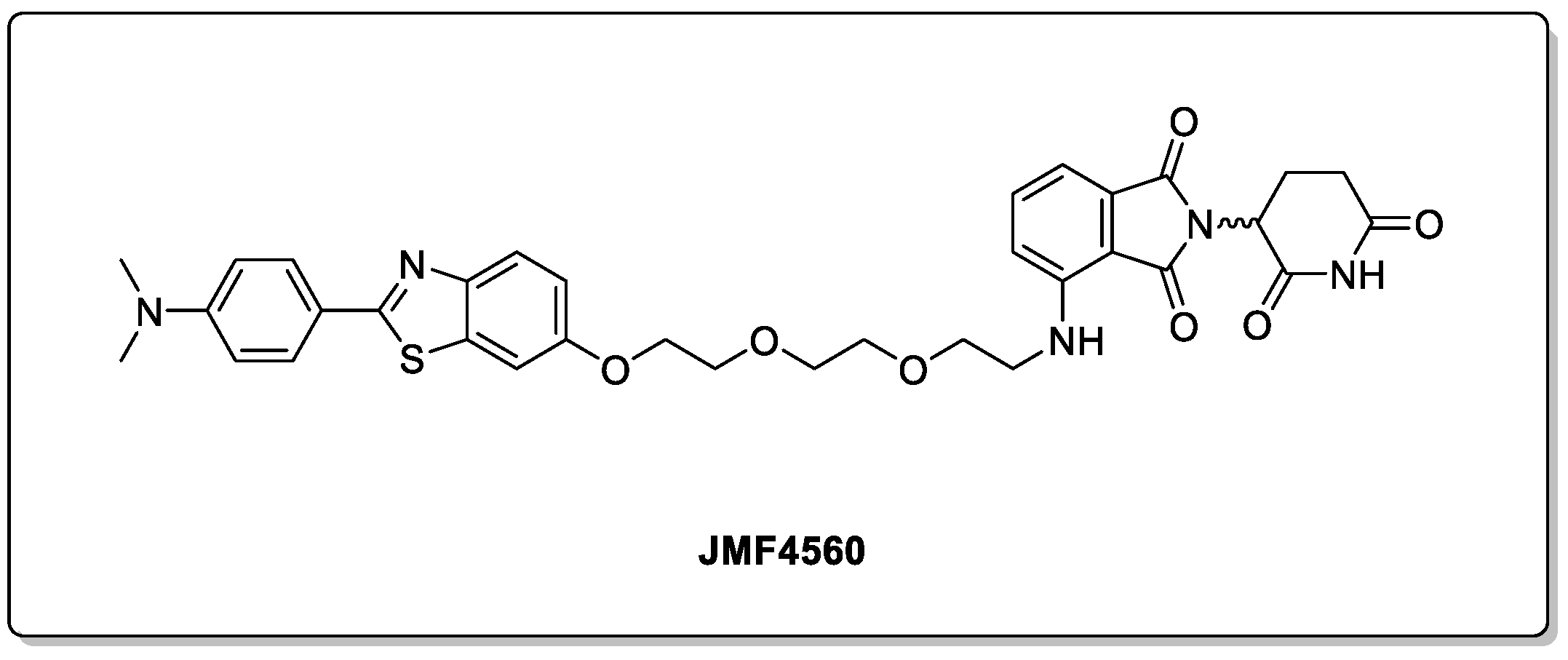
2.2. Huntingtin and HD
2.3. LRRK2
2.4. Alpha-Synuclein
2.5. C-TDP-43
3. Antibody PROTACs
4. The Potential of New Degrader Technology
5. Hydrophobic Tags
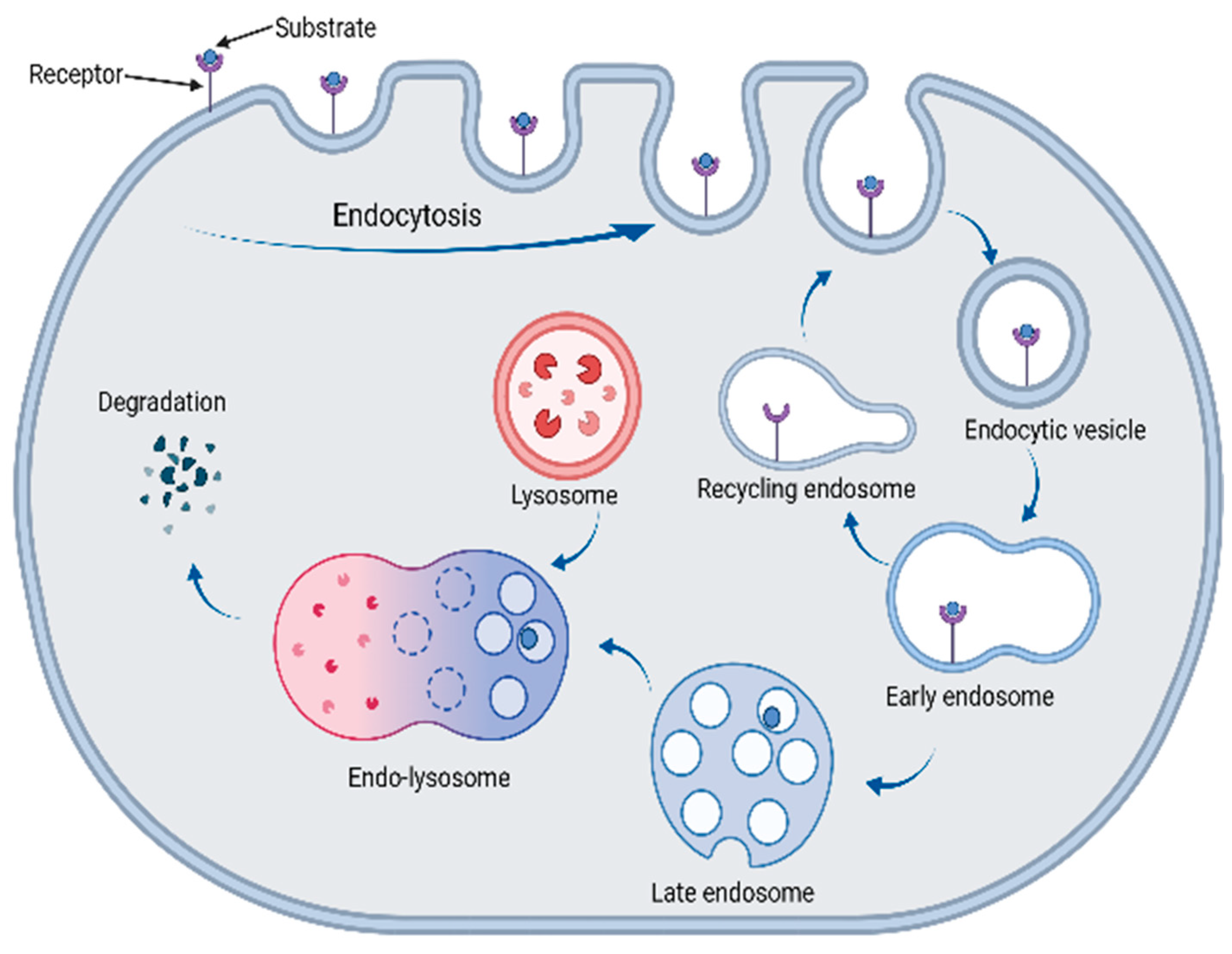
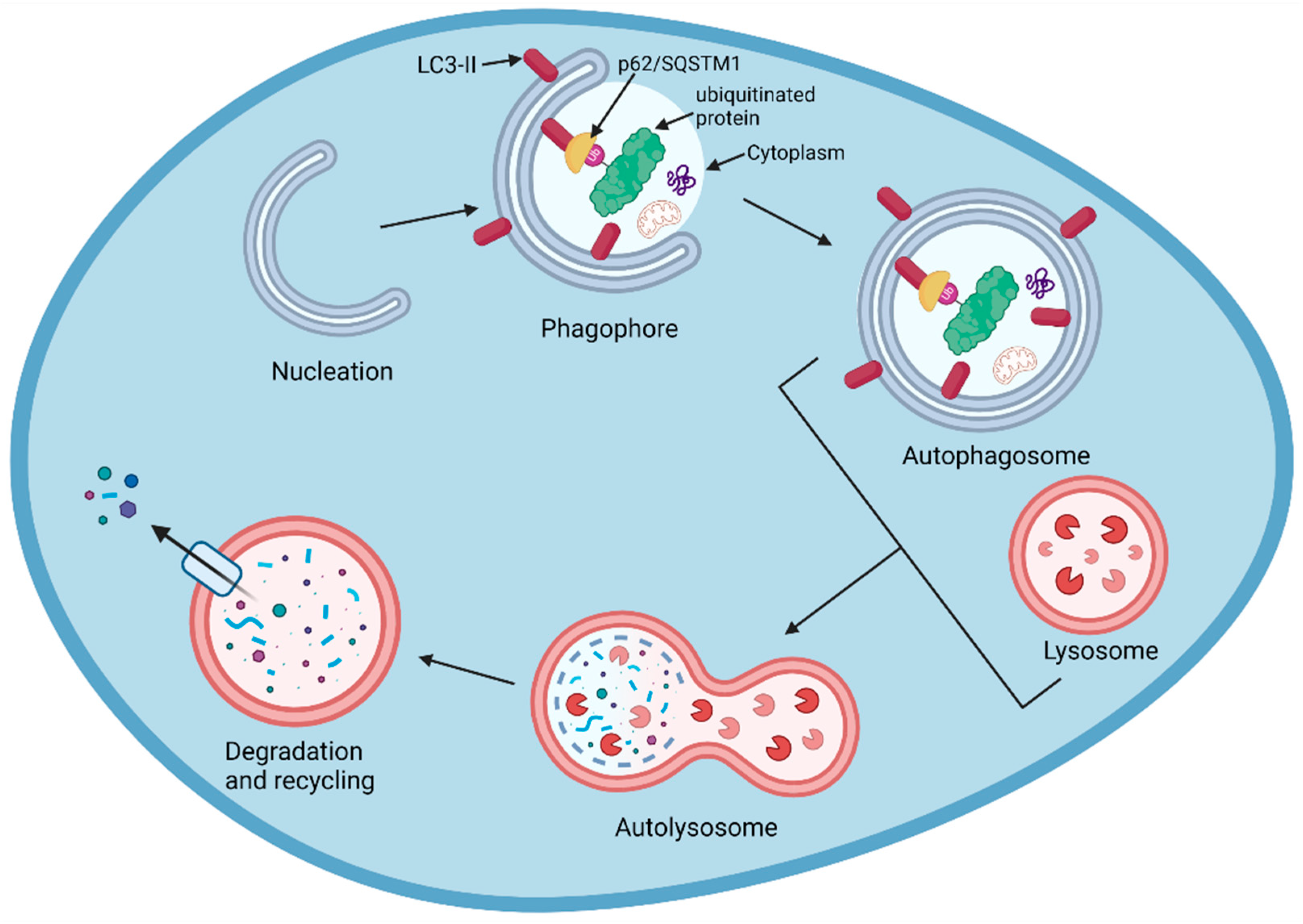
6. The Lysosomal Pathway
7. Lysosomal-Based Degraders
7.1. LYTACs
7.2. AUTACs
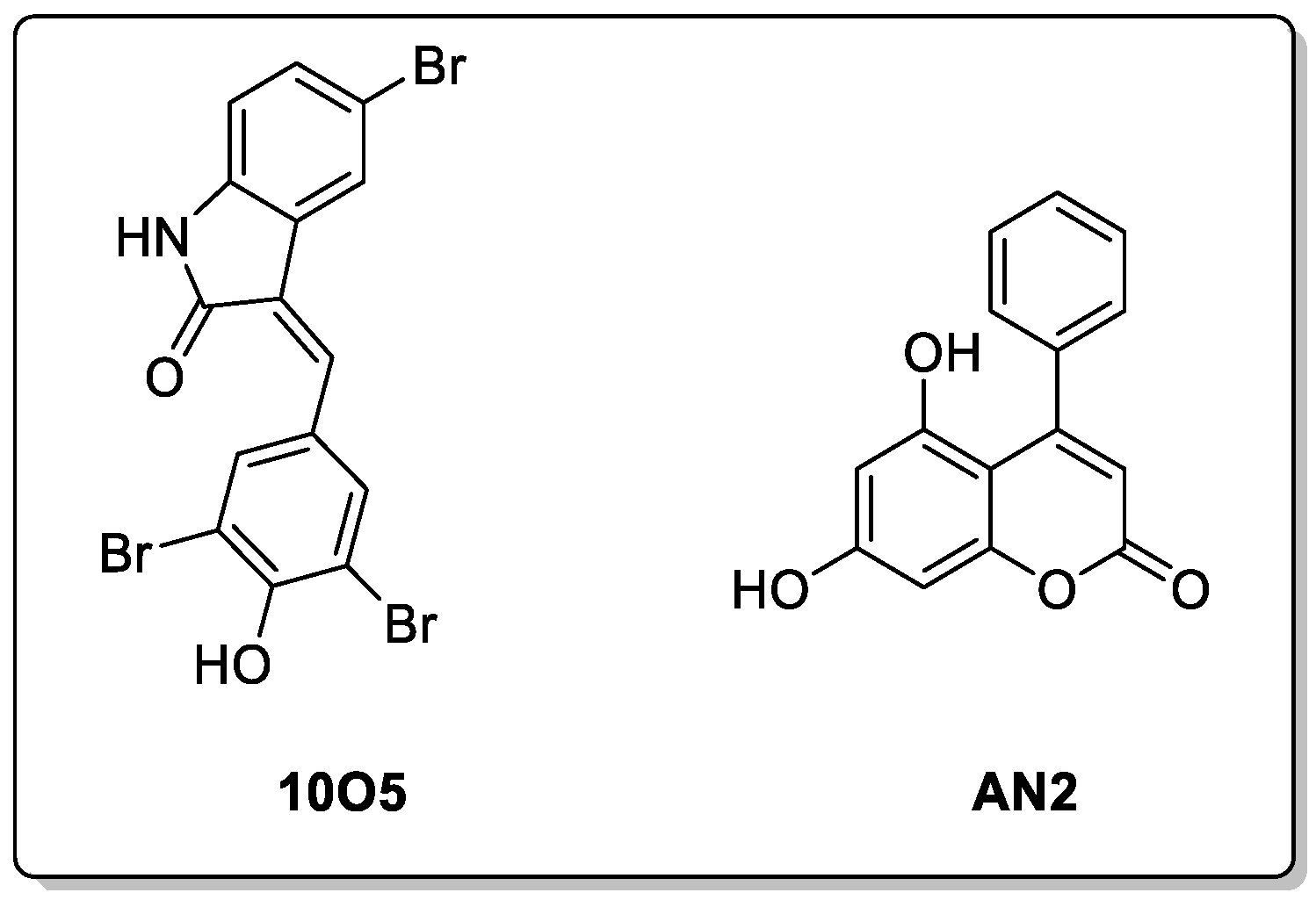
7.3. Molecular Glues and ATTECs
7.4. Chaperone-Mediated Autophagy Degraders
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gribkoff, V.K.; Kaczmarek, L.K. The Need for New Approaches in CNS Drug Discovery: Why Drugs Have Failed, and What Can Be Done to Improve Outcomes. Neuropharmacology 2017, 120, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Roy, S. Gene-Based Therapies for Neurodegenerative Diseases. Nat. Neurosci. 2021, 24, 297–311. [Google Scholar] [CrossRef]
- Perneczky, R.; Jessen, F.; Grimmer, T.; Levin, J.; Flöel, A.; Peters, O.; Froelich, L. Anti-Amyloid Antibody Therapies in Alzheimer’s Disease. Brain 2023, 146, 842–849. [Google Scholar] [CrossRef]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC Targeted Protein Degraders: The Past Is Prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Churcher, I. Protac-Induced Protein Degradation in Drug Discovery: Breaking the Rules or Just Making New Ones? J. Med. Chem. 2018, 61, 444–452. [Google Scholar] [CrossRef]
- Cromm, P.M.; Crews, C.M. Targeted Protein Degradation: From Chemical Biology to Drug Discovery. Cell Chem. Biol. 2017, 24, 1181–1190. [Google Scholar] [CrossRef]
- Raina, K.; Crews, C.M. Targeted Protein Knockdown Using Small Molecule Degraders. Curr. Opin. Chem. Biol. 2017, 39, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.C.; Crews, C.M. Induced Protein Degradation: An Emerging Drug Discovery Paradigm. Nat. Rev. Drug Discov. 2017, 16, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Bondeson, D.P.; Smith, B.E.; Burslem, G.M.; Buhimschi, A.D.; Hines, J.; Jaime-Figueroa, S.; Wang, J.; Hamman, B.D.; Ishchenko, A.; Crews, C.M. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem. Biol. 2018, 25, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Wager, T.T.; Hou, X.; Verhoest, P.R.; Villalobos, A. Central Nervous System Multiparameter Optimization Desirability: Application in Drug Discovery. ACS Chem. Neurosci. 2016, 7, 767–775. [Google Scholar] [CrossRef]
- Tashima, T. Proteolysis-Targeting Chimera (PROTAC) Delivery into the Brain across the Blood-Brain Barrier. Antibodies 2023, 12, 43. [Google Scholar] [CrossRef]
- Kargbo, R.B. Treatment of Alzheimer’s by PROTAC-Tau Protein Degradation. ACS Med. Chem. Lett. 2019, 10, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kalogeropulou, A.F.; Domingos, S.; Makukhin, N.; Nirujogi, R.S.; Singh, F.; Shpiro, N.; Saalfrank, A.; Sammler, E.; Ganley, I.G.; et al. Discovery of XL01126: A Potent, Fast, Cooperative, Selective, Oral Bioavailable and Blood Brain Barrier Penetrant PROTAC Degrader of Leucine Rich Repeat Kinase 2. J. Am. Chem. Soc. 2022, 144, 16930–16952. [Google Scholar] [CrossRef]
- Clark, D.E. Rapid Calculation of Polar Molecular Surface Area and Its Application to the Prediction of Transport Phenomena. 1. Prediction of Intestinal Absorption. J. Pharm. Sci. 1999, 88, 807–814. [Google Scholar] [CrossRef]
- Troy Bemis, M.A.; La Clair, J.J.; Burkart, M.D. Unraveling the Role of Linker Design in Proteolysis Targeting Chimeras. J. Med. Chem. 2021, 64, 8042–8052. [Google Scholar] [CrossRef]
- García Jiménez, D.; Rossi Sebastiano, M.; Vallaro, M.; Mileo, V.; Pizzirani, D.; Moretti, E.; Ermondi, G.; Caron, G. Designing Soluble PROTACs: Strategies and Preliminary Guidelines. J. Med. Chem. 2022, 65, 12639–12649. [Google Scholar] [CrossRef]
- Bulatov, E.; Ciulli, A. Targeting Cullin–RING E3 Ubiquitin Ligases for Drug Discovery: Structure, Assembly and Small-Molecule Modulation. Biochem. J. 2015, 467, 365–386. [Google Scholar] [CrossRef]
- Collins, I.; Wang, H.; Caldwell, J.J.; Chopra, R. Chemical Approaches to Targeted Protein Degradation through Modulation of the Ubiquitin–Proteasome Pathway. Biochem. J. 2017, 474, 1127–1147. [Google Scholar] [CrossRef] [PubMed]
- Steinebach, C.; Kehm, H.; Lindner, S.; Vu, L.P.; Köpff, S.; López Mármol, Á.; Weiler, C.; Wagner, K.G.; Reichenzeller, M.; Krönke, J.; et al. PROTAC-Mediated Crosstalk between E3 Ligases. Chem. Commun. 2019, 55, 1821–1824. [Google Scholar] [CrossRef]
- Maneiro, M.; de Vita, E.; Conole, D.; Kounde, C.S.; Zhang, Q.; Tate, E.W. PROTACs, Molecular Glues and Bifunctionals from Bench to Bedside: Unlocking the Clinical Potential of Catalytic Drugs. In Progress in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2021; pp. 67–190. ISBN 9780323850568. [Google Scholar]
- Lescouzères, L.; Bomont, P. E3 Ubiquitin Ligases in Neurological Diseases: Focus on Gigaxonin and Autophagy. Front. Physiol. 2020, 11, 1022. [Google Scholar] [CrossRef]
- Jevtić, P.; Haakonsen, D.L.; Rapé, M. An E3 Ligase Guide to the Galaxy of Small-Molecule-Induced Protein Degradation. Cell Chem. Biol. 2021, 28, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Farrell, K.; Jarome, T.J. Is PROTAC Technology Really a Game Changer for Central Nervous System Drug Discovery? Expert Opin. Drug Discov. 2021, 16, 833–840. [Google Scholar] [CrossRef]
- Schapira, M.; Calabrese, M.F.; Bullock, A.N.; Crews, C.M. Targeted Protein Degradation: Expanding the Toolbox. Nat. Rev. Drug Discov. 2019, 18, 949–963. [Google Scholar] [CrossRef]
- Liu, L.; Damerell, D.R.; Koukouflis, L.; Tong, Y.; Marsden, B.D.; Schapira, M. UbiHub: A Data Hub for the Explorers of Ubiquitination Pathways. Bioinformatics 2019, 35, 2882–2884. [Google Scholar] [CrossRef]
- Paiva, S.-L.; Crews, C.M. Targeted Protein Degradation: Elements of PROTAC Design. Curr. Opin. Chem. Biol. 2019, 50, 111–119. [Google Scholar] [CrossRef]
- Maple, H.J.; Clayden, N.; Baron, A.; Stacey, C.; Felix, R. Developing Degraders: Principles and Perspectives on Design and Chemical Space. Medchemcomm 2019, 10, 1755–1764. [Google Scholar] [CrossRef]
- Nowak, R.P.; Deangelo, S.L.; Buckley, D.; He, Z.; Donovan, K.A.; An, J.; Safaee, N.; Jedrychowski, M.P.; Ponthier, C.M.; Ishoey, M.; et al. Plasticity in Binding Confers Selectivity in Ligand-Induced Protein Degradation. Nat. Chem. Biol. 2018, 14, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric Molecules That Target Proteins to the Skp1-Cullin-F Box Complex for Ubiquitination and Degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef] [PubMed]
- Klein, V.G.; Bond, A.G.; Craigon, C.; Lokey, R.S.; Ciulli, A. Amide-to-Ester Substitution as a Strategy for Optimizing PROTAC Permeability and Cellular Activity. J. Med. Chem. 2021, 64, 18082–18101. [Google Scholar] [CrossRef]
- Klein, V.G.; Townsend, C.E.; Testa, A.; Zengerle, M.; Maniaci, C.; Hughes, S.J.; Chan, K.-H.; Ciulli, A.; Lokey, R.S. Understanding and Improving the Membrane Permeability of VH032-Based PROTACs. ACS Med. Chem. Lett. 2020, 11, 1732–1738. [Google Scholar] [CrossRef]
- Yang, J.; Chang, Y.; Tien, C.-Y.J.; Wang, Z.; Zhou, Y.; Zhang, P.; Huang, W.; Vo, J.; Apel, I.J.; Wang, C.; et al. Discovery of a Highly Potent and Selective Dual PROTAC Degrader of CDK12 and CDK13. J. Med. Chem. 2022, 65, 11066–11083. [Google Scholar] [CrossRef]
- Hendrick, C.E.; Jorgensen, J.R.; Chaudhry, C.; Strambeanu, I.I.; Brazeau, J.-F.; Schiffer, J.; Shi, Z.; Venable, J.D.; Wolkenberg, S.E. Direct-to-Biology Accelerates PROTAC Synthesis and the Evaluation of Linker Effects on Permeability and Degradation. ACS Med. Chem. Lett. 2022, 13, 1182–1190. [Google Scholar] [CrossRef] [PubMed]
- Trial of ARV-110 in Patients with Metastatic Castration Resistant Prostate Cancer—Full Text View—ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/ct2/show/NCT03888612 (accessed on 9 February 2023).
- A Phase 1/2 Trial of ARV-471 Alone and in Combination with Palbociclib (IBRANCE®) in Patients With ER+/HER2- Locally Advanced or Metastatic Breast Cancer—Full Text View—ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/ct2/show/NCT04072952 (accessed on 9 February 2023).
- Sun, X.; Gao, H.; Yang, Y.; He, M.; Wu, Y.; Song, Y.; Tong, Y.; Rao, Y. PROTACs: Great Opportunities for Academia and Industry. Signal Transduct. Target Ther. 2019, 4, 64. [Google Scholar] [CrossRef]
- Chu, T.T.; Gao, N.; Li, Q.Q.; Chen, P.G.; Yang, X.F.; Chen, Y.X.; Zhao, Y.F.; Li, Y.M. Specific Knockdown of Endogenous Tau Protein by Peptide-Directed Ubiquitin-Proteasome Degradation. Cell Chem. Biol. 2016, 23, 453–461. [Google Scholar] [CrossRef]
- Jansson, B. Models for the Transfer of Drugs from the Nasal Cavity to the Central Nervous System; Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Lu, M.; Liu, T.; Jiao, Q.; Ji, J.; Tao, M.; Liu, Y.; You, Q.; Jiang, Z. Discovery of a Keap1-Dependent Peptide PROTAC to Knockdown Tau by Ubiquitination-Proteasome Degradation Pathway. Eur. J. Med. Chem. 2018, 146, 251–259. [Google Scholar] [CrossRef]
- Chien, D.T.; Bahri, S.; Szardenings, A.K.; Walsh, J.C.; Mu, F.; Su, M.Y.; Shankle, W.R.; Elizarov, A.; Kolb, H.C. Early Clinical PET Imaging Results with the Novel PHF-Tau Radioligand [F-18]-T807. J. Alzheimer Dis. 2013, 34, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.F.; Arteaga, J.; Chen, G.; Gangadharmath, U.; Gomez, L.F.; Kasi, D.; Lam, C.; Liang, Q.; Liu, C.; Mocharla, V.P.; et al. [18F]T807, a Novel Tau Positron Emission Tomography Imaging Agent for Alzheimer’s Disease. Alzheimer Dement. 2013, 9, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Ferguson, F.M.; Cai, Q.; Donovan, K.A.; Nandi, G.; Patnaik, D.; Zhang, T.; Huang, H.T.; Lucente, D.E.; Dickerson, B.C.; et al. Targeted Degradation of Aberrant Tau in Frontotemporal Dementia Patient-Derived Neuronal Cell Models. Elife 2019, 8, e45457. [Google Scholar] [CrossRef]
- Silva, M.C.; Nandi, G.; Donovan, K.A.; Cai, Q.; Berry, B.C.; Nowak, R.P.; Fischer, E.S.; Gray, N.S.; Ferguson, F.M.; Haggarty, S.J. Discovery and Optimization of Tau Targeted Protein Degraders Enabled by Patient Induced Pluripotent Stem Cells-Derived Neuronal Models of Tauopathy. Front. Cell. Neurosci. 2022, 16, 51. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, Q.; Jiang, T.; Li, S.; Ye, J.; Zheng, J.; Wang, X.; Liu, Y.; Deng, M.; Ke, D.; et al. A Novel Small-Molecule PROTAC Selectively Promotes Tau Clearance to Improve Cognitive Functions in Alzheimer-like Models. Theranostics 2021, 11, 5279–5295. [Google Scholar] [CrossRef]
- Ferguson, F.M.; Gray, N.S.; Haggarty, S.J.; Telo Baptista Lima Da Silva, M.C. Targeted Degraders of Aberrant Tau Based on the PET Tracer PBB3. US Patent US20230133538A1, 4 May 2023. [Google Scholar]
- Gray, N.S.; Haggarty, S.J.; Cai, Q.; Telo Baptista Lima Da Silva, M.C.; Zhang, T.; Ferguson, F.M. Compounds for Tau Protein Degradation. European Patent WO2019014429A1, 17 January 2019. [Google Scholar]
- Arvinas Arvinas to Present Preclinical Tau-Directed PROTAC® Protein Degrader Data at Alzheimer’s Association International Conference—Arvinas. Available online: www.arvinas.com (accessed on 21 April 2022).
- Tomoshige, S.; Nomura, S.; Ohgane, K.; Hashimoto, Y.; Ishikawa, M. Discovery of Small Molecules That Induce the Degradation of Huntingtin. Angew. Chem. Int. Ed. 2017, 56, 11530–11533. [Google Scholar] [CrossRef] [PubMed]
- Tomoshige, S.; Ishikawa, M. PROTACs and Other Chemical Protein Degradation Technologies for the Treatment of Neurodegenerative Disorders. Angew. Chem. Int. Ed. 2021, 60, 3346–3354. [Google Scholar] [CrossRef]
- Tomoshige, S.; Nomura, S.; Ohgane, K.; Hashimoto, Y.; Ishikawa, M. Degradation of Huntingtin Mediated by a Hybrid Molecule Composed of IAP Antagonist Linked to Phenyldiazenyl Benzothiazole Derivative. Bioorg. Med. Chem. Lett. 2018, 28, 707–710. [Google Scholar] [CrossRef]
- An, S.; Fu, L. Small-Molecule PROTACs: An Emerging and Promising Approach for the Development of Targeted Therapy Drugs. EBioMedicine 2018, 36, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Toure, M.; Crews, C.M. Small-Molecule PROTACS: New Approaches to Protein Degradation. Angew. Chem. Int. Ed. 2016, 55, 1966–1973. [Google Scholar] [CrossRef]
- Li, J.Q.; Tan, L.; Yu, J.T. The Role of the LRRK2 Gene in Parkinsonism. Mol. Neurodegener. 2014, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- West, A.B.; Moore, D.J.; Choi, C.; Andrabi, S.A.; Li, X.; Dikeman, D.; Biskup, S.; Zhang, Z.; Lim, K.L.; Dawson, V.L.; et al. Parkinson’s Disease-Associated Mutations in LRRK2 Link Enhanced GTP-Binding and Kinase Activities to Neuronal Toxicity. Hum. Mol. Genet. 2007, 16, 223–232. [Google Scholar] [CrossRef]
- Konstantinidou, M.; Oun, A.; Pathak, P.; Zhang, B.; Wang, Z.; ter Brake, F.; Dolga, A.M.; Kortholt, A.; Dömling, A. The Tale of Proteolysis Targeting Chimeras (PROTACs) for Leucine-Rich Repeat Kinase 2 (LRRK2). ChemMedChem 2021, 16, 959–965. [Google Scholar] [CrossRef]
- Araujo, E.; Berlin, M.; Sparks, S.M.; Wang, J.; Zhang, W. Selective Modulators of Mutant LRRK2 Proteolysis and Associated Methods of Use. US Patent US20220064168A1, 3 March 2022. [Google Scholar]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.Y.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. α-Synuclein in Lewy Bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef]
- Qu, J.; Ren, X.; Xue, F.; He, Y.; Zhang, R.; Zheng, Y.; Huang, H.; Wang, W.; Zhang, J. Specific Knockdown of α-Synuclein by Peptide-Directed Proteasome Degradation Rescued Its Associated Neurotoxicity. Cell Chem. Biol. 2020, 27, 751–762. [Google Scholar] [CrossRef]
- Kargbo, R.B. PROTAC Compounds Targeting α-Synuclein Protein for Treating Neurogenerative Disorders: Alzheimer’s and Parkinson’s Diseases. ACS Med. Chem. Lett. 2020, 11, 1086–1087. [Google Scholar] [CrossRef]
- Crew, A.P.; Dong, H.; Berlin, M.; Sparks, S.M. Proteolysis Targeting Chimeric (PROTAC) Compound with E3 Ubiquitin Ligase Binding Activity and Targeting Alpha-Synuclein Protein for Treating Neurodegenerative Diseases. European Patent WO2020041331A1, 27 February 2020. [Google Scholar]
- Jo, M.; Lee, S.; Jeon, Y.M.; Kim, S.; Kwon, Y.; Kim, H.J. The Role of TDP-43 Propagation in Neurodegenerative Diseases: Integrating Insights from Clinical and Experimental Studies. Exp. Mol. Med. 2020, 52, 1652–1662. [Google Scholar] [CrossRef]
- Tseng, Y.-L.; Lu, P.-C.; Lee, C.-C.; He, R.-Y.; Huang, Y.-A.; Tseng, Y.-C.; Cheng, T.-J.R.; Huang, J.J.-T.; Fang, J.-M. Degradation of Neurodegenerative Disease-Associated TDP-43 Aggregates and Oligomers via a Proteolysis-Targeting Chimera. J. Biomed. Sci. 2023, 30, 27. [Google Scholar] [CrossRef] [PubMed]
- Guenette, R.G.; Yang, S.W.; Min, J.; Pei, B.; Potts, P.R. Target and Tissue Selectivity of PROTAC Degraders. Chem. Soc. Rev. 2022, 51, 5740–5756. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, C.; Holubowska, A.; Schwedhelm-Domeyer, N.; Mitkovski, M.; Lee, S.J.; Kannan, M.; Matz, A.; Vadhvani, M.; Stegmüller, J. Loss of the Neuron-Specific F-Box Protein FBXO41 Models an Ataxia-Like Phenotype in Mice with Neuronal Migration Defects and Degeneration in the Cerebellum. J. Neurosci. 2015, 35, 8701. [Google Scholar] [CrossRef]
- Balastik, M.; Ferraguti, F.; Pires-da Silva, A.; Tae, H.L.; Alvarez-Bolado, G.; Kun, P.L.; Gruss, P. Deficiency in Ubiquitin Ligase TRIM2 Causes Accumulation of Neurofilament Light Chain and Neurodegeneration. Proc. Natl. Acad. Sci. USA 2008, 105, 12016–12021. [Google Scholar] [CrossRef] [PubMed]
- Menon, S.; Boyer, N.P.; Winkle, C.C.; McClain, L.M.; Hanlin, C.C.; Pandey, D.; Rothenfußer, S.; Taylor, A.M.; Gupton, S.L. The E3 Ubiquitin Ligase TRIM9 Is a Filopodia Off Switch Required for Netrin-Dependent Axon Guidance. Dev. Cell 2015, 35, 698–712. [Google Scholar] [CrossRef]
- Wang, C.; Zheng, C.; Wang, H.; Zhang, L.; Liu, Z.; Xu, P. The State of the Art of PROTAC Technologies for Drug Discovery. Eur. J. Med. Chem. 2022, 235, 114290. [Google Scholar] [CrossRef]
- Zolot, R.S.; Basu, S.; Million, R.P. Antibody–Drug Conjugates. Nat. Rev. Drug Discov. 2013, 12, 259–260. [Google Scholar] [CrossRef]
- Maneiro, M.; Forte, N.; Shchepinova, M.M.; Kounde, S.C.; Chudasama, V.; Richard Baker, J.; Tate, W.E. Antibody–PROTAC Conjugates Enable HER2-Dependent Targeted Protein Degradation of BRD4. ACS Chem. Biol. 2020, 15, 1306–1312. [Google Scholar] [CrossRef]
- Dragovich, P.S.; Adhikari, P.; Blake, R.A.; Blaquiere, N.; Chen, J.; Cheng, Y.X.; den Besten, W.; Han, J.; Hartman, S.J.; He, J.; et al. Antibody-Mediated Delivery of Chimeric Protein Degraders Which Target Estrogen Receptor Alpha (ERα). Bioorg. Med. Chem. Lett. 2020, 30, 126907. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ji, J.; Tong, Y.; Zhu, Y.; Dou, J.; Zhang, X.; Xu, S.; Zhu, T.; Xu, X.; You, Q.; et al. Non-Small Molecule PROTACs (NSM-PROTACs): Protein Degradation Kaleidoscope. Acta Pharm. Sin. B 2022, 12, 2990–3005. [Google Scholar] [CrossRef] [PubMed]
- Pillow, T.H.; Adhikari, P.; Blake, R.A.; Chen, J.; Del Rosario, G.; Deshmukh, G.; Figueroa, I.; Gascoigne, K.E.; Kamath, A.V.; Kaufman, S.; et al. Antibody Conjugation of a Chimeric BET Degrader Enables In Vivo Activity. ChemMedChem 2020, 15, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Cotton, D.A.; Nguyen, P.D.; Gramespacher, A.J.; Seiple, B.I.; Wells, A.J. Development of Antibody-Based PROTACs for the Degradation of the Cell-Surface Immune Checkpoint Protein PD-L1. J. Am. Chem. Soc. 2021, 143, 593–598. [Google Scholar] [CrossRef]
- Zebisch, M.; Xu, Y.; Krastev, C.; Macdonald, B.T.; Chen, M.; Gilbert, R.J.C.; He, X.; Jones, E.Y. Structural and Molecular Basis of ZNRF3/RNF43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin. Nat. Commun. 2013, 4, 2787. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, N.; An, Z. Engineering Antibody and Protein Therapeutics to Cross the Blood–Brain Barrier. Antib. Ther. 2022, 5, 311–331. [Google Scholar] [CrossRef]
- Ding, Y.; Fei, Y.; Lu, B. Emerging New Concepts of Degrader Technologies. Trends Pharmacol. Sci. 2020, 41, 464–474. [Google Scholar] [CrossRef]
- Alabi, S.B.; Crews, C.M. Major Advances in Targeted Protein Degradation: PROTACs, LYTACs, and MADTACs. J. Biol. Chem. 2021, 296, 100647. [Google Scholar] [CrossRef]
- Zhang, L.; Riley-Gillis, B.; Vijay, P.; Shen, Y. Acquired Resistance to BET-ProTACS (Proteolysis-Targeting Chimeras) Caused by Genomic Alterations in Core Components of E3 Ligase Complexes. Mol. Cancer Ther. 2019, 18, 1302–1311. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, X.; Feng, F.; Liu, W.; Sun, H. Degradation of Proteins by PROTACs and Other Strategies. Acta Pharm. Sin. B 2020, 10, 207–238. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347. [Google Scholar] [CrossRef]
- Odenthal, J.; Takes, R.; Friedl, P. Plasticity of Tumor Cell Invasion: Governance by Growth Factors and Cytokines. Carcinogenesis 2016, 37, 1117–1128. [Google Scholar] [CrossRef]
- Gustafson, J.L.; Neklesa, T.K.; Cox, C.S.; Roth, A.G.; Buckley, D.L.; Tae, H.S.; Sundberg, T.B.; Blake, D.; Ohn Hines, J.; Mcdonnell, D.P.; et al. Hot Paper Small-Molecule-Mediated Degradation of the Androgen Receptor through Hydrophobic Tagging. Angew. Chem. 2015, 54, 9659–9662. [Google Scholar] [CrossRef] [PubMed]
- Hyun, S.; Shin, D.; Park, S.J.; Choi, J.W.; Kim, H. Chemical-Mediated Targeted Protein Degradation in Neurodegenerative Diseases. Life 2021, 11, 607. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Hassan, M.I. Targeted Protein Degraders March towards the Clinic for Neurodegenerative Diseases. Ageing Res. Rev. 2022, 78, 101616. [Google Scholar] [CrossRef] [PubMed]
- Hirai, K.; Yamashita, H.; Tomoshige, S.; Mishima, Y.; Niwa, T.; Ohgane, K.; Ishii, M.; Kanamitsu, K.; Ikemi, Y.; Nakagawa, S.; et al. Conversion of a PROTAC Mutant Huntingtin Degrader into Small-Molecule Hydrophobic Tags Focusing on Drug-like Properties. ACS Med. Chem. Lett. 2022, 2022, 402. [Google Scholar] [CrossRef]
- Gao, N.; Chu, T.-T.; Li, Q.-Q.; Lim, Y.-J.; Qiu, T.; Ma, M.-R.; Hu, Z.-W.; Yang, X.-F.; Chen, Y.-X.; Zhao, Y.-F.; et al. Hydrophobic Tagging-Mediated Degradation of Alzheimer’s Disease Related Tau. RSC Adv. 2017, 7, 40362–40366. [Google Scholar] [CrossRef]
- Gao, N.; Huang, Y.P.; Chu, T.T.; Li, Q.Q.; Zhou, B.; Chen, Y.X.; Zhao, Y.F.; Li, Y.M. TDP-43 Specific Reduction Induced by Di-Hydrophobic Tags Conjugated Peptides. Bioorg. Chem. 2019, 84, 254–259. [Google Scholar] [CrossRef]
- Pei, J.; Wang, G.; Feng, L.; Zhang, J.; Jiang, T.; Sun, Q.; Ouyang, L. Targeting Lysosomal Degradation Pathways: New Strategies and Techniques for Drug Discovery. J. Med. Chem. 2021, 64, 3507. [Google Scholar] [CrossRef]
- Cooper, G.M. Endocytosis. In The Cell: A Molecular Approach; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Hu, Y.-B.; Dammer, E.B.; Ren, R.-J.; Wang, G. The endosomal-lysosomal system: From acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 2015, 4, 18. [Google Scholar] [CrossRef]
- Zaffagnini, G.; Martens, S. Mechanisms of Selective Autophagy. J. Mol. Biol. 2016, 428, 1714–1724. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. P62/SQSTM1 Binds Directly to Atg8/LC3 to Facilitate Degradation of Ubiquitinated Protein Aggregates by Autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The Machinery of Macroautophagy 24 The Machinery of Macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and Molecular Mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the Pathogenesis of Disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Klionsky, D.J. Protein Turnover Via Autophagy: Implications for Metabolism. Annu. Rev. Nutr. 2007, 27, 19–40. [Google Scholar] [CrossRef] [PubMed]
- Banik, S.M.; Pedram, K.; Wisnovsky, S.; Ahn, G.; Riley, N.M.; Bertozzi, C.R. Lysosome-Targeting Chimaeras for Degradation of Extracellular Proteins. Nature 2020, 584, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Teng, P.; Montgomery, N.T.; Li, X.; Tang, W. Development of Triantennary N-Acetylgalactosamine Conjugates as Degraders for Extracellular Proteins. ACS Cent. Sci. 2021, 7, 499–506. [Google Scholar] [CrossRef]
- Zhu, W.; Gao, M.; Chen, B.; Tan, T.; Cao, H.; Liu, L. The Synthesis of Mannose-6-Phosphate Using Polyphosphate-Dependent Mannose Kinase. Catalysts 2019, 9, 250. [Google Scholar] [CrossRef]
- Van Regenmortel, M.H.V. Antigenicity and Immunogenicity of Synthetic Peptides. Biologicals 2001, 29, 209–213. [Google Scholar] [CrossRef]
- Lin, J.; Jin, J.; Shen, Y.; Zhang, L.; Gong, G.; Bian, H.; Chen, H.; Nagle, D.G.; Wu, Y.; Zhang, W.; et al. Emerging Protein Degradation Strategies: Expanding the Scope to Extracellular and Membrane Proteins. Theranostics 2021, 11, 8337–8349. [Google Scholar] [CrossRef]
- Takahashi, D.; Moriyama, J.; Nakamura, T.; Miki, E.; Takahashi, E.; Sato, A.; Akaike, T.; Itto-Nakama, K.; Arimoto, H. AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol. Cell 2019, 76, 797–810. [Google Scholar] [CrossRef]
- Kirkin, V.; McEwan, D.G.; Novak, I.; Dikic, I. A Role for Ubiquitin in Selective Autophagy. Mol. Cell 2009, 34, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Arimoto, H. Targeting Selective Autophagy by AUTAC Degraders. Autophagy 2020, 16, 765–766. [Google Scholar] [CrossRef] [PubMed]
- Hoon Ji, C.; Yeon Kim, H.; Ju Lee, M.; Jung Heo, A.; Youngjae Park, D.; Lim, S.; Shin, S.; Seung Yang, W.; An Jung, C.; Young Kim, K.; et al. The AUTOTAC Chemical Biology Platform for Targeted Protein Degradation via the Autophagy-Lysosome System. Nat. Commun. 2022, 13, 904. [Google Scholar] [CrossRef]
- Li, H.; Dong, J.; Cai, M.; Xu, Z.; Cheng, X.-D.; Qin, J.-J. Protein Degradation Technology: A Strategic Paradigm Shift in Drug Discovery. J. Hematol. Oncol. 2021, 14, 138. [Google Scholar] [CrossRef]
- Dong, G.; Ding, Y.; He, S.; Sheng, C. Molecular Glues for Targeted Protein Degradation: From Serendipity to Rational Discovery. J. Med. Chem. 2021, 64, 10606–10620. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Aguilar, A.; Liu, Z.; Yang, C.-Y.; Wang, S. Simple Structural Modifications Converting a Bona Fide MDM2 PROTAC Degrader into a Molecular Glue Molecule: A Cautionary Tale in the Design of PROTAC Degraders. J. Med. Chem. 2019, 62, 9471–9487. [Google Scholar] [CrossRef] [PubMed]
- Ishoey, M.; Chorn, S.; Singh, N.; Jaeger, M.G.; Brand, M.; Paulk, J.; Bauer, S.; Erb, M.A.; Parapatics, K.; Müller, A.C.; et al. Translation Termination Factor GSPT1 Is a Phenotypically Relevant Off-Target of Heterobifunctional Phthalimide Degraders. ACS Chem. Biol. 2018, 13, 553–560. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, C.; Ding, Y.; Fei, Y.; Lu, B. ATTEC: A Potential New Approach to Target Proteinopathies. Autophagy 2020, 16, 185. [Google Scholar] [CrossRef]
- Li, Z.; Wang, C.; Wang, Z.; Zhu, C.; Li, J.; Sha, T.; Ma, L.; Gao, C.; Yang, Y.; Sun, Y.; et al. Allele-Selective Lowering of Mutant HTT Protein by HTT–LC3 Linker Compounds. Nature 2019, 575, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Mehlem, A.; Hagberg, C.E.; Muhl, L.; Eriksson, U.; Falkevall, A. Imaging of Neutral Lipids by Oil Red O for Analyzing the Metabolic Status in Health and Disease. Nat. Protoc. 2013, 8, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Chen, N.; Wang, Z.; Luo, S.; Ding, Y.; Lu, B. Degradation of Lipid Droplets by Chimeric Autophagy-Tethering Compounds. Cell Res. 2021, 31, 965–979. [Google Scholar] [CrossRef]
- Fu, Y.; Lu, B. Targeting Lipid Droplets for Autophagic Degradation by ATTEC. Autophagy 2021, 17, 4486–4488. [Google Scholar] [CrossRef]
- Kaushik, S.; Maria Cuervo, A. The Coming of Age of Chaperone-Mediated Autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Jin, W.Y.; Lu, J.; Wang, J.; Wang, Y.T. Rapid and Reversible Knockdown of Endogenous Proteins by Peptide-Directed Lysosomal Degradation. Nat. Neurosci. 2014, 17, 471–480. [Google Scholar] [CrossRef]
- Shaltiel-Karyo, R.; Frenkel-Pinter, M.; Egoz-Matia, N.; Frydman-Marom, A.; Shalev, D.E.; Segal, D.; Gazit, E. Inhibiting α-Synuclein Oligomerization by Stable Cell-Penetrating β-Synuclein Fragments Recovers Phenotype of Parkinson’s Disease Model Flies. PLoS ONE 2010, 5, e13863. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, B.a.I.; Lewis, H.L.; Jones, D.H.; Ward, S.E. Central Nervous System Targeted Protein Degraders. Biomolecules 2023, 13, 1164. https://doi.org/10.3390/biom13081164
Thomas BaI, Lewis HL, Jones DH, Ward SE. Central Nervous System Targeted Protein Degraders. Biomolecules. 2023; 13(8):1164. https://doi.org/10.3390/biom13081164
Chicago/Turabian StyleThomas, Bedwyr ab Ion, H. Lois Lewis, D. Heulyn Jones, and Simon E. Ward. 2023. "Central Nervous System Targeted Protein Degraders" Biomolecules 13, no. 8: 1164. https://doi.org/10.3390/biom13081164
APA StyleThomas, B. a. I., Lewis, H. L., Jones, D. H., & Ward, S. E. (2023). Central Nervous System Targeted Protein Degraders. Biomolecules, 13(8), 1164. https://doi.org/10.3390/biom13081164





