Fatty Acid-Activated Proton Transport by Bisaryl Anion Transporters Depolarises Mitochondria and Reduces the Viability of MDA-MB-231 Breast Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemistry
2.1.1. General Procedure for the Synthesis of Bisaryl Squaramides
2.1.2. General Procedure for the Synthesis of Bisaryl Amides
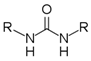
2.1.3. General Procedure for the Synthesis of Bisaryl Diureas
- Bis({[3-chloro-5-(trifluoromethyl)phenyl]amino})cyclobut-3-ene-1,2-dione (S1). 1H NMR (500 MHz, CD3OD) δ 7.78 (s, 2H), 7.71 (s, 2H), 7.37 (s, 2H). 13C NMR (125 MHz, CD3OD) δ 184.2 (2C), 167.2 (2C), 142.2 (2C), 137.1 (2C), 134.1 (q, J = 33 Hz, 2C), 124.5 (q, J = 271 Hz, 2C), 123.2 (2C), 121.0 (2C), 115.1 (q, J = 4 Hz, 2C). HRMS (ESI): m/z [M + H]+ calculated for C18H8Cl2F6N2O2: 468.9940, found: 468.9937.
- Bis({[4-(trifluoromethoxy)phenyl]amino})cyclobut-3-ene-1,2-dione (S3). 1H NMR (500 MHz, DMSO-d6) δ 10.5 (br s, 2H), 7.59 (d, J = 8.5 Hz, 4H), 7.35 (d, J = 8 Hz, 4H) 13C NMR (125 MHz, DMSO-d6) δ 181.7 (2C), 165.7 (2C), 143.8 (2C), 137.9 (2C), 122.2 (4C), 120.1 (q, J = 255 Hz, 2H), 120.0 (4C). HRMS (ESI): m/z [M + H]+ calculated for C18H10F6N2O4: 433.0618, found: 433.0623.
- Bis(phenylamino)cyclobut-3-ene-1,2-dione (S4). 1H and 13C NMR are in agreement with previously reported data [14]. HRMS (ESI): m/z [M + H]+ calculated for C16H12N2O2: 265.0972, found: 265.0958.
- 3-Chloro-N-[3-chloro-5-(trifluoromethyl)phenyl]-5-(trifluoromethyl)benzamide (A1). 1H NMR (500 MHz, CDCl3) δ 8.10 (br s, 1H), 8.04 (s, 1H), 7.99 (s, 1H), 7.98 (s, 1H), 7.82 (s, 1H), 7.77 (s, 1H), 7.43 (s, 1H). 13C NMR (125 MHz, CDCl3) δ 163.1 (1C), 138.8 (1C), 136.5 (1C), 136.1 (1C), 135.8 (1C), 133.1 (q, J = 34 Hz, 1C), 132.8 (q, J = 33 Hz, 1C), 130.8 (1C), 129.3 (q, J = 4 Hz, 1C), 123.47 (1C), 123.46 (1C), 122.9 (q, J = 271 Hz, 1C), 122.7 (q, J = 272 Hz, 1C), 122.1 (m, 2C), 115.3 (q, J = 4 Hz, 1C). HRMS (ESI): m/z [M + H]+ calculated for C15H7Cl2F6NO: 401.9882 found: 401.9883.
- 4-(Trifluoromethyl)-N-[4-(trifluoromethyl)phenyl]benzamide (A2). 1H and 13C NMR and HRMS are in agreement with previously reported data [19].
- 4-(Trifluoromethoxy)-N-[4-(trifluoromethoxy)phenyl]benzamide (A3). 1H NMR (500 MHz, DMSO-d6) δ 10.53 (s, 1H), 8.08 (AA’BB’, 2H), 7.88 (AA’BB’, 2H), 7.54 (d, J = 9 Hz, 2H), 7.38 (d, J = 9 Hz, 2H). 13C NMR (125 MHz, DMSO-d6) δ 164.5 (1C), 151.9 (1C), 145.7 (1C), 136.2 (1C), 132.9 (1C), 129.0 (2C), 121.9 (2C), 121.5 (2C), 120.8 (2C), 120.4 (q, J = 256 Hz, 1C), 120.3 (q, J = 257 Hz, 1C). HRMS (ESI): m/z [M + H]+ calculated for C15H9F6NO3: 366.0559 found: 366.0562.
- N-Phenylbenzamide (A4). 1H and 13C NMR and HRMS are in agreement with previously reported data [19].
- 3-[3-Chloro-5-(trifluoromethyl)phenyl]-1-[6-({[3-chloro-5-(trifluoromethyl)phenyl]carbamoyl}camino)hexyl]urea (D1). 1H NMR (400 MHz, DMSO-d6) δ 8.97 (s, 2H), 7.76 (s, 2H), 7.75 (s, 2H), 7.27 (s, 2H), 6.40 (t, J = 5.6 Hz, 2H), 3.08 (q, J = 6.2 Hz, 4H), 1.44 (m, 4H), 1.29 (m, 4H). 13C NMR (100 MHz, DMSO-d6) δ 154.7 (2C), 143.0 (2C), 134.1 (2C), 130.9 (q, J = 32 Hz, 2C), 123.4 (q, J = 271 Hz, 2C), 120.3 (2C), 116.7 (2C), 112.3 (2C), 39.1 (2C), 29.6 (2C), 26.1 (2C). HRMS (ESI): m/z [M − H]− calculated for C22H22Cl2F6N4O2: 557.0953 found: 557.0953.
- 3-[4-(Trifluoromethyl)phenyl]-1-[6-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)hexyl]urea (D2). 1H NMR (400 MHz, DMSO-d6) δ 8.86 (s, 2H), 7.58 (d, J = 8.9 Hz, 4H), 7.54 (d, J = 8.9 Hz, 4H), 6.30 (t, J = 5.5 Hz, 2H), 3.09 (q, J = 6.6 Hz, 4H), 1.43 (m, 4H), 1.31 (m, 4H). 13C NMR (100 MHz, DMSO-d6) δ 154.8 (2C), 144.3 (2C), 125.9 (2C), 124.7 (q, J = 269 Hz, 2C), 120.8 (q, J = 32 Hz, 2C), 117.1 (2C), 39.0 (2C), 29.6 (2C), 26.1 (2C). HRMS (ESI): m/z [M − H]− calculated for C22H24F6N4O2: 489.1732 found: 489.1727.
- 3-[4-(Trifluoromethoxy)phenyl]-1-[6-({[4-(trifluoromethoxy)phenyl]carbamoyl}amino)hexyl]urea (D3). 1H NMR (400 MHz, DMSO-d6) δ 8.60 (s, 2H), 7.47 (d, J = 9.2 Hz, 4H), 7.19 (d, J = 8.4 Hz, 4H), 6.18 (t, J = 5.6 Hz, 2H), 3.07 (q, J = 6.7 Hz, 4H), 1.43 (m, 4H), 1.30 (m, 4H). 13C NMR (100 MHz, DMSO-d6) δ 155.2 (2C), 142.0 (2C), 140.0 (2C), 121.6 (2C), 120.3 (q, J = 254 Hz, 2C), 118.7 (2C), 39.1 (2C), 29.7 (2C), 26.2 (2C). HRMS (ESI): m/z [M − H]− calculated for C22H24F6N4O4: 521.1629 found: 521.1630.
- 3-Phenyl-1-{6-[(phenylcarbamoyl)amino]hexyl}urea (D4). 1H and 13C NMR and HRMS are in agreement with previously reported data [20].
2.2. Biology
2.2.1. Cell Lines and Culture Conditions
2.2.2. MTS Cell Viability Assay
2.2.3. JC-1 Mitochondrial Membrane Potential Assay
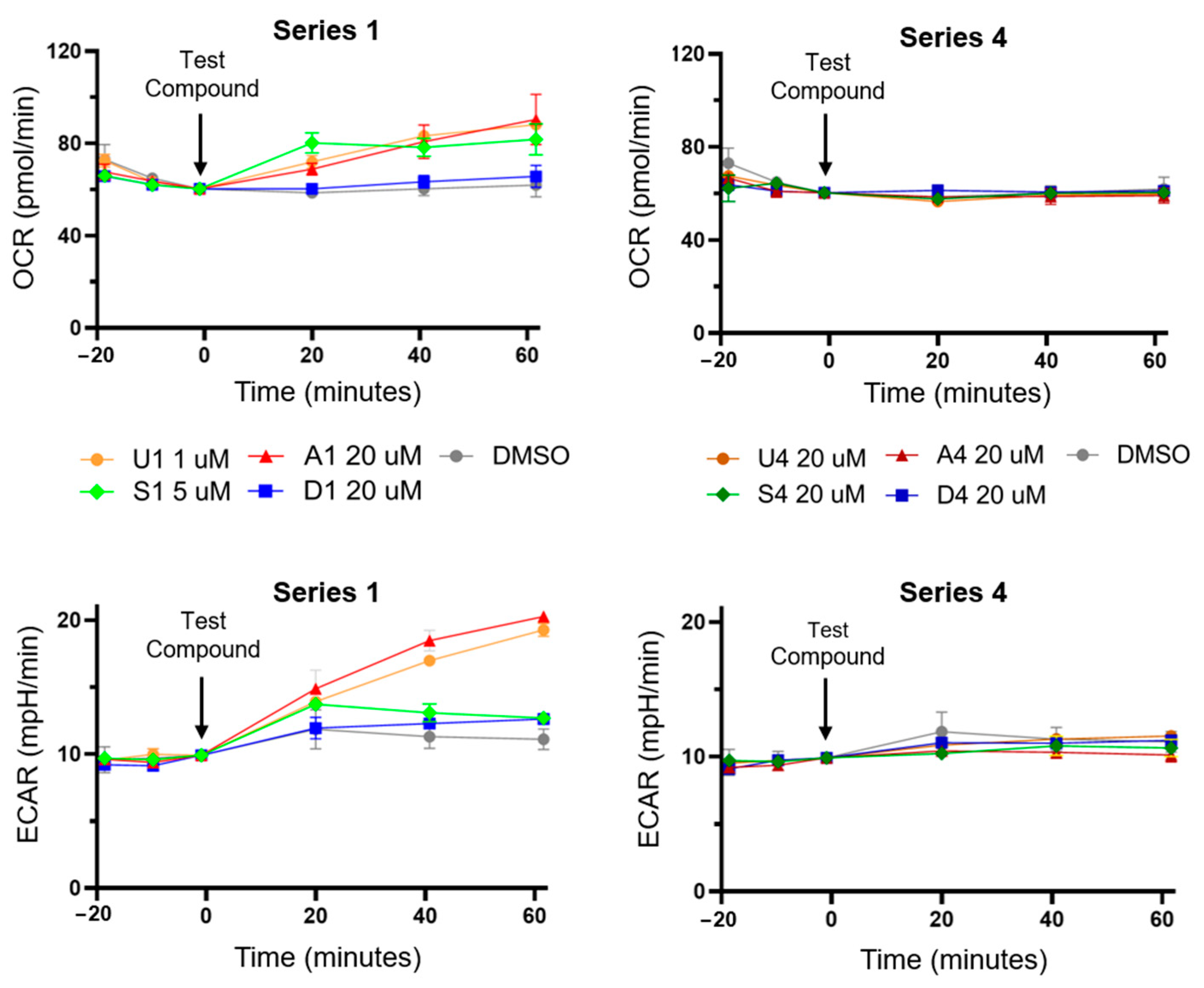
2.2.4. Seahorse XFe24 Analyser Assay
2.2.5. Statistical Analysis
3. Results and Discussion
3.1. Compound Library Design and Synthesis
3.2. Bisaryl Anion Transporter Effects in MDA-MB-231 Cells
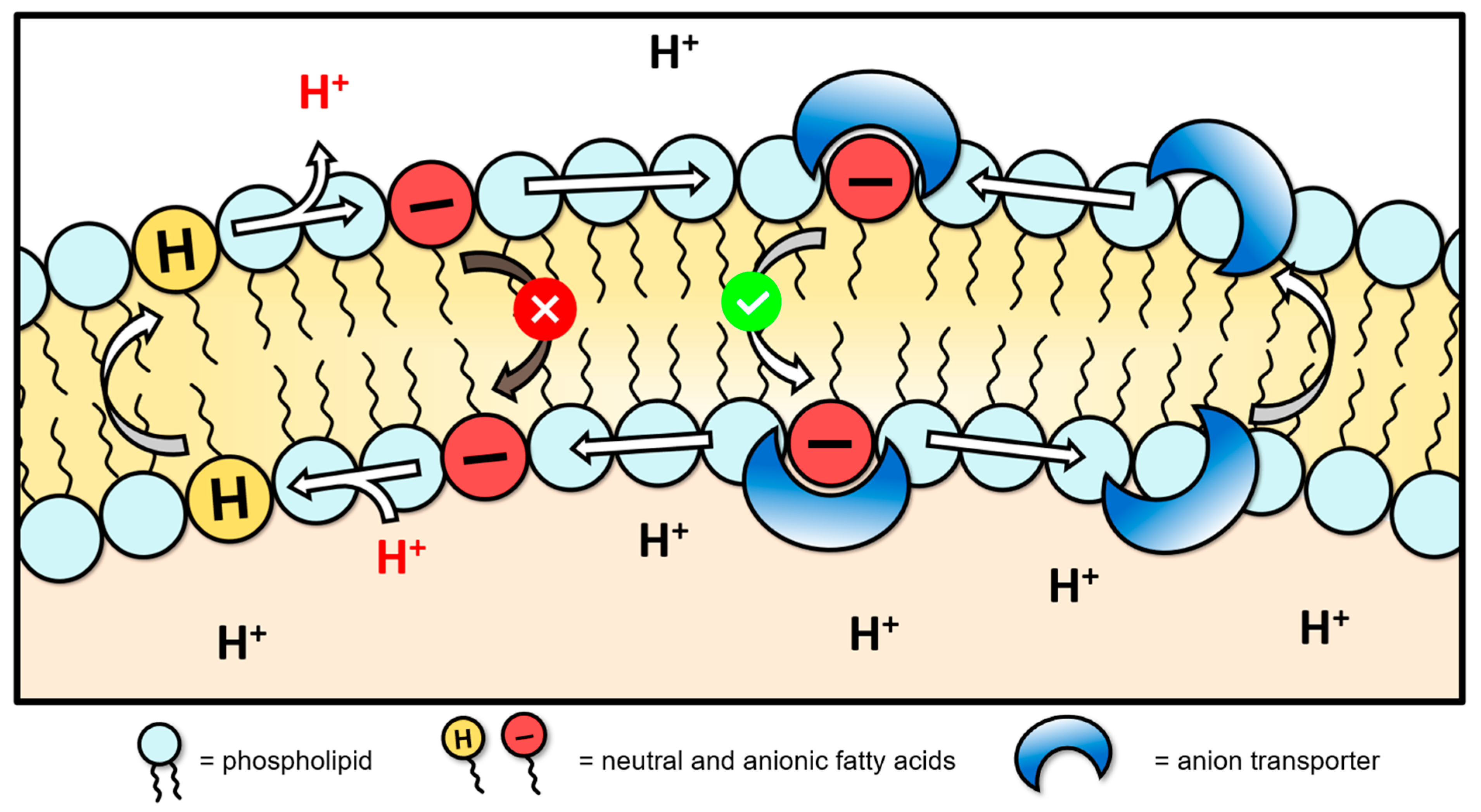
3.3. HPTS Proton Transport Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fulda, S.; Galluzzi, L.; Kroemer, G. Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 2010, 9, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.; Johnson, E.; Byrne, F.L. Exploring the therapeutic potential of mitochondrial uncouplers in cancer. Mol. Metab. 2021, 51, 101222. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.M. The Mechanism of Oxidative Phosphorylation. In The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, UK, 2000. [Google Scholar]
- Papa, S.; Martino, P.L.; Capitanio, G.; Gaballo, A.; De Rasmo, D.; Signorile, A.; Petruzzella, V. The oxidative phosphorylation system in mammalian mitochondria. Adv. Exp. Med. Biol. 2012, 942, 3–37. [Google Scholar] [CrossRef]
- Childress, E.S.; Alexopoulos, S.J.; Hoehn, K.L.; Santos, W.L. Small Molecule Mitochondrial Uncouplers and Their Therapeutic Potential. J. Med. Chem. 2018, 61, 4641–4655. [Google Scholar] [CrossRef] [PubMed]
- Figarola, J.L.; Singhal, J.; Singhal, S.; Kusari, J.; Riggs, A. Bioenergetic modulation with the mitochondria uncouplers SR4 and niclosamide prevents proliferation and growth of treatment-naïve and vemurafenib-resistant melanomas. Oncotarget 2018, 9, 36945–36965. [Google Scholar]
- Lim, M.L.; Minamikawa, T.; Nagley, P. The protonophore CCCP induces mitochondrial permeability transition without cytochrome c release in human osteosarcoma cells. FEBS Lett. 2001, 503, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, J.; Feng, X.; Zhu, M.; Hoffmann, S.; Hsu, A.; Qian, K.; Huang, D.; Zhao, F.; Liu, W.; et al. Mitochondria-targeting fluorescent molecules for high efficiency cancer growth inhibition and imaging. Chem. Sci. 2019, 10, 7946–7951. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Gale, P.A. Small-Molecule Uncoupling Protein Mimics: Synthetic Anion Receptors as Fatty Acid-Activated Proton Transporters. J. Am. Chem. Soc. 2016, 138, 16508–16514. [Google Scholar] [CrossRef]
- Davis, J.T.; Gale, P.A.; Quesada, R. Advances in anion transport and supramolecular medicinal chemistry. Chem. Soc. Rev. 2020, 49, 6056–6086. [Google Scholar] [CrossRef]
- York, E.; McNaughton, D.A.; Roseblade, A.; Cranfield, C.G.; Gale, P.A.; Rawling, T. Structure–Activity Relationship and Mechanistic Studies of Bisaryl Urea Anticancer Agents Indicate Mitochondrial Uncoupling by a Fatty Acid-Activated Mechanism. ACS Chem. Biol. 2022, 17, 2065–2073. [Google Scholar] [CrossRef]
- Penzo, D.; Tagliapietra, C.; Colonna, R.; Petronilli, V.; Bernardi, P. Effects of fatty acids on mitochondria: Implications for cell death. Biochim. Biophys. Acta Bioenerg. 2002, 1555, 160–165. [Google Scholar] [CrossRef]
- Schenkel, L.C.; Bakovic, M. Formation and Regulation of Mitochondrial Membranes. Int. J. Cell Biol. 2014, 2014, 1–13. [Google Scholar] [CrossRef]
- Busschaert, N.; Kirby, I.L.; Young, S.; Coles, S.J.; Horton, P.N.; Light, M.E.; Gale, P.A. Squaramides as potent transmembrane anion transporters. Angew. Chem. Int. Ed. Engl. 2012, 51, 4426–4430. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-H.; Park, S.-H.; Howe, E.N.W.; Hyun, J.Y.; Chen, L.-J.; Hwang, I.; Vargas-Zuñiga, G.; Busschaert, N.; Gale, P.A.; Sessler, J.L.; et al. Determinants of Ion-Transporter Cancer Cell Death. Chem 2019, 5, 2079–2098. [Google Scholar] [CrossRef]
- Andrews, N.J.; Haynes, C.J.E.; Light, M.E.; Moore, S.J.; Tong, C.C.; Davis, J.T.; Harrell, W.A., Jr.; Gale, P.A. Structurally simple lipid bilayer transport agents for chloride and bicarbonate. Chem. Sci. 2011, 2, 256–260. [Google Scholar] [CrossRef]
- Cushman, M.; Georg, G.I.; Holzgrabe, U.; Wang, S. Absolute Quantitative 1H NMR Spectroscopy for Compound Purity Determination. J. Med. Chem. 2014, 57, 9219. [Google Scholar] [CrossRef]
- Amendola, V.; Fabbrizzi, L.; Mosca, L.; Schmidtchen, F.-P. Urea-, Squaramide-, and Sulfonamide-Based Anion Receptors: A Thermodynamic Study. Chem. Eur. J. 2011, 17, 5972–5981. [Google Scholar] [CrossRef]
- Sheetal; Sharma, A.K.; Shaifali; Bhattacherjee, D.; Sharma, N.; Giri, K.; Das, P. Supported-Pd catalyzed tandem approach for N-arylbenzamides synthesis. Mol. Catal. 2021, 516, 111948. [Google Scholar] [CrossRef]
- Skrylkova, A.S.; Egorov, D.M.; Tarabanov, R.V. Reaction of Hexamethylene Diisocyanate with Amines. Russ. J. Gen. Chem. 2022, 92, 2033–2041. [Google Scholar] [CrossRef]
- Bao, X.; Wu, X.; Berry, S.N.; Howe, E.N.W.; Chang, Y.-T.; Gale, P.A. Fluorescent squaramides as anion receptors and transmembrane anion transporters. Chem. Comm. 2018, 54, 1363–1366. [Google Scholar] [CrossRef] [PubMed]
- Marques, I.; Costa, P.M.R.; Miranda, M.Q.; Busschaert, N.; Howe, E.N.W.; Clarke, H.J.; Haynes, C.J.E.; Kirby, I.L.; Rodilla, A.M.; Pérez-Tomás, R.; et al. Full elucidation of the transmembrane anion transport mechanism of squaramides using in silico investigations. Phys. Chem. Chem. Phys. 2018, 20, 20796–20811. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.N.W.; Gale, P.A. Fatty Acid Fueled Transmembrane Chloride Transport. J. Am. Chem. Soc. 2019, 141, 10654–10660. [Google Scholar] [CrossRef] [PubMed]
- Hansch, C.; Leo, A.; Unger, S.H.; Kim, K.H.; Nikaitani, D.; Lien, E.J. Aromatic substituent constants for structure-activity correlations. J. Med. Chem. 1973, 16, 1207–1216. [Google Scholar] [CrossRef]
- Rostami, A.; Colin, A.; Li, X.Y.; Chudzinski, M.G.; Lough, A.J.; Taylor, M.S. N,N′-Diarylsquaramides: General, High-Yielding Synthesis and Applications in Colorimetric Anion Sensing. J. Org. Chem. 2010, 75, 3983–3992. [Google Scholar] [CrossRef]
- MacMillan, D.S.; Murray, J.; Sneddon, H.F.; Jamieson, C.; Watson, A.J.B. Evaluation of alternative solvents in common amide coupling reactions: Replacement of dichloromethane and N,N-dimethylformamide. Green Chem. 2013, 15, 596–600. [Google Scholar] [CrossRef]
- Gilchrist, A.M.; Wang, P.; Carreira-Barral, I.; Alonso-Carrillo, D.; Wu, X.; Quesada, R.; Gale, P.A. Supramolecular methods: The 8-hydroxypyrene-1,3,6-trisulfonic acid (HPTS) transport assay. Supramol Chem. 2021, 33, 325–344. [Google Scholar] [CrossRef]
- Gensure, R.H.; Zeidel, M.L.; Hill, W.G. Lipid raft components cholesterol and sphingomyelin increase H+/OH− permeability of phosphatidylcholine membranes. Biochem. J. 2006, 398, 485–495. [Google Scholar] [CrossRef]
- van Hoogevest, P.; Wendel, A. The use of natural and synthetic phospholipids as pharmaceutical excipients. Eur. J. Lipid Sci. Technol. 2014, 116, 1088–1107. [Google Scholar] [CrossRef]
- Amendola, V.; Bergamaschi, G.; Boiocchi, M.; Fabbrizzi, L.; Milani, M. The Squaramide versus Urea Contest for Anion Recognition. Chem. Eur. J. 2010, 16, 4368–4380. [Google Scholar] [CrossRef]
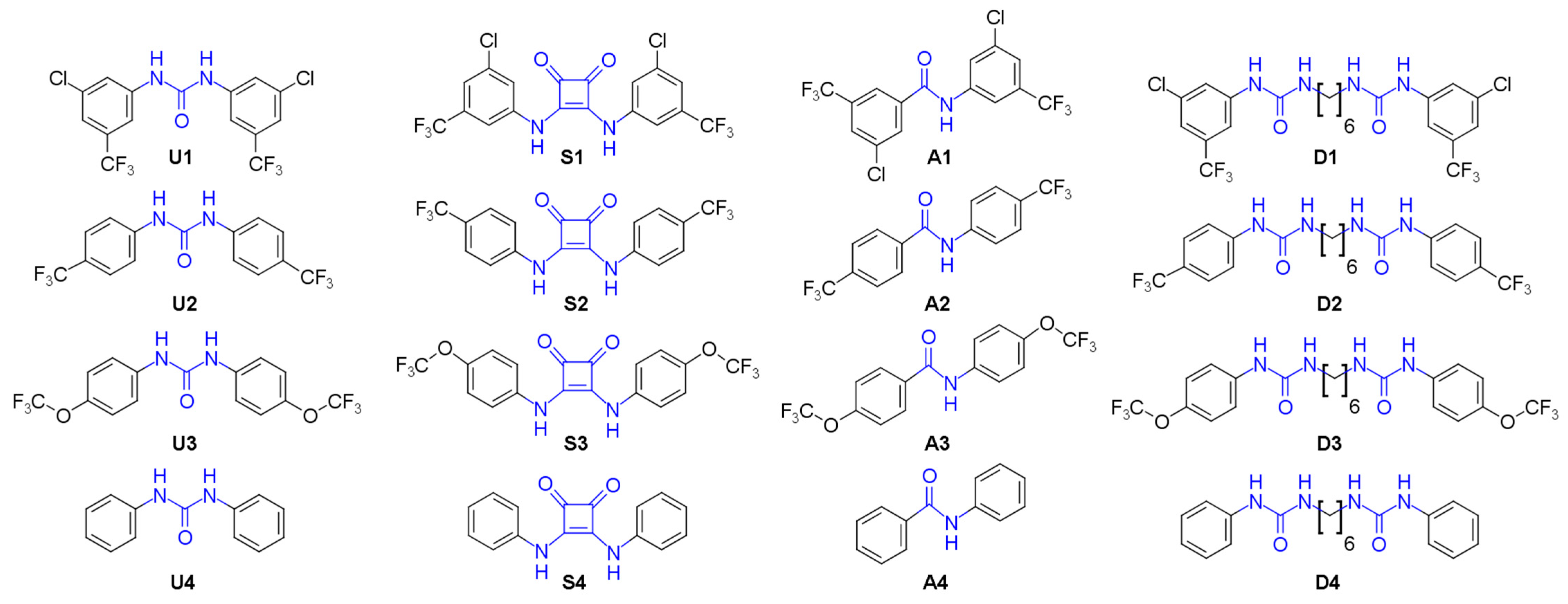


 U1–4 | 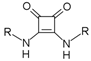 S1–4 |  A1–4 | 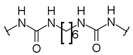 D1–4 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Substitution Pattern | R | IC50 (μM) | IC50 (μM) | IC50 (μM) | IC50 (μM) | ||||
| MTS | JC-1 | MTS | JC-1 | MTS | JC-1 | MTS | JC-1 | ||
| Series 1 | 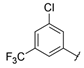 | 0.37 ± 0.1 | 0.26 ± 0.1 | 4.15 ± 1.4 | 1.73 ± 0.1 | >20 a | 7.30 ± 1.0 | 10.5 ± 1.1 | 8.1 ± 1.2 |
| Series 2 | 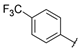 | 0.84 ± 0.1 | 2.26 ± 0.4 | 7.28 ± 0.9 | 3.34 ± 0.9 | - b | 19.7 ± 5.9 | - b | >50 a |
| Series 3 | 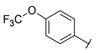 | >100 a | >100 a | - b | - b | 5.64 ± 1.1 | - b | - b | >50 a |
| Series 4 | 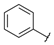 | - b | >100 a | - b | - b | - b | - b | - b | >100 a |
| Compound a | EC50 (Utd, mol%) | n (Utd) | EC50 (BSA, mol%) | n (BSA) | EC50 (OA, mol%) | n (OA) | Activation Factor |
|---|---|---|---|---|---|---|---|
| U1 | 0.0046 ± 4 × 10−4 | 0.82 ± 0.07 | 0.06 ± 0.004 | 1.24 ± 0.09 | 0.003 ± 1 × 10−4 | 0.8 ± 0.04 | 20 |
| A1 | 0.02 ± 2 × 10−4 | 2.00 ± 0.07 | 0.12 ± 0.005 | 1.74 ± 0.08 | 0.02 ± 0.001 | 1.44 ± 0.2 | 6 |
| D1 | 0.04 ± 0.003 | 0.91 ± 0.08 | 0.15 ± 0.02 | 0.58 ± 0.05 | 0.04 ± 0.001 | 0.66 ± 0.05 | 3.75 |
| S1 | 0.007 ± 6 × 10−5 | 1.63 ± 0.02 | 0.02 ± 0.001 | 1.38 ± 0.11 | 0.01 ± 0.001 | 1.24 ± 0.01 | 2 |
| S4 | 0.15 ± 0.006 | 1.09 ± 0.09 | 0.89 ± 0.009 | 1.67 ± 0.05 | 0.13 ± 0.008 | 0.74 ± 0.04 | 6.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
York, E.; McNaughton, D.A.; Duman, M.-N.; Gale, P.A.; Rawling, T. Fatty Acid-Activated Proton Transport by Bisaryl Anion Transporters Depolarises Mitochondria and Reduces the Viability of MDA-MB-231 Breast Cancer Cells. Biomolecules 2023, 13, 1202. https://doi.org/10.3390/biom13081202
York E, McNaughton DA, Duman M-N, Gale PA, Rawling T. Fatty Acid-Activated Proton Transport by Bisaryl Anion Transporters Depolarises Mitochondria and Reduces the Viability of MDA-MB-231 Breast Cancer Cells. Biomolecules. 2023; 13(8):1202. https://doi.org/10.3390/biom13081202
Chicago/Turabian StyleYork, Edward, Daniel A. McNaughton, Meryem-Nur Duman, Philip A. Gale, and Tristan Rawling. 2023. "Fatty Acid-Activated Proton Transport by Bisaryl Anion Transporters Depolarises Mitochondria and Reduces the Viability of MDA-MB-231 Breast Cancer Cells" Biomolecules 13, no. 8: 1202. https://doi.org/10.3390/biom13081202
APA StyleYork, E., McNaughton, D. A., Duman, M.-N., Gale, P. A., & Rawling, T. (2023). Fatty Acid-Activated Proton Transport by Bisaryl Anion Transporters Depolarises Mitochondria and Reduces the Viability of MDA-MB-231 Breast Cancer Cells. Biomolecules, 13(8), 1202. https://doi.org/10.3390/biom13081202






