Long-Term Culturing of FreeStyle 293-F Cells Affects Immunoglobulin G Glycome Composition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfections
2.2. Calculation of Total Cell Age
2.3. Isolation of IgG
2.4. IgG Glycan Preparation
2.5. Analysis of IgG Glycome Using HILIC-UPLC Method
2.6. RNA Isolation and RNA Sequencing
2.7. Quantitative Real-Time PCR (qPCR)
2.8. Protein Extraction and Western Blot Analysis
2.9. Statistical Analysis
3. Results
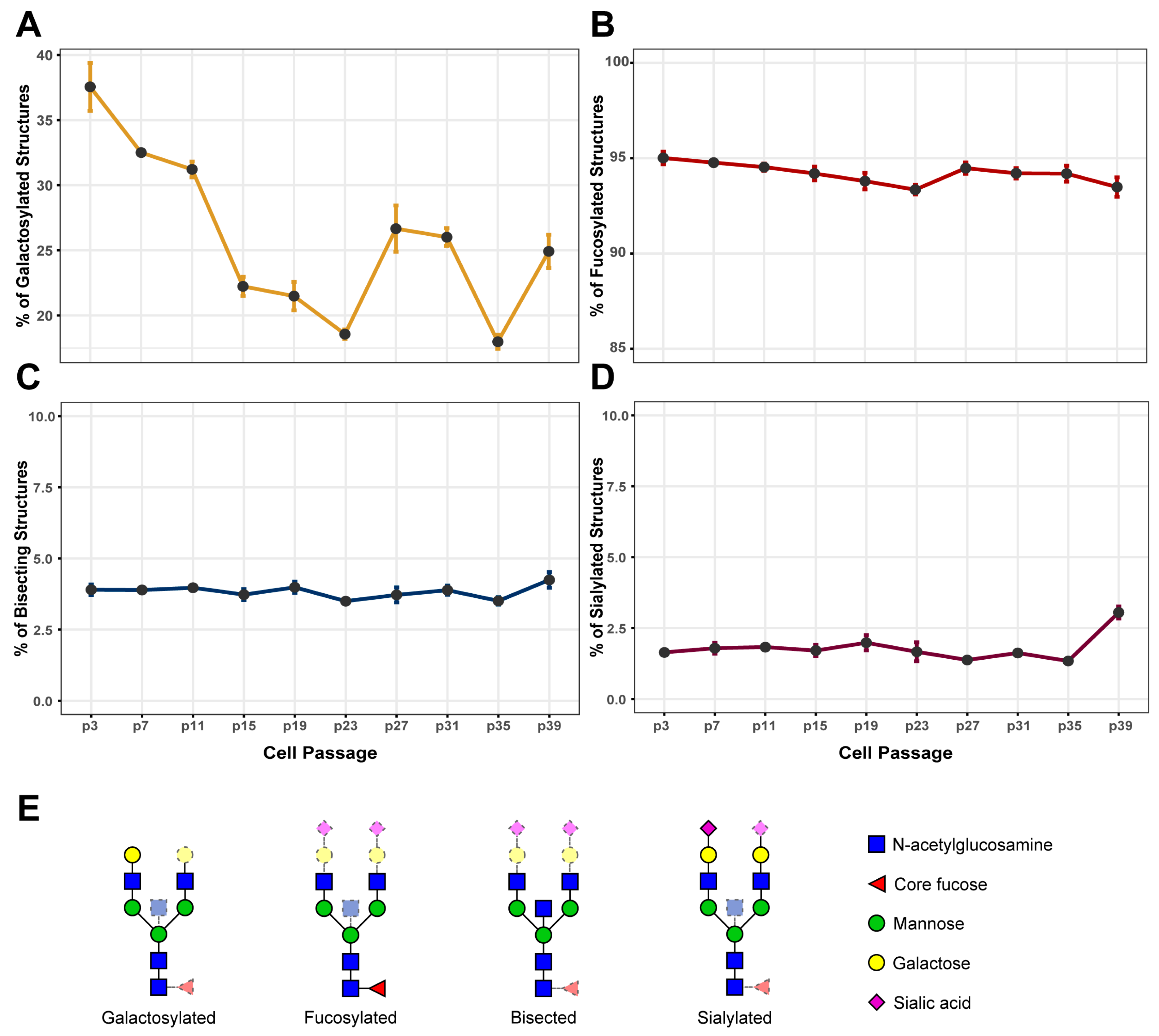
3.1. IgG Galactosylation Changes Extensively during Cell Culturing
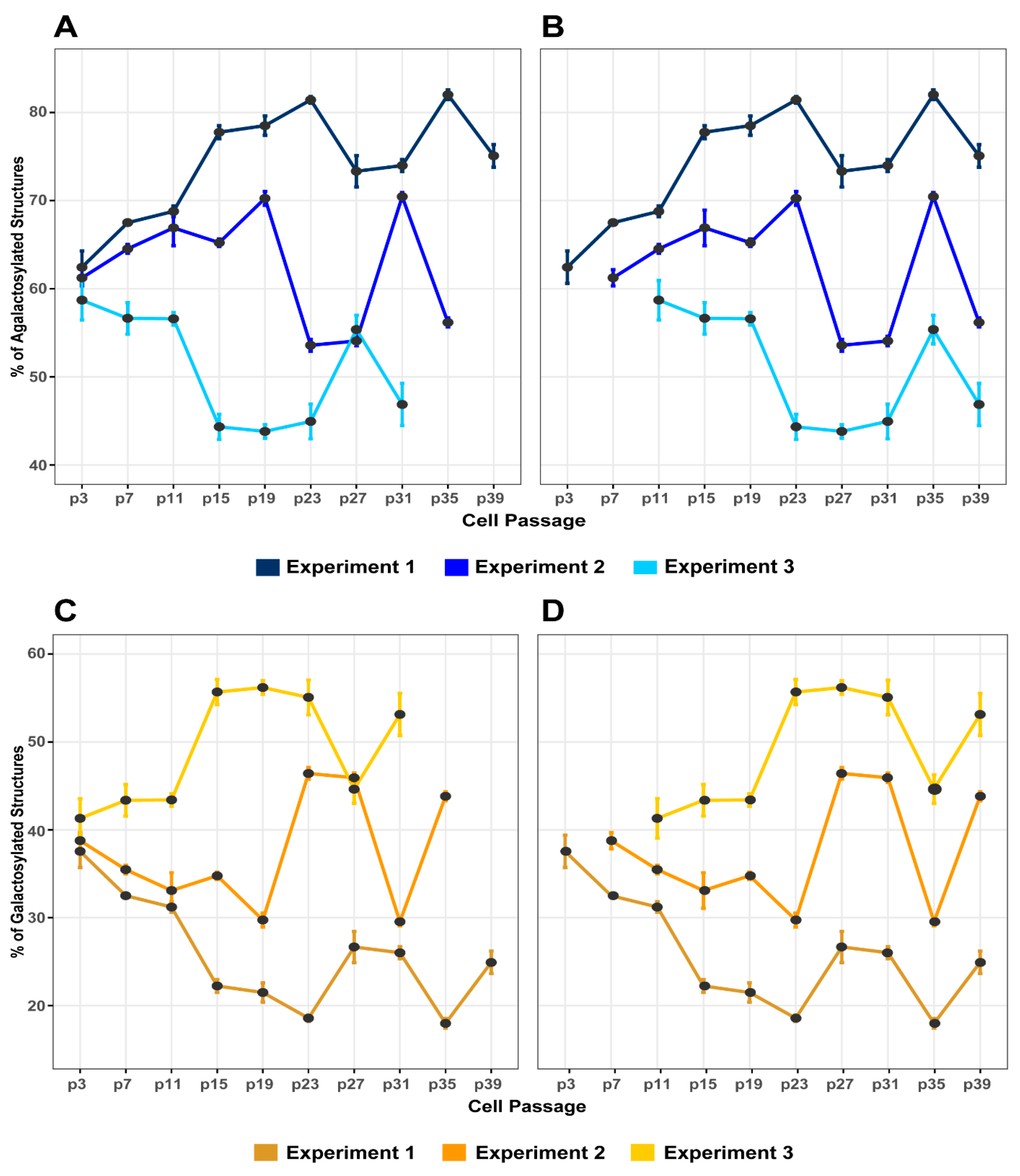
3.2. The Variability of IgG Glycome Composition during Cell Culturing Replicates between Independent Experiments
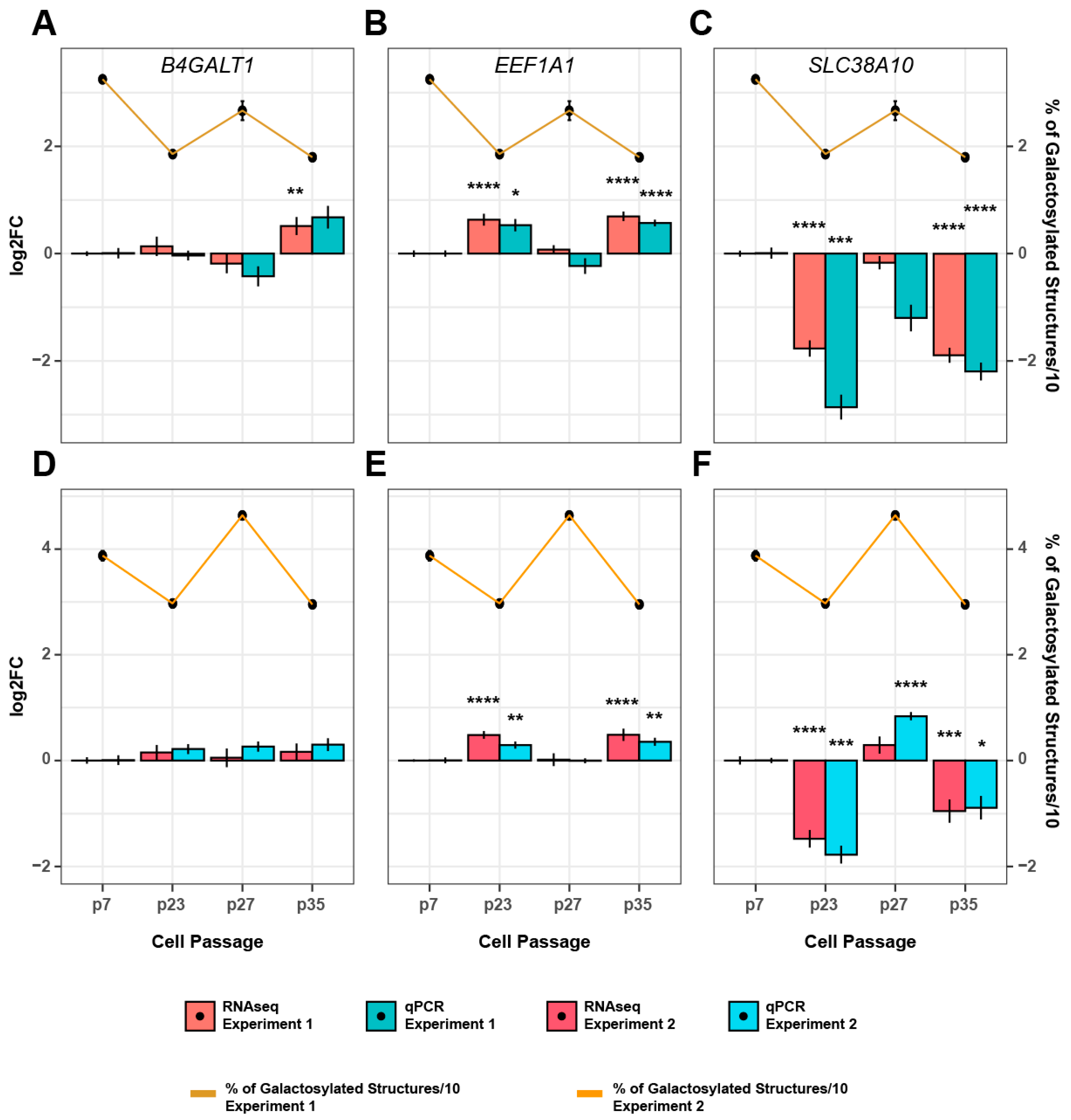
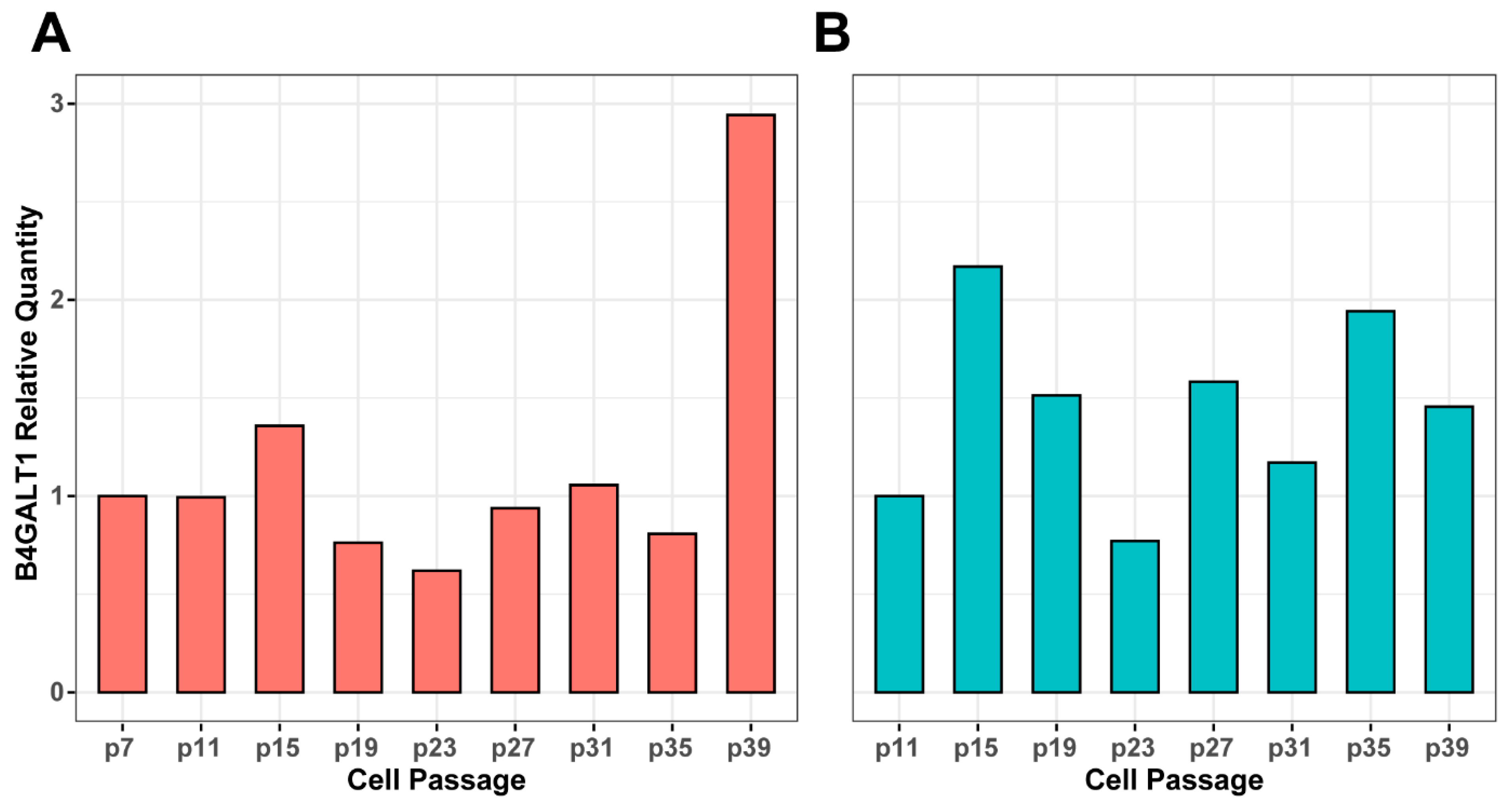
3.3. Genetic Background of IgG Glycosylation Changes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, H.S.; Qi, Y.; Im, W. Effects of N-Glycosylation on Protein Conformation and Dynamics: Protein Data Bank Analysis and Molecular Dynamics Simulation Study. Sci. Rep. 2015, 5, 8926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in Health and Disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global View of Human Protein Glycosylation Pathways and Functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Liu, W.; Li, Y.; Ma, B.; Shang, S.; Tan, Z. Impact of N-Linked Glycosylation on Therapeutic Proteins. Molecules 2022, 27, 8859. [Google Scholar] [CrossRef] [PubMed]
- Dammen-Brower, K.; Epler, P.; Zhu, S.; Bernstein, Z.J.; Stabach, P.R.; Braddock, D.T.; Spangler, J.B.; Yarema, K.J. Strategies for Glycoengineering Therapeutic Proteins. Front. Chem. 2022, 10, 863118. [Google Scholar] [CrossRef]
- Narimatsu, Y.; Büll, C.; Chen, Y.H.; Wandall, H.H.; Yang, Z.; Clausen, H. Genetic Glycoengineering in Mammalian Cells. J. Biol. Chem. 2021, 296, 100448. [Google Scholar] [CrossRef]
- Ma, B.; Guan, X.; Li, Y.; Shang, S.; Li, J.; Tan, Z. Protein Glycoengineering: An Approach for Improving Protein Properties. Front. Chem. 2020, 8, 622. [Google Scholar] [CrossRef]
- Geisler, C.; Mabashi-Asazuma, H.; Jarvis, D.L. Overview and History of Glyco-Engineering in Insect Expression Systems. Methods Mol. Biol. 2015, 1321, 131–152. [Google Scholar] [CrossRef]
- Loos, A.; Steinkellner, H. IgG-Fc Glycoengineering in Non-Mammalian Expression Hosts. Arch. Biochem. Biophys. 2012, 526, 167–173. [Google Scholar] [CrossRef] [Green Version]
- Piirainen, M.A.; de Ruijter, J.C.; Koskela, E.V.; Frey, A.D. Glycoengineering of Yeasts from the Perspective of Glycosylation Efficiency. New Biotechnol. 2014, 31, 532–537. [Google Scholar] [CrossRef] [Green Version]
- Yates, L.E.; Mills, D.C.; DeLisa, M.P. Bacterial Glycoengineering as a Biosynthetic Route to Customized Glycomolecules. Adv. Biochem. Eng. Biotechnol. 2021, 175, 167–200. [Google Scholar] [CrossRef] [PubMed]
- Malm, M.; Kuo, C.-C.; Moradi Barzadd, M.; Mebrahtu, A.; Wistbacka, N.; Razavi, R.; Volk, A.-L.; Lundqvist, M.; Kotol, D.; Edfors, F.; et al. Harnessing Secretory Pathway Differences between HEK293 and CHO to Rescue Production of Difficult to Express Proteins. Metab. Eng. 2022, 72, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.; Spearman, M. The Choice of Mammalian Cell Host and Possibilities for Glycosylation Engineering. Curr. Opin. Biotechnol. 2014, 30, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Goh, J.B.; Ng, S.K. Impact of Host Cell Line Choice on Glycan Profile. Crit. Rev. Biotechnol. 2018, 38, 851–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, L.A.; Hatton, D.; Field, R.; Dickson, A.J. Determination of Chinese Hamster Ovary Cell Line Stability and Recombinant Antibody Expression during Long-Term Culture. Biotechnol. Bioeng. 2012, 109, 2093–2103. [Google Scholar] [CrossRef] [PubMed]
- Vink, T.; Oudshoorn-Dickmann, M.; Roza, M.; Reitsma, J.J.; de Jong, R.N. A Simple, Robust and Highly Efficient Transient Expression System for Producing Antibodies. Methods 2014, 65, 5–10. [Google Scholar] [CrossRef]
- Tan, E.; Chin, C.S.H.; Lim, Z.F.S.; Ng, S.K. HEK293 Cell Line as a Platform to Produce Recombinant Proteins and Viral Vectors. Front. Bioeng. Biotechnol. 2021, 9, 796991. [Google Scholar] [CrossRef]
- Ren, W.-W.; Jin, Z.-C.; Dong, W.; Kitajima, T.; Gao, X.-D.; Fujita, M. Glycoengineering of HEK293 Cells to Produce High-Mannose-Type N-Glycan Structures. J. Biochem. 2019, 166, 245–258. [Google Scholar] [CrossRef]
- Huang, Y.F.; Aoki, K.; Akase, S.; Ishihara, M.; Liu, Y.S.; Yang, G.; Kizuka, Y.; Mizumoto, S.; Tiemeyer, M.; Gao, X.D.; et al. Global Mapping of Glycosylation Pathways in Human-Derived Cells. Dev. Cell 2021, 56, 1195–1209. [Google Scholar] [CrossRef]
- Liu, C.; Dalby, B.; Chen, W.; Kilzer, J.M.; Chiou, H.C. Transient Transfection Factors for High-Level Recombinant Protein Production in Suspension Cultured Mammalian Cells. Mol. Biotechnol. 2008, 39, 141–153. [Google Scholar] [CrossRef]
- Chiou, H.C.; Vasu, S.; Liu, C.Y.; Cisneros, I.; Jones, M.B.; Zmuda, J.F. Scalable Transient Protein Expression. Methods Mol. Biol. 2014, 1104, 35–55. [Google Scholar] [CrossRef] [PubMed]
- Hua, L.; Liu, Y.; Zhen, S.; Wan, D.; Cao, J.; Gao, X. Expression and Biochemical Characterization of Recombinant Human Epididymis Protein 4. Protein Expr. Purif. 2014, 102, 52–62. [Google Scholar] [CrossRef]
- Faravelli, S.; Campioni, M.; Palamini, M.; Canciani, A.; Chiapparino, A.; Forneris, F. Optimized Recombinant Production of Secreted Proteins Using Human Embryonic Kidney (HEK293) Cells Grown in Suspension. Bio-Protocol 2021, 11, e3998. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, G.; Plomp, R.; Koeleman, C.A.M.; Visser, R.; Von Horsten, H.H.; Sandig, V.; Rispens, T.; Wuhrer, M.; Vidarsson, G. Multi-Level Glyco-Engineering Techniques to Generate IgG with Defined Fc-Glycans. Sci. Rep. 2016, 6, 36964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.B.; Liu, C.Y.; Vasu, S.; Cisneros, I.; Chiou, H.; Zmuda, J.F. Mammalian Transient Transfection System. Genet. Eng. Biotechnol. News 2012, 32, 50–51. [Google Scholar] [CrossRef]
- Jain, N.K.; Barkowski-Clark, S.; Altman, R.; Johnson, K.; Sun, F.; Zmuda, J.; Liu, C.Y.; Kita, A.; Schulz, R.; Neill, A.; et al. A High Density CHO-S Transient Transfection System: Comparison of ExpiCHO and Expi293. Protein Expr. Purif. 2017, 134, 38–46. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, N.K.; Dwiwedi, P.; Charan, J.; Kaur, R.; Sidhu, P.; Chugh, V.K. Monoclonal Antibodies: A Review. Curr. Clin. Pharmacol. 2018, 13, 85–99. [Google Scholar] [CrossRef]
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.-J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of Therapeutic Antibodies for the Treatment of Diseases. J. Biomed. Sci. 2020, 27, 1–30. [Google Scholar] [CrossRef]
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG Subclasses and Allotypes: From Structure to Effector Functions. Front. Immunol. 2014, 5, 520. [Google Scholar] [CrossRef] [Green Version]
- de Taeye, S.W.; Rispens, T.; Vidarsson, G. The Ligands for Human IgG and Their Effector Functions. Antibodies 2019, 8, 30. [Google Scholar] [CrossRef] [Green Version]
- Cobb, B.A. The History of IgG Glycosylation and Where We Are Now. Glycobiology 2021, 30, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Shkunnikova, S.; Mijakovac, A.; Sironic, L.; Hanic, M.; Lauc, G.; Kavur, M.M. IgG Glycans in Health and Disease: Prediction, Intervention, Prognosis, and Therapy. Biotechnol. Adv. 2023, 67, 108169. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.X.; Tong, X.; Li, C.; Giddens, J.P.; Li, T. Glycoengineering of Antibodies for Modulating Functions. Annu. Rev. Biochem. 2019, 88, 433–459. [Google Scholar] [CrossRef]
- Masuda, K.; Kubota, T.; Kaneko, E.; Iida, S.; Wakitani, M.; Kobayashi-Natsume, Y.; Kubota, A.; Shitara, K.; Nakamura, K. Enhanced Binding Affinity for FcγRIIIa of Fucose-Negative Antibody Is Sufficient to Induce Maximal Antibody-Dependent Cellular Cytotoxicity. Mol. Immunol. 2007, 44, 3122–3131. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.L.; Lai, J.; Keck, R.; O’Connell, L.Y.; Hong, K.; Gloria Meng, Y.; Weikert, S.H.A.; Presta, L.G. Lack of Fucose on Human IgG1 N-Linked Oligosaccharide Improves Binding to Human FcγRIII and Antibody-Dependent Cellular Toxicity. J. Biol. Chem. 2002, 277, 26733–26740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Illidge, T.; Cheadle, E.J.; Donaghy, C.; Honeychurch, J. Update on Obinutuzumab in the Treatment of B-Cell Malignancies. Expert Opin. Biol. Ther. 2014, 14, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Jiang, L.; Pan, L.Z.; Labarre, M.J.; Anderson, D.; Reff, M. Expression of GnTIII in a Recombinant Anti-CD20 CHO Production Cell Line: Expression of Antibodies with Altered Glycoforms Leads to an Increase in ADCC through Higher Affinity for FcγRIII. Biotechnol. Bioeng. 2001, 74, 288–294. [Google Scholar] [CrossRef]
- Kaneko, Y.; Nimmerjahn, F.; Ravetch, J.V. Anti-Inflammatory Activity of Immunoglobulin G Resulting from Fc Sialylation. Science 2006, 313, 670–673. [Google Scholar] [CrossRef] [Green Version]
- Anthony, R.M.; Nimmerjahn, F.; Ashline, D.J.; Reinhold, V.N.; Paulson, J.C.; Ravetch, J.V. Recapitulation of IVIG Anti-Inflammatory Activity with a Recombinant IgG Fc. Science 2008, 320, 373–376. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Lou, Y.; Zhang, Y.; Liu, S.; Li, J.; Tao, J. Sialylated Immunoglobulin G: A Promising Diagnostic and Therapeutic Strategy for Autoimmune Diseases. Theranostics 2021, 11, 5430–5446. [Google Scholar] [CrossRef]
- Krištić, J.; Vučković, F.; Menni, C.; Klarić, L.; Keser, T.; Beceheli, I.; Pučić-Baković, M.; Novokmet, M.; Mangino, M.; Thaqi, K.; et al. Glycans Are a Novel Biomarker of Chronological and Biological Ages. J. Gerontol.-Ser. A Biol. Sci. Med. Sci. 2014, 69, 779–789. [Google Scholar] [CrossRef] [Green Version]
- Gudelj, I.; Lauc, G.; Pezer, M. Immunoglobulin G Glycosylation in Aging and Diseases. Cell. Immunol. 2018, 333, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Jain, R.; Budholiya, N.; Rathore, A.S. Long-Term Culturing of CHO Cells: Phenotypic Drift and Quality Attributes of the Expressed Monoclonal Antibody. Biotechnol. Lett. 2023, 45, 357–370. [Google Scholar] [CrossRef]
- Mijakovac, A.; Miškec, K.; Krištić, J.; Vičić Bočkor, V.; Tadić, V.; Bošković, M.; Lauc, G.; Zoldoš, V.; Vojta, A. A Transient Expression System with Stably Integrated CRISPR-DCas9 Fusions for Regulation of Genes Involved in Immunoglobulin G Glycosylation. Cris. J. 2022, 5, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. Image to ImageJ: 25 Years of Image Analysis. Nature Methods. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Harvey, D.J.; Merry, A.H.; Royle, L.; Campbell, M.P.; Dwek, R.A.; Rudd, P.M. Proposal for a Standard System for Drawing Structural Diagrams of N- and O-Linked Carbohydrates and Related Compounds. Proteomics 2009, 9, 3796–3801. [Google Scholar] [CrossRef]
- Lauc, G.; Huffman, J.E.; Pučić, M.; Zgaga, L.; Adamczyk, B.; Mužinić, A.; Novokmet, M.; Polašek, O.; Gornik, O.; Krištić, J.; et al. Loci Associated with N-Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers. PLoS Genet. 2013, 9, e1003225. [Google Scholar] [CrossRef]
- Klarić, L.; Tsepilov, Y.A.; Stanton, C.M.; Mangino, M.; Sikka, T.T.; Esko, T.; Pakhomov, E.; Salo, P.; Deelen, J.; McGurnaghan, S.J.; et al. Glycosylation of Immunoglobulin G Is Regulated by a Large Network of Genes Pleiotropic with Inflammatory Diseases. Sci. Adv. 2020, 6, eaax0301. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Klarić, L.; Sharapov, S.; Mangino, M.; Ning, Z.; Wu, D.; Trbojević-Akmačić, I.; Pučić-Baković, M.; Rudan, I.; Polašek, O.; et al. Multivariate Discovery and Replication of Five Novel Loci Associated with Immunoglobulin G N-Glycosylation. Nat. Commun. 2017, 8, 447. [Google Scholar] [CrossRef] [Green Version]
- Shadrina, A.S.; Zlobin, A.S.; Zaytseva, O.O.; Klarić, L.; Sharapov, S.Z.; Pakhomov, E.D.; Perola, M.; Esko, T.; Hayward, C.; Wilson, J.F.; et al. Multivariate Genome-Wide Analysis of Immunoglobulin G N-Glycosylation Identifies New Loci Pleiotropic with Immune Function. Hum. Mol. Genet. 2021, 30, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Frkatović-Hodžić, A.; Miškec, K.; Mijakovac, A.; Nostaeva, A.; Sharapov, S.Z.; Landini, A.; Haller, T.; van den Akker, E.; Sharma, S.; Cuadrat, R.R.C.; et al. Mapping of the Gene Network That Regulates Glycan Clock of Aging. medRxiv 2023. [Google Scholar] [CrossRef]
- Štambuk, J.; Nakić, N.; Vučković, F.; Pučić-Baković, M.; Razdorov, G.; Trbojević-Akmačić, I.; Novokmet, M.; Keser, T.; Vilaj, M.; Štambuk, T.; et al. Global Variability of the Human IgG Glycome. Aging (Albany N. Y.) 2020, 12, 15222–15259. [Google Scholar] [CrossRef] [PubMed]
- Pučić, M.; Knežević, A.; Vidič, J.; Adamczyk, B.; Novokmet, M.; Polašek, O.; Gornik, O.; Šupraha-Goreta, S.; Wormald, M.R.; Redžic, I.; et al. High Throughput Isolation and Glycosylation Analysis of IgG-Variability and Heritability of the IgG Glycome in Three Isolated Human Populations. Mol. Cell. Proteom. 2011, 10, M111.010090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivarsson, M.; Villiger, T.K.; Morbidelli, M.; Soos, M. Evaluating the Impact of Cell Culture Process Parameters on Monoclonal Antibody N-Glycosylation. J. Biotechnol. 2014, 188, 88–96. [Google Scholar] [CrossRef]
- Ehret, J.; Zimmermann, M.; Eichhorn, T.; Zimmer, A. Impact of Cell Culture Media Additives on IgG Glycosylation Produced in Chinese Hamster Ovary Cells. Biotechnol. Bioeng. 2019, 116, 816–830. [Google Scholar] [CrossRef] [Green Version]
- Majid, F.A.A.; Butler, M.; Al-Rubeai, M. Glycosylation of an Immunoglobulin Produced From a Murine Hybridoma Cell Line: The Effect of Culture Mode and the Anti-Apoptotic Gene, Bcl-2. Biotechnol. Bioeng. 2007, 97, 156–169. [Google Scholar] [CrossRef]
- Hossler, P.; Khattak, S.F.; Li, Z.J. Optimal and Consistent Protein Glycosylation in Mammalian Cell Culture. Glycobiology 2009, 19, 936–949. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wang, J.; Fan, L.; Chen, X.; Hu, D.; Deng, X.; Fai Poon, H.; Wang, H.; Liu, X.; Tan, W.-S. Galactose Supplementation Enhance Sialylation of Recombinant Fc-Fusion Protein in CHO Cell: An Insight into the Role of Galactosylation in Sialylation. World J. Microbiol. Biotechnol. 2015, 31, 1147–1156. [Google Scholar] [CrossRef]
- Pacis, E.; Yu, M.; Autsen, J.; Bayer, R.; Li, F. Effects of Cell Culture Conditions on Antibody N-Linked Glycosylation-What Affects High Mannose 5 Glycoform. Biotechnol. Bioeng. 2011, 108, 2348–2358. [Google Scholar] [CrossRef]
- Grainger, R.K.; James, D.C. CHO Cell Line Specific Prediction and Control of Recombinant Monoclonal Antibody N-Glycosylation. Biotechnol. Bioeng. 2013, 110, 2970–2983. [Google Scholar] [CrossRef]
- Krištić, J.; Lauc, G.; Pezer, M. Immunoglobulin G Glycans—Biomarkers and Molecular Effectors of Aging. Clin. Chim. Acta 2022, 535, 30–45. [Google Scholar] [CrossRef]
- Taniguchi, N.; Honke, K.; Fukuda, M.; Narimatsu, H.; Yamaguchi, Y.; Angata, T. Handbook of Glycosyltransferases and Related Genes, 2nd ed.; Taniguchi, N., Honke, K., Fukuda, M., Narimatsu, H., Yamaguchi, Y., Angata, T., Eds.; Springer: Tokyo, Japan, 2014; Volumes 1–2, ISBN 9784431542407. [Google Scholar]
- Mijakovac, A.; Jurić, J.; Kohrt, W.M.; Krištić, J.; Kifer, D.; Gavin, K.M.; Miškec, K.; Frkatović, A.; Vučković, F.; Pezer, M.; et al. Effects of Estradiol on Immunoglobulin G Glycosylation: Mapping of the Downstream Signaling Mechanism. Front. Immunol. 2021, 12, 680227. [Google Scholar] [CrossRef] [PubMed]
- Wahl, A.; van den Akker, E.; Klaric, L.; Štambuk, J.; Benedetti, E.; Plomp, R.; Razdorov, G.; Trbojević-Akmačić, I.; Deelen, J.; van Heemst, D.; et al. Genome-Wide Association Study on Immunoglobulin G Glycosylation Patterns. Front. Immunol. 2018, 9, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dekkers, G.; Rispens, T.; Vidarsson, G. Novel Concepts of Altered Immunoglobulin G Galactosylation in Autoimmune Diseases. Front. Immunol. 2018, 9, 553. [Google Scholar] [CrossRef] [Green Version]
- van Osch, T.L.J.; Nouta, J.; Derksen, N.I.L.; van Mierlo, G.; van der Schoot, C.E.; Wuhrer, M.; Rispens, T.; Vidarsson, G. Fc Galactosylation Promotes Hexamerization of Human IgG1, Leading to Enhanced Classical Complement Activation. J. Immunol. 2021, 207, 1545–1554. [Google Scholar] [CrossRef]
- Dekkers, G.; Treffers, L.; Plomp, R.; Bentlage, A.E.H.; de Boer, M.; Koeleman, C.A.M.; Lissenberg-Thunnissen, S.N.; Visser, R.; Brouwer, M.; Mok, J.Y.; et al. Decoding the Human Immunoglobulin G-Glycan Repertoire Reveals a Spectrum of Fc-Receptor- and Complement-Mediated-Effector Activities. Front. Immunol. 2017, 8, 877. [Google Scholar] [CrossRef]
- Peschke, B.; Keller, C.W.; Weber, P.; Quast, I.; Lünemann, J.D. Fc-Galactosylation of Human Immunoglobulin Gamma Isotypes Improves C1q Binding and Enhances Complement-Dependent Cytotoxicity. Front. Immunol. 2017, 8, 646. [Google Scholar] [CrossRef] [Green Version]
- Wei, B.; Gao, X.; Cadang, L.; Izadi, S.; Liu, P.; Zhang, H.-M.; Hecht, E.; Shim, J.; Magill, G.; Pabon, J.R.; et al. Fc Galactosylation Follows Consecutive Reaction Kinetics and Enhances Immunoglobulin G Hexamerization for Complement Activation. MAbs 2021, 13, 1893427. [Google Scholar] [CrossRef]
- Nairn, A.V.; York, W.S.; Harris, K.; Hall, E.M.; Pierce, J.M.; Moremen, K.W. Regulation of Glycan Structures in Animal Tissues: Transcript Profiling of Glycan-Related Genes. J. Biol. Chem. 2008, 283, 17298–17313. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.T.B.; Lin, J.; Tay, S.J.; Mariati; Yeo, J.; Nguyen-Khuong, T.; Yang, Y. Multiplexed Engineering Glycosyltransferase Genes in CHO Cells via Targeted Integration for Producing Antibodies with Diverse Complex-Type N-Glycans. Sci. Rep. 2021, 11, 12969. [Google Scholar] [CrossRef] [PubMed]
- Timoshchuk, A.; Sharapov, S.; Aulchenko, Y.S. Twelve Years of Genome-Wide Association Studies of Human Protein N-Glycosylation. Engineering 2023, in press. [Google Scholar] [CrossRef]
- Schulz, I.; Engel, C.; Niestroj, A.J.; Kehlen, A.; Rahfeld, J.-U.; Kleinschmidt, M.; Lehmann, K.; Roßner, S.; Demuth, H.-U. A Non-Canonical Function of Eukaryotic Elongation Factor 1A1: Regulation of Interleukin-6 Expression. Biochim. Biophys. Acta 2014, 1843, 965–975. [Google Scholar] [CrossRef]
- Abbas, W.; Kumar, A.; Herbein, G. The EEF1A Proteins: At the Crossroads of Oncogenesis, Apoptosis, and Viral Infections. Front. Oncol. 2015, 5, 75. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Wu, X.; Zhang, X.; Chen, C.; Chen, H.; She, F. CagA Orchestrates EEF1A1 and PKCδ to Induce Interleukin-6 Expression in Helicobacter Pylori-Infected Gastric Epithelial Cells. Gut Pathog. 2020, 12, 31. [Google Scholar] [CrossRef] [PubMed]
- Migliaccio, N.; Martucci, N.M.; Ruggiero, I.; Sanges, C.; Ohkubo, S.; Lamberti, A.; Agostinelli, E.; Arcari, P. Ser/Thr Kinases and Polyamines in the Regulation of Non-Canonical Functions of Elongation Factor 1A. Amino Acids 2016, 48, 2339–2352. [Google Scholar] [CrossRef] [PubMed]
| Experiment 1 | Experiment 2 | |||||
|---|---|---|---|---|---|---|
| Gene | 7–23 | 23–27 | 27–35 | 7–23 | 23–27 | 27–35 |
| SAT1 | 3.789 | 0.558 | 1.397 | 3.432 | 0.194 | 4.102 |
| INSIG1 | 2.683 | 0.391 | 2.455 | 2.352 | 0.365 | 2.253 |
| PIM1 | 3.354 | 0.434 | 1.727 | 2.627 | 0.325 | 2.389 |
| CRYAB | 10.416 | 0.190 | 2.582 | 6.680 | 0.104 | 5.267 |
| GOLT1B | 2.937 | 0.390 | 1.771 | 2.173 | 0.477 | 1.843 |
| FUS | 0.405 | 1.792 | 0.430 | 0.440 | 2.487 | 0.555 |
| LINC00342 | 0.155 | 3.137 | 0.237 | 0.355 | 2.437 | 0.427 |
| APPAT | 0.204 | 2.907 | 0.231 | 0.281 | 3.986 | 0.295 |
| PLAU | 3.276 | 0.422 | 1.560 | 2.554 | 0.271 | 2.251 |
| MT-CYB | 0.230 | 3.167 | 0.185 | 0.417 | 2.555 | 0.480 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lukšić, F.; Mijakovac, A.; Josipović, G.; Vičić Bočkor, V.; Krištić, J.; Cindrić, A.; Vinicki, M.; Rokić, F.; Vugrek, O.; Lauc, G.; et al. Long-Term Culturing of FreeStyle 293-F Cells Affects Immunoglobulin G Glycome Composition. Biomolecules 2023, 13, 1245. https://doi.org/10.3390/biom13081245
Lukšić F, Mijakovac A, Josipović G, Vičić Bočkor V, Krištić J, Cindrić A, Vinicki M, Rokić F, Vugrek O, Lauc G, et al. Long-Term Culturing of FreeStyle 293-F Cells Affects Immunoglobulin G Glycome Composition. Biomolecules. 2023; 13(8):1245. https://doi.org/10.3390/biom13081245
Chicago/Turabian StyleLukšić, Fran, Anika Mijakovac, Goran Josipović, Vedrana Vičić Bočkor, Jasminka Krištić, Ana Cindrić, Martina Vinicki, Filip Rokić, Oliver Vugrek, Gordan Lauc, and et al. 2023. "Long-Term Culturing of FreeStyle 293-F Cells Affects Immunoglobulin G Glycome Composition" Biomolecules 13, no. 8: 1245. https://doi.org/10.3390/biom13081245
APA StyleLukšić, F., Mijakovac, A., Josipović, G., Vičić Bočkor, V., Krištić, J., Cindrić, A., Vinicki, M., Rokić, F., Vugrek, O., Lauc, G., & Zoldoš, V. (2023). Long-Term Culturing of FreeStyle 293-F Cells Affects Immunoglobulin G Glycome Composition. Biomolecules, 13(8), 1245. https://doi.org/10.3390/biom13081245






