Mass Spectrometry Imaging of In Vitro Cryptosporidium parvum-Infected Cells and Host Tissue
Abstract
:1. Introduction
2. Experimental Section
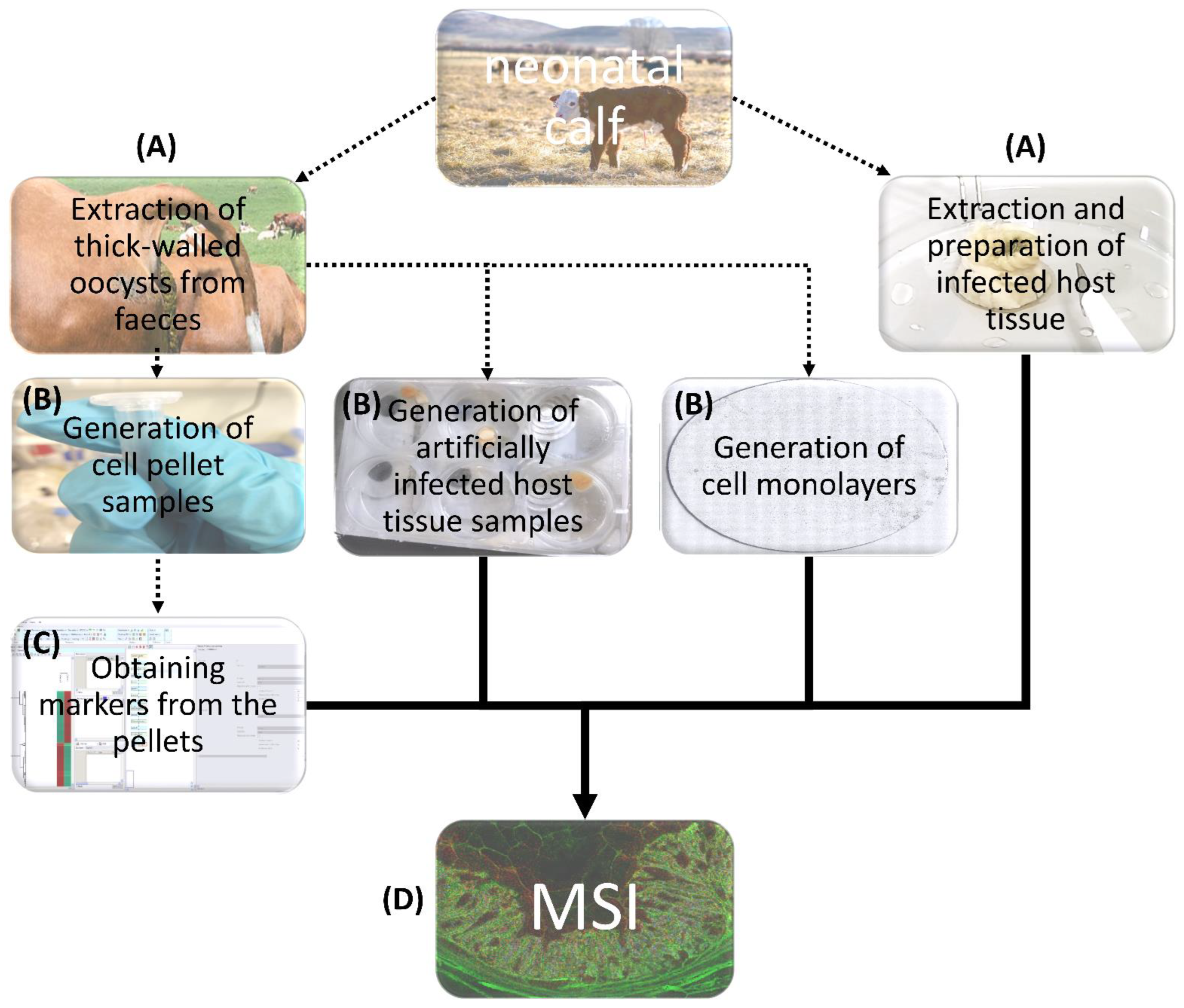
2.1. Parasites
2.2. Cell Culture
2.3. Host Cell Infection
2.4. Preparation of Cell Monolayers and Parasite Pellets
2.5. Preparation of C. parvum-Infected Bovine Intestinal Cryo-Sections
2.6. MALDI-MS Sample Preparation
2.7. Metabolite Extraction
2.8. UHPLC-MS/MS Analysis
2.9. MALDI-MS(I)
2.10. Data Processing
3. Results and Discussion
3.1. MALDI-MSI Measurements of Cell Pellets
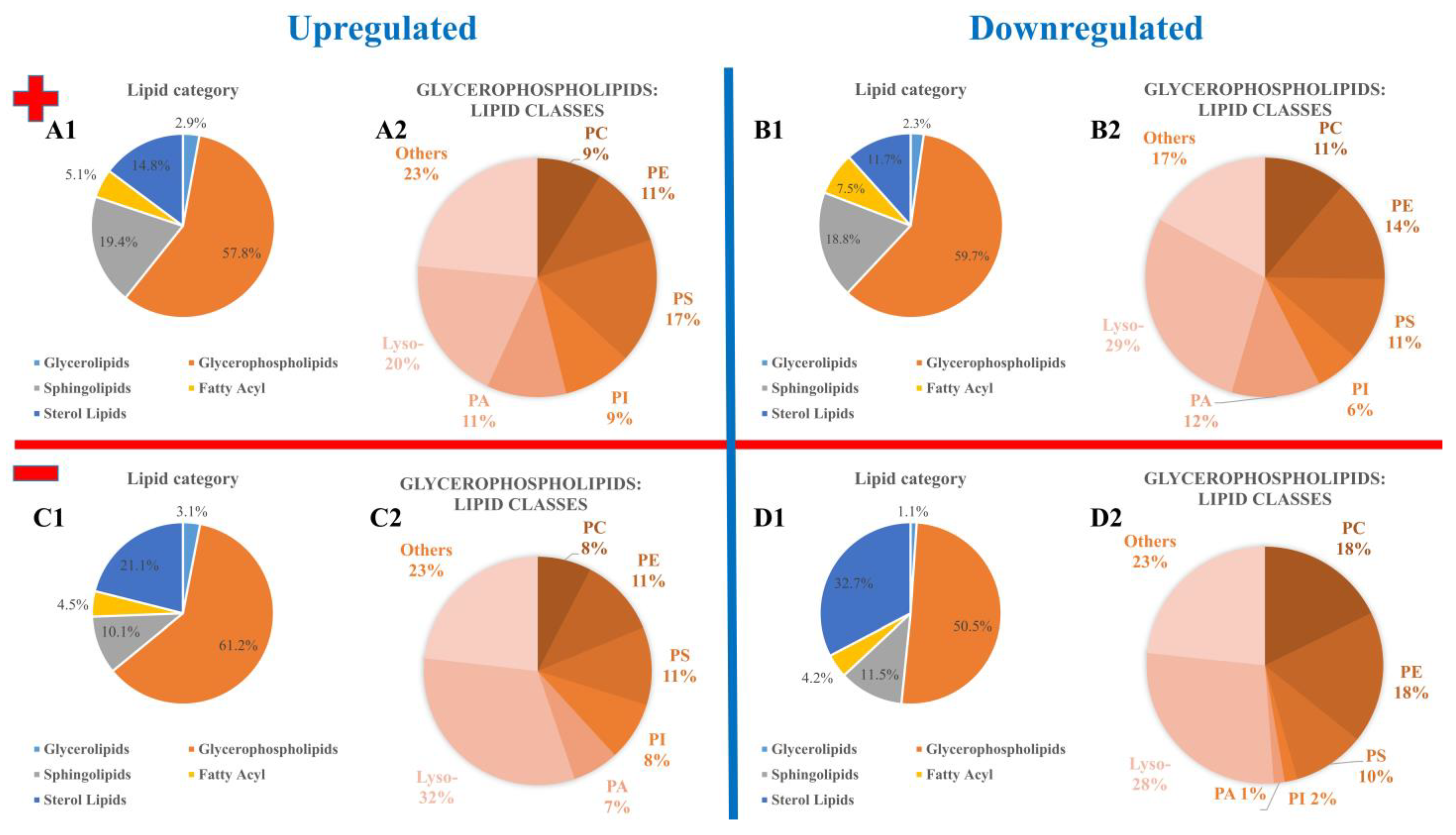
3.2. Annotation and Verification of Markers
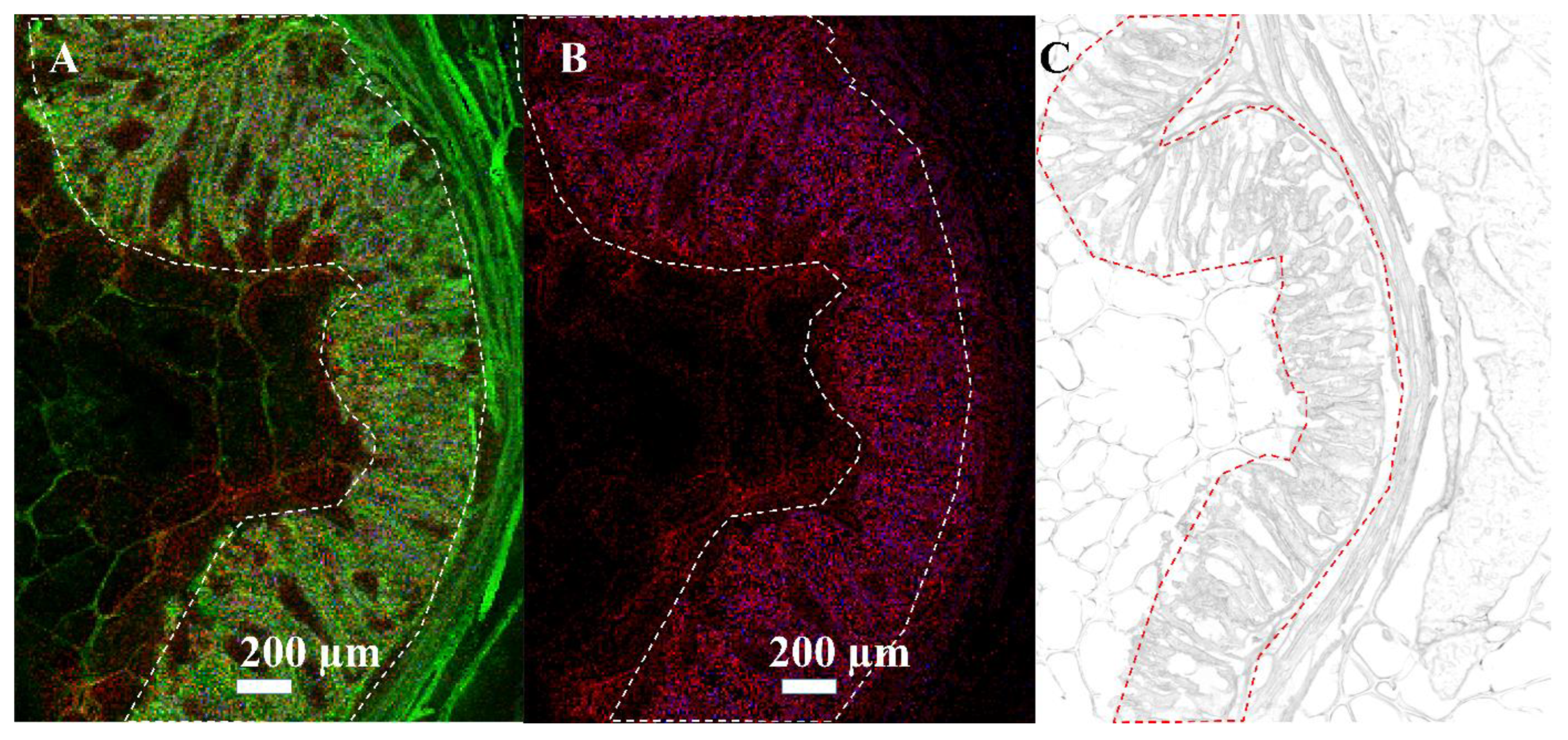
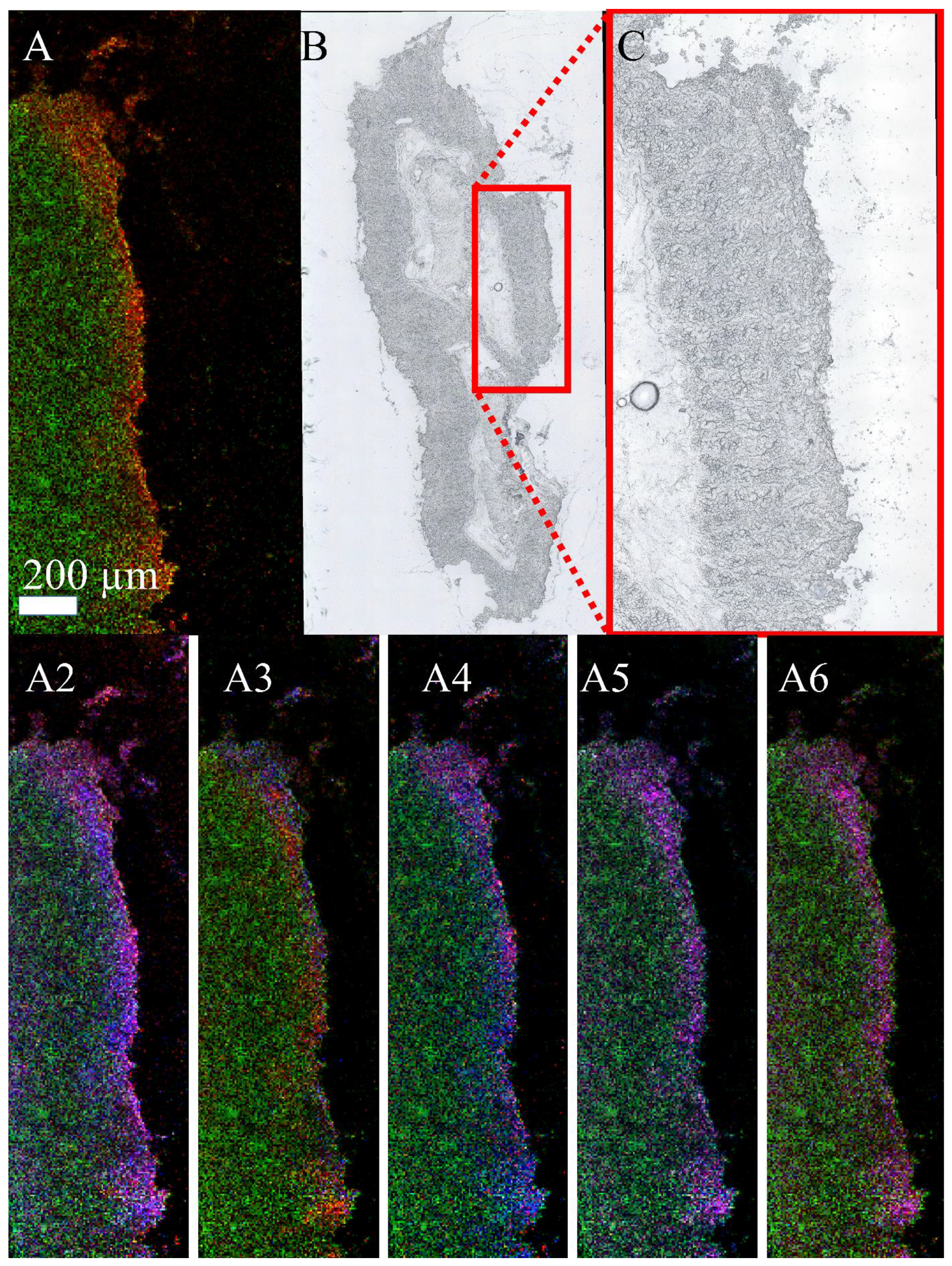
3.3. Mass Spectrometry Imaging of C. parvum-Infected Cell Layers and Intestinal Tissue at High Lateral Resolution
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gerace, E.; Lo Presti, V.D.; Biondo, C. Cryptosporidium infection: Epidemiology, pathogenesis, and differential diagnosis. Eur. J. Microbiol. Immunol. 2019, 9, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Tzipori, S.; Ward, H. Cryptosporidiosis: Biology, pathogenesis and disease. Microbes Infect. 2002, 4, 1047–1058. [Google Scholar] [CrossRef]
- Minghui, R.; WHO. Ending the Neglect to Attain the Sustainable Development Goals: A Road Map for Neglected Tropical Diseases 2021−2030; WHO: Geneva, Switzerland, 2020; 196p. [Google Scholar]
- Khalil, I.A.; Troeger, C.; Rao, P.C.; Blacker, B.F.; Brown, A.; Brewer, T.G.; Colombara, D.V.; De Hostos, E.L.; Engmann, C.; Guerrant, R.L.; et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: A meta-analyses study. Lancet Glob. Health 2018, 6, E758–E768. [Google Scholar] [CrossRef] [Green Version]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Oza, S.; Hogan, D.; Chu, Y.; Perin, J.; Zhu, J.; Lawn, J.E.; Cousens, S.; Mathers, C.; Black, R.E. Global, regional, and national causes of under-5 mortality in 2000-15: An updated systematic analysis with implications for the sustainable development goals. Lancet 2016, 388, 3027–3035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diptyanusa, A.; Sari, I.P. Treatment of human intestinal cryptosporidiosis: A review of published clinical trials. Int. J. Parasitol.-Drugs Drug Resist. 2021, 17, 128–138. [Google Scholar] [CrossRef]
- Checkley, W.; White, A.C.; Jaganath, D.; Arrowood, M.J.; Chalmers, R.M.; Chen, X.M.; Fayer, R.; Griffiths, J.K.; Guerrant, R.L.; Hedstrom, L.; et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 2015, 15, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Fox, L.M.; Saravolatz, L.D. Nitazoxanide: A new thiazolide antiparasitic agent. Clin. Infect. Dis. 2005, 40, 1173–1180. [Google Scholar] [CrossRef]
- Rider, S.D.; Zhu, G. Cryptosporidium: Genomic and biochemical features. Exp. Parasitol. 2010, 124, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.Y.; Roellig, D.M.; Guo, Y.Q.; Li, N.; Frace, M.A.; Tang, K.; Zhang, L.X.; Feng, Y.Y.; Xiao, L.H. Evolution of mitosome metabolism and invasion-related proteins in Cryptosporidium. BMC Genom. 2016, 17, 1006. [Google Scholar] [CrossRef] [Green Version]
- Velez, J.; Velasquez, Z.; Silva, L.M.R.; Gartner, U.; Failing, K.; Daugschies, A.; Mazurek, S.; Hermosilla, C.; Taubert, A. Metabolic Signatures of Cryptosporidium parvum-Infected HCT-8 cells and impact of selected metabolic inhibitors on C. parvum Infection under physioxia and hyperoxia. Biology 2021, 10, 60. [Google Scholar] [CrossRef]
- Velez, J.; Silva, L.M.R.; Gaertner, U.; Daugschies, A.; Mazurek, S.; Hermosilla, C.; Taubert, A. First metabolic insights into ex vivo Cryptosporidium parvum-Infected bovine small intestinal xxplants studied under physioxic conditions. Biology 2021, 10, 963. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, M.L.; Owens, J.H.; Kelty, C.A. Characterization of Cryptosporidium parvum by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 2000, 66, 4720–4724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snelling, W.J.; Lin, Q.S.; Moore, J.E.; Millar, B.C.; Tosini, F.; Pozio, E.; Dooley, J.S.G.; Lowery, C.J. Proteomics analysis and protein expression during sporozoite excystation of Cryptosporidium parvum (Coccidia, Apicomplexa). Mol. Cell. Proteom. 2007, 6, 346–355. [Google Scholar] [CrossRef] [Green Version]
- Li, D.F.; Cui, Z.H.; Wang, L.Y.; Zhang, K.H.; Cao, L.T.; Zheng, S.J.; Zhang, L.X. Tandem mass tag (TMT)-based proteomic analysis of Cryptosporidium andersoni oocysts before and after excystation. Parasites Vectors 2021, 14, 608. [Google Scholar] [CrossRef]
- Mauzy, M.J.; Enomoto, S.; Lancto, C.A.; Abrahamsen, M.S.; Rutherford, M.S. The Cryptosporidium parvum transcriptome during in vitro development. PLoS ONE 2012, 7, e31715. [Google Scholar] [CrossRef] [Green Version]
- Matos, L.V.S.; McEvoy, J.; Tzipori, S.; Bresciani, K.D.S.; Widmer, G. The transcriptome of Cryptosporidium oocysts and intracellular stages. Sci. Rep. 2019, 9, 7856. [Google Scholar] [CrossRef] [Green Version]
- Spengler, B. De novo sequencing, peptide composition analysis, and composition-based sequencing: A new strategy employing accurate mass determination by Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 2004, 15, 703–714. [Google Scholar] [CrossRef] [Green Version]
- Spengler, B. Accurate mass as a bioinformatic parameter in data-to-knowledge conversion: Fourier transform ion cyclotron resonance mass spectrometry for peptide de novo sequencing. Eur. J. Mass Spectrom. 2007, 13, 83–87. [Google Scholar] [CrossRef] [Green Version]
- Spengler, B.; Hester, A. Mass-based classification (MBC) of peptides: Highly accurate precursor ion mass values can be used to directly recognize peptide phosphorylation. J. Am. Soc. Mass Spectrom. 2008, 19, 1808–1812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kompauer, M.; Heiles, S.; Spengler, B. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-mu m lateral resolution. Nat. Methods 2017, 14, 90–96. [Google Scholar] [CrossRef]
- Shahiduzzaman, M.; Dyachenko, V.; Obwaller, A.; Unglaube, S.; Daugschies, A. Combination of cell culture and quantitative PCR for screening of drugs against Cryptosporidium parvum. Vet. Parasitol. 2009, 162, 271–277. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.X.; van Dommelen, J.; van der Heijden, R.; Spijksma, G.; Reijmers, T.H.; Wang, M.; Slee, E.; Lu, X.; Xu, G.W.; van der Greef, J.; et al. RPLC-Ion-Trap-FTMS method for lipid profiling of plasma: Method validation and application to p53 mutant mouse model. J. Proteome Res. 2008, 7, 4982–4991. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.A.; Kompauer, M.; Strupat, K.; Heiles, S.; Spengler, B. Implementation of a high-repetition-rate laser in an AP-SMALDI MSI system for enhanced measurement performance. J. Am. Soc. Mass Spectrom. 2021, 32, 465–472. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Kadesch, P.; Hollubarsch, T.; Gerbig, S.; Schneider, L.; Silva, L.M.R.; Hermosilla, C.; Taubert, A.; Spengler, B. Intracellular parasites Toxoplasma gondii and Besnoitia besnoiti, unveiled in single host cells using AP-SMALDI MS imaging. J. Am. Soc. Mass Spectrom. 2020, 31, 1815–1824. [Google Scholar] [CrossRef]
- Anschütz, N.H.; Gerbig, S.; Ventura, A.M.P.; Silva, L.M.R.; Larrazabal, C.; Hermosilla, C.; Taubert, A.; Spengler, B. Atmospheric-pressure scanning microprobe matrix-assisted laser desorption/ionization mass spectrometry imaging of Neospora caninum-infected cell monolayers. Anal. Sci. Adv. 2022, 3, 244–254. [Google Scholar] [CrossRef]
- Paschke, C.; Leisner, A.; Hester, A.; Maass, K.; Guenther, S.; Bouschen, W.; Spengler, B. Mirion-A software package for automatic processing of mass spectrometric images. J. Am. Soc. Mass Spectrom. 2013, 24, 1296–1306. [Google Scholar] [CrossRef] [PubMed]
- The LIPID MAPS® Lipidomics Gateway. Available online: https://www.lipidmaps.org/ (accessed on 7 July 2022).
- Koelmel, J.P.; Kroeger, N.M.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Cochran, J.A.; Beecher, C.W.W.; Garrett, T.J.; Yost, R.A. LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinform. 2017, 18, 331. [Google Scholar] [CrossRef] [PubMed]
- Kessner, D.; Chambers, M.; Burke, R.; Agusand, D.; Mallick, P. Proteo Wizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Mitschler, R.R.; Welti, R.; Upton, S.J. A comparative-study of lipid compositions of Cryptosporidium-parvum (apicomplexa) and madin-darby bovine kidney-cells. J. Eukaryot. Microbiol. 1994, 41, 8–12. [Google Scholar] [CrossRef]
- Besteiro, S.; Bertrand-Michel, J.; Lebrun, M.; Vial, H.; Dubremetz, J.F. Lipidomic analysis of Toxoplasma gondii tachyzoites rhoptries: Further insights into the role of cholesterol. Biochem. J. 2008, 415, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Welti, R.; Mui, E.; Sparks, A.; Wernimont, S.; Isaac, G.; Kirisits, M.; Roth, M.; Roberts, C.W.; Botte, C.; Marechal, E.; et al. Lipidomic analysis of Toxoplasma gondii reveals unusual polar lipids. Biochemistry 2007, 46, 13882–13890. [Google Scholar] [CrossRef] [Green Version]
- Botte, C.Y.; Yamaryo-Botte, Y.; Rupasinghe, T.W.T.; Mullin, K.A.; MacRae, J.I.; Spurck, T.P.; Kalanon, M.; Shears, M.J.; Coppel, R.L.; Crellin, P.K.; et al. Atypical lipid composition in the purified relict plastid (apicoplast) of malaria parasites. Proc. Natl. Acad. Sci. USA 2013, 110, 7506–7511. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Zahn, M.M.; Coppens, I.; Joiner, K.A.; Voelker, D.R. Selective disruption of phosphatidylcholine metabolism of the intracellular parasite Toxoplasma gondii arrests its growth. J. Biol. Chem. 2005, 280, 16345–16353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldarelli, S.A.; Duckert, J.F.; Wein, S.; Calas, M.; Perigaud, C.; Vial, H.; Peyrottes, S. Synthesis and evaluation of bis-thiazolium salts as potential antimalarial drugs. Chemmedchem 2010, 5, 1102–1109. [Google Scholar] [CrossRef]
- Itoe, M.A.; Sampaio, J.L.; Cabal, G.G.; Real, E.; Zuzarte-Luis, V.; March, S.; Bhatia, S.N.; Frischknecht, F.; Thiele, C.; Shevchenko, A.; et al. Host cell phosphatidylcholine is a key mediator of malaria parasite survival during liver stage infection. Cell Host Microbe 2014, 16, 778–786. [Google Scholar] [CrossRef] [Green Version]
- Bobenchik, A.M.; Witola, W.H.; Augagneur, Y.; Lochlainn, L.N.; Garg, A.; Pachikara, N.; Choi, J.Y.; Zhao, Y.O.; Usmani-Brown, S.; Lee, A.; et al. Plasmodium falciparum phosphoethanolamine methyltransferase is essential for malaria transmission. Proc. Natl. Acad. Sci. USA 2013, 110, 18262–18267. [Google Scholar] [CrossRef]
- Richmond, G.S.; Smith, T.K. The role and characterization of phospholipase A(1) in mediating lysophosphatidylcholine synthesis in Trypanosoma brucei. Biochem. J. 2007, 405, 319–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheokand, P.K.; Narwal, M.; Thakur, V.; Mohmmed, A. GlmS mediated knock-down of a phospholipase expedite alternate pathway to generate phosphocholine required for phosphatidylcholine synthesis in Plasmodium falciparum. Biochem. J. 2021, 478, 3429–3444. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.B. Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 2004, 9, 229–235. [Google Scholar] [CrossRef]
- Seron, K.; Dzierszinski, F.; Tomavo, S. Molecular cloning, functional complementation in Saccharomyces cerevisiae and enzymatic properties of phosphatidylinositol synthase from the protozoan parasite Toxoplasma gondii. Eur. J. Biochem. 2000, 267, 6571–6579. [Google Scholar] [CrossRef] [Green Version]
- Hublin, J.; Ryan, U.; Trengove, R.; Maker, G. Metabolomic profiling of faecal extracts from cryptosporidium parvum infection in experimental mouse models. PLoS ONE 2013, 8, e77803. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anschütz, N.H.; Gerbig, S.; Ghezellou, P.; Silva, L.M.R.; Vélez, J.D.; Hermosilla, C.R.; Taubert, A.; Spengler, B. Mass Spectrometry Imaging of In Vitro Cryptosporidium parvum-Infected Cells and Host Tissue. Biomolecules 2023, 13, 1200. https://doi.org/10.3390/biom13081200
Anschütz NH, Gerbig S, Ghezellou P, Silva LMR, Vélez JD, Hermosilla CR, Taubert A, Spengler B. Mass Spectrometry Imaging of In Vitro Cryptosporidium parvum-Infected Cells and Host Tissue. Biomolecules. 2023; 13(8):1200. https://doi.org/10.3390/biom13081200
Chicago/Turabian StyleAnschütz, Nils H., Stefanie Gerbig, Parviz Ghezellou, Liliana M. R. Silva, Juan Diego Vélez, Carlos R. Hermosilla, Anja Taubert, and Bernhard Spengler. 2023. "Mass Spectrometry Imaging of In Vitro Cryptosporidium parvum-Infected Cells and Host Tissue" Biomolecules 13, no. 8: 1200. https://doi.org/10.3390/biom13081200
APA StyleAnschütz, N. H., Gerbig, S., Ghezellou, P., Silva, L. M. R., Vélez, J. D., Hermosilla, C. R., Taubert, A., & Spengler, B. (2023). Mass Spectrometry Imaging of In Vitro Cryptosporidium parvum-Infected Cells and Host Tissue. Biomolecules, 13(8), 1200. https://doi.org/10.3390/biom13081200








