Aggregation of an Amyloidogenic Peptide on Gold Surfaces
Abstract
:1. Introduction
2. Model and Methodology
3. Results
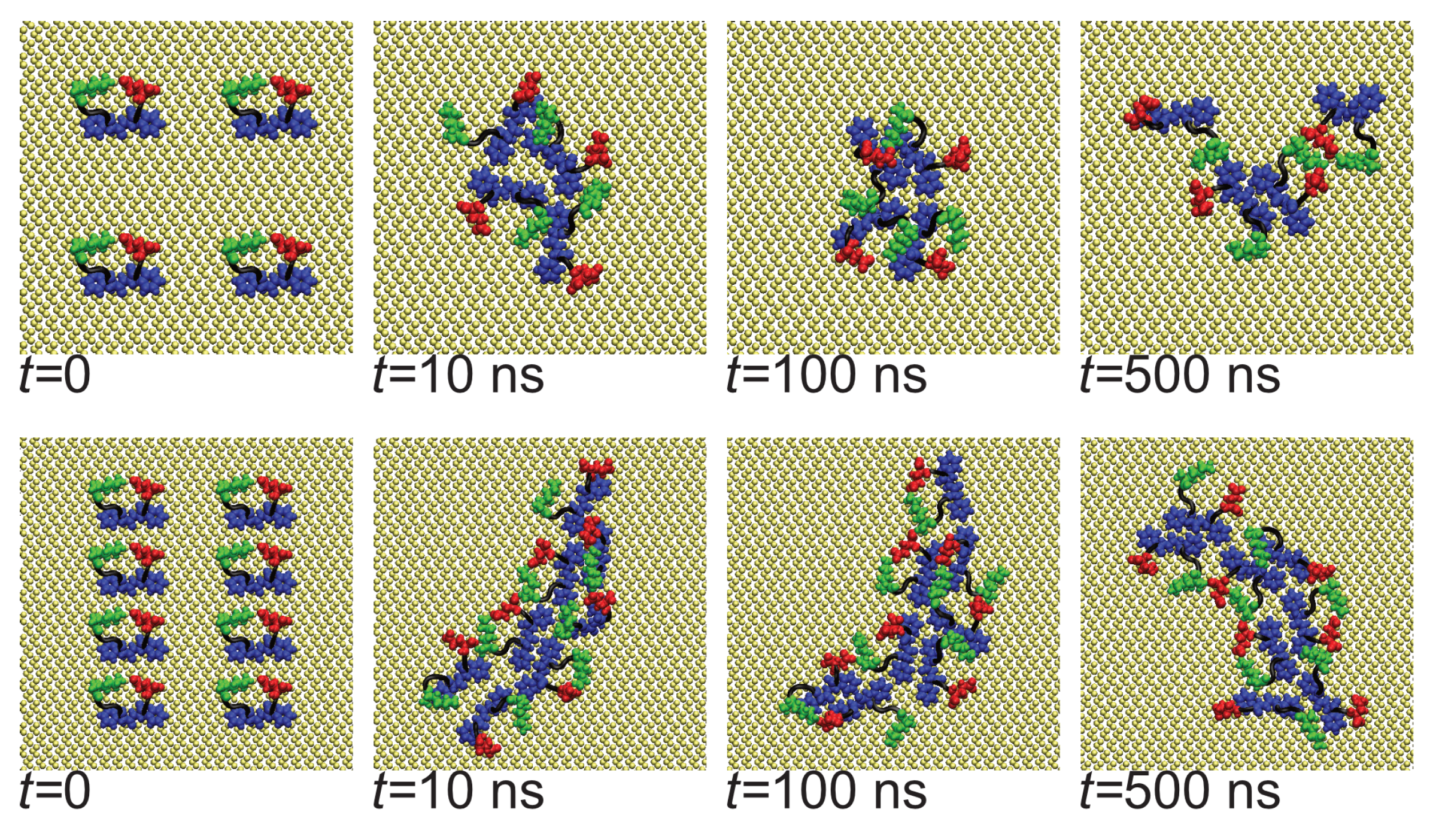
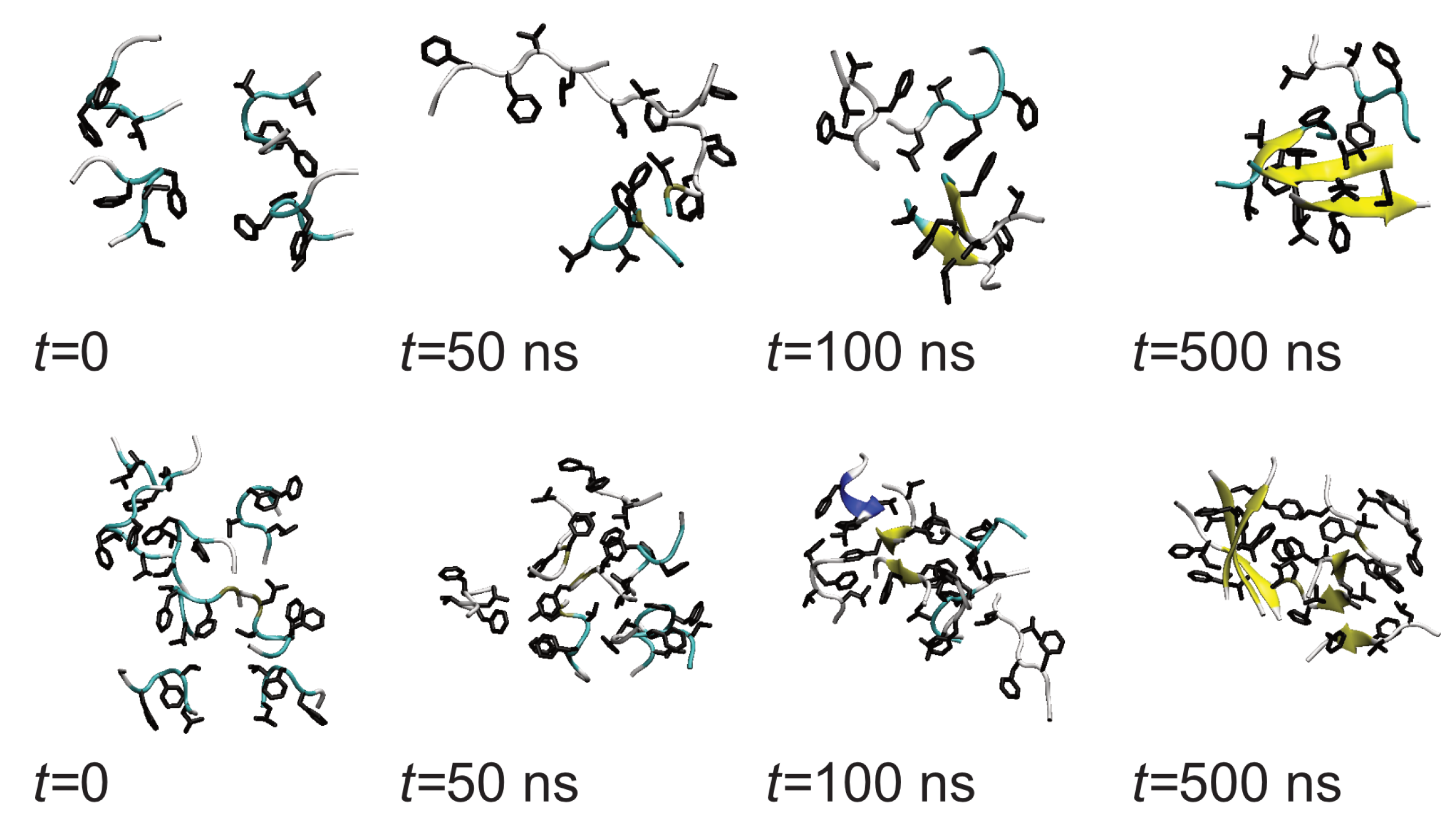
3.1. Oligomerization of A(16–22) on Au111 Surfaces and in Solution
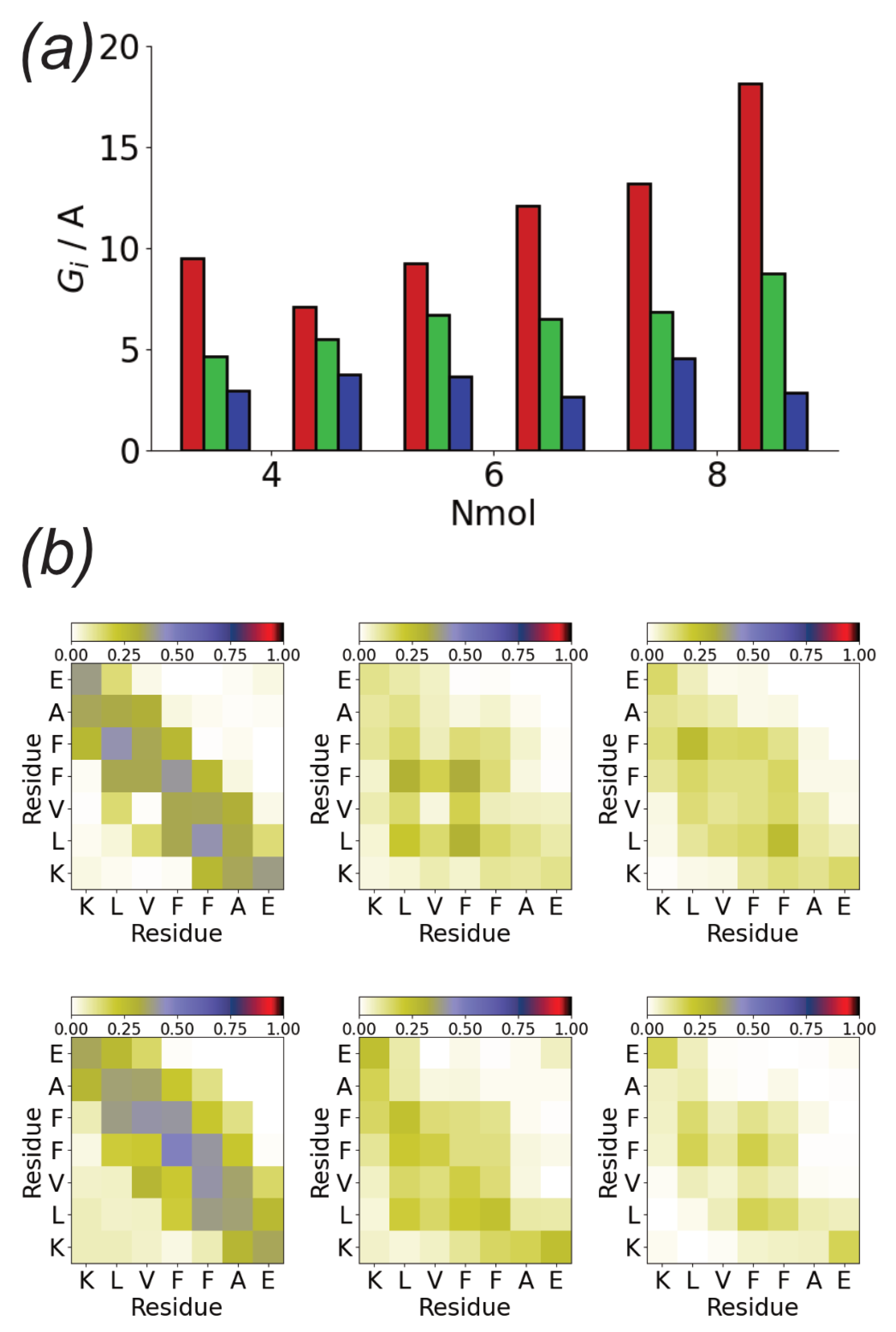
3.2. Surfaces Drive Formation of Linear Aggregates
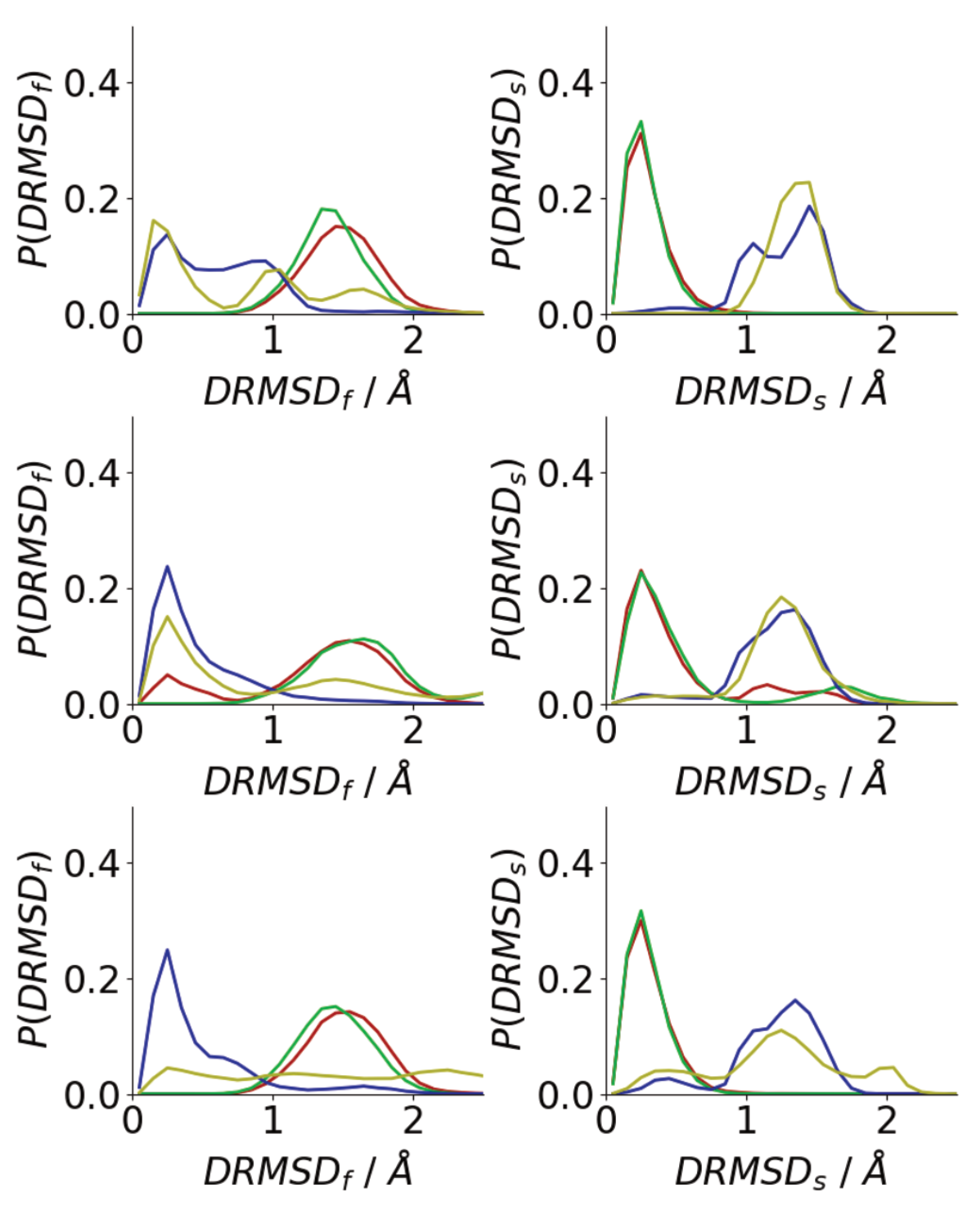
3.3. Oligomerization of A(16–22) on Surfaces Does Not Favour Formation of Fibril-Like Conformations
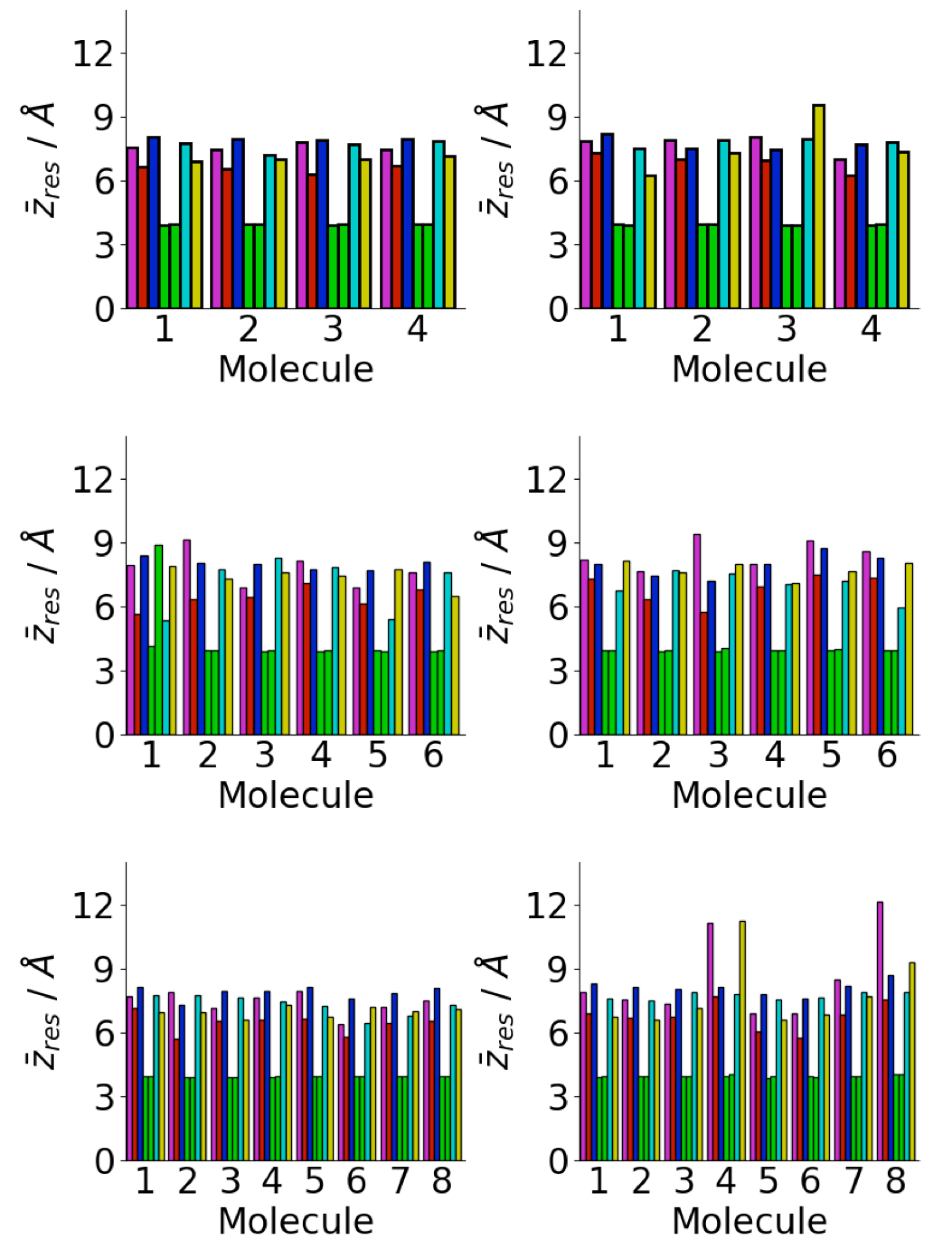
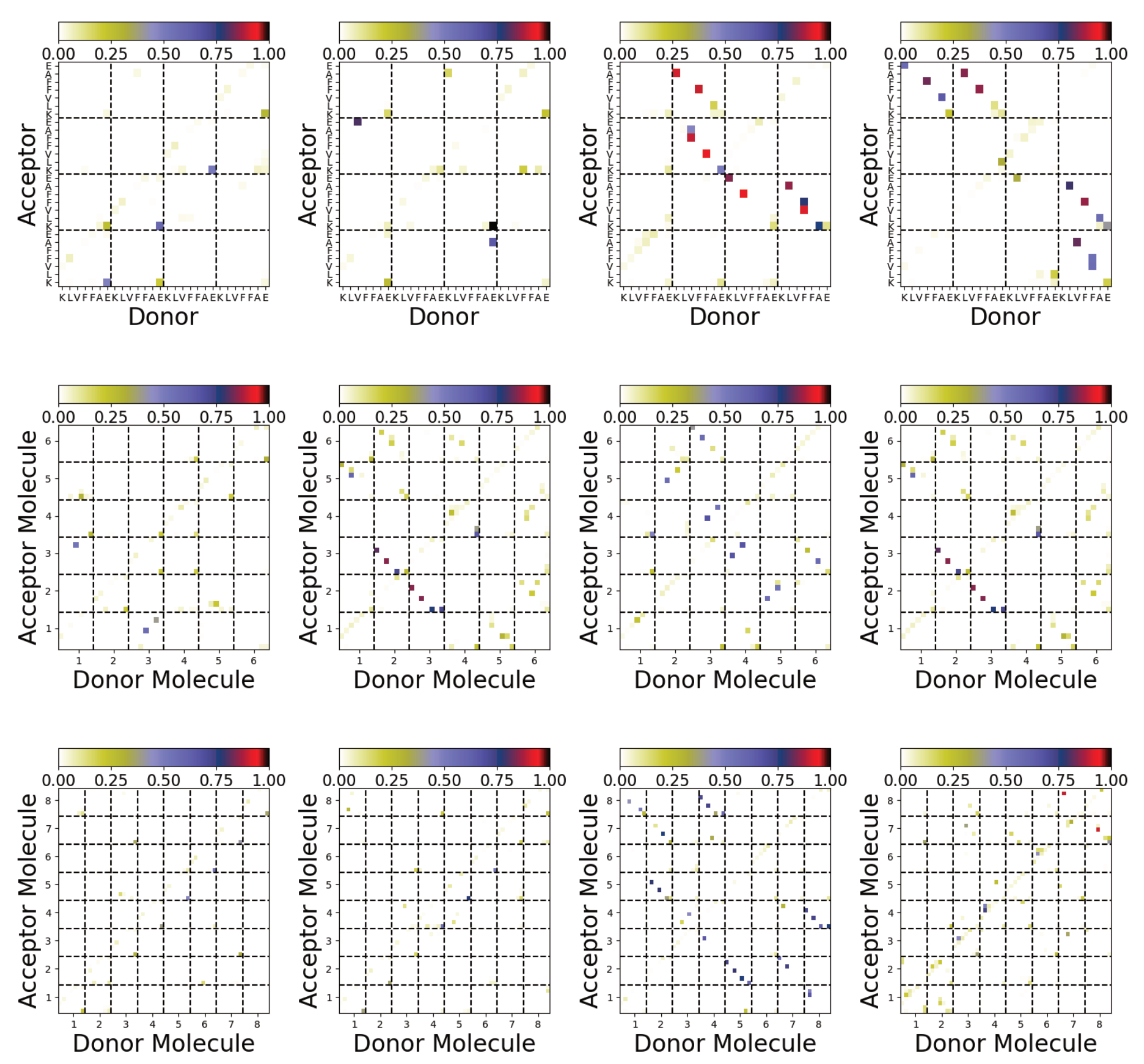
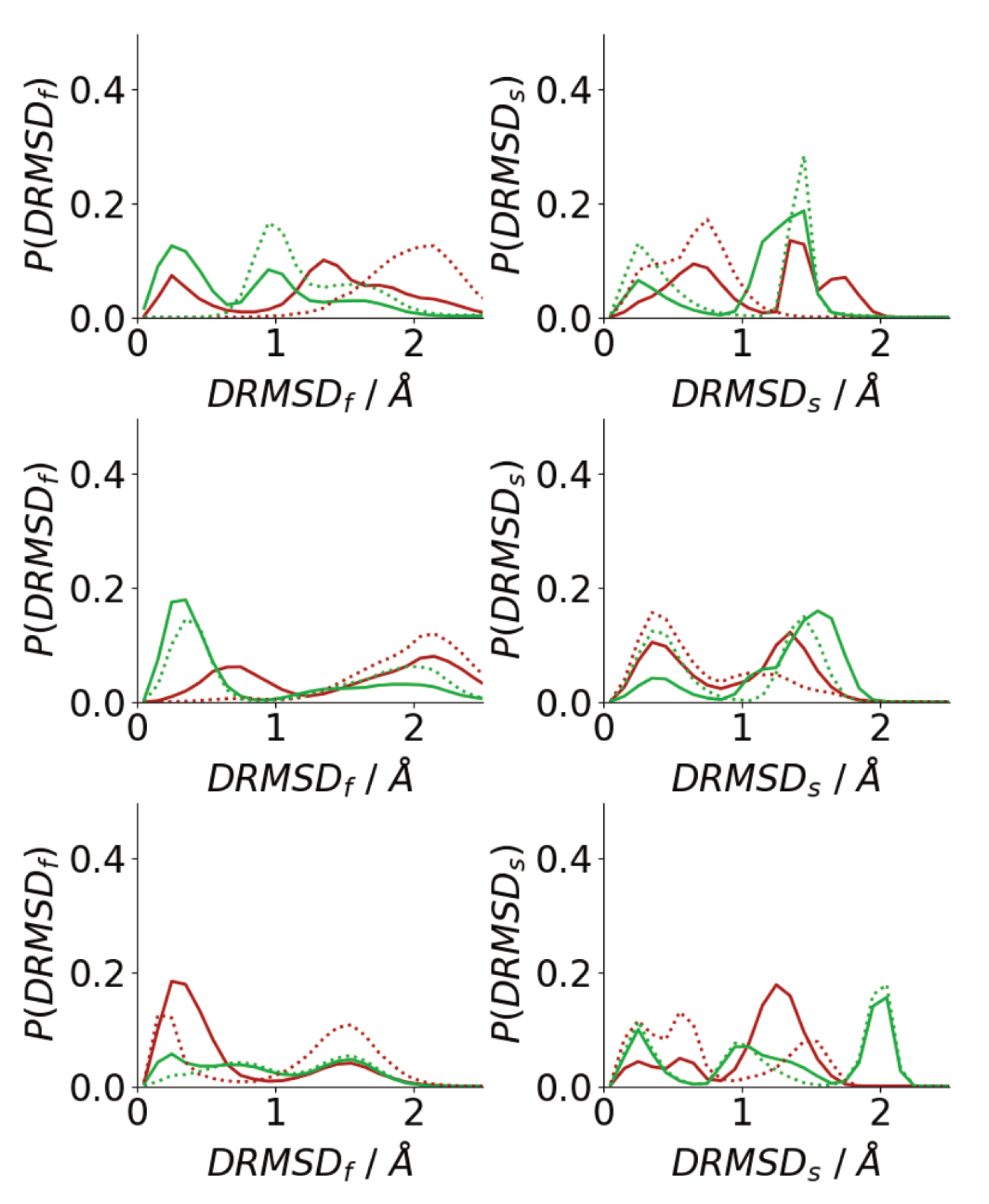
3.4. Difference between Oligomers Formed on Surface and Adsorbed from Solution
4. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Time Evolution of Oligomer Size Distributions

References
- Ke, P.C.; Zhou, R.; Serpell, L.C.; Riek, R.; Knowles, T.P.J.; Lashuel, H.A.; Gazit, E.; Hamley, I.W.; Davis, T.P.; Fändrich, M.; et al. Half a Century of Amyloids: Past, Present and Future. Chem. Soc. Rev. 2020, 49, 5473–5509. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G. A Century of Alzheimer’s Disease. Science 2006, 314, 777–781. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The Amyloid State and Its Association with Protein Misfolding Diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Sacchettini, J.C.; Kelly, J.W. Therapeutic Strategies for Human Amyloid Diseases. Nat. Rev. Drug Discov. 2002, 1, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Mezzenga, R. Amyloid Fibrils as Building Blocks for Natural and Artificial Functional Materials. Adv. Mater. 2016, 28, 6546–6561. [Google Scholar] [CrossRef] [PubMed]
- Gebbink, M.F.; Claessen, D.; Bouma, B.; Dijkhuizen, L.; Wösten, H.A.B. Amyloids—A Functional Coat for Microorganisms. Nat. Rev. Microbiol. 2005, 3, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Erskine, E.E.; MacPhee, C.E.; Stanley-Wall, N.R. Functional Amyloid and Other Protein Fibres in the Biofilm Matrix. J. Mol. Biol. 2018, 430, 3642–3656. [Google Scholar] [CrossRef]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.R.; Simon, R.; Schubert, D.; et al. Functional Amyloids As Natural Storage of Peptide Hormones in Pituitary Secretory Granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef]
- Li, C.; Qin, R.; Liu, R.; Miao, S.; Yang, P. Functional Amyloid Materials at Surfaces/Interfaces. Biomater. Sci. 2018, 6, 462–472. [Google Scholar] [CrossRef]
- Kasai, S.; Ohga, Y.; Mochizuki, M.; Nishi, N.; Kadoya, Y.; Nomizu, M. Multifunctional Peptide Fibrils for Biomedical Materials. Biopolym. Pept. Sci. Sect. 2004, 76, 27–33. [Google Scholar] [CrossRef]
- Liang, Y.; Guo, P.; Pingali, S.V.; Pabit, S.; Thiyagarajan, P.; Berland, K.M.; Lynn, D.G. Light Harvesting Antenna on an Amyloid Scaffold. Chem. Commun. 2008, 6522–6524. [Google Scholar] [CrossRef]
- Burke, K.A.; Yates, E.A.; Legleiter, J. Biophysical Insights into How Surfaces, Including Lipid Membranes, Modulate Protein Aggregation Related to Neurodegeneration. Front. Neurol. 2013, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Grundmeier, G. Amyloid Aggregation at Solid-Liquid Interfaces: Perspectives of Studies Using Model Surfaces. Appl. Surf. Sci. 2020, 506, 144991. [Google Scholar] [CrossRef]
- Linse, S.; Cabaleiro-Lago, C.; Xue, W.F.; Lynch, I.; Lindman, S.; Thulin, E.; Radford, S.E.; Dawson, K.A. Nucleation of Protein Fibrillation by Nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 8691–8696. [Google Scholar] [CrossRef] [PubMed]
- John, T.; Gladytz, A.; Kubeil, C.; Martin, L.L.; Risselada, H.J.; Abel, B. Impact of Nanoparticles on Amyloid Peptide and Protein Aggregation: A Review with a Focus on Gold Nanoparticles. Nanoscale 2018, 10, 20894–20913. [Google Scholar] [CrossRef] [PubMed]
- Jean, L.; Lee, C.F.; Lee, C.; Shaw, M.; Vaux, D.J. Competing Discrete Interfacial Effects Are Critical for Amyloidogenesis. FASEB J. 2010, 24, 309–317. [Google Scholar] [CrossRef]
- Auer, S.; Trovato, A.; Vendruscolo, M. A Condensation-Ordering Mechanism in Nanoparticle-Catalyzed Peptide Aggregation. PLoS Comput. Biol. 2009, 5, e1000458. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Lynch, I.; Ejtehadi, M.R.; Monopoli, M.P.; Bombelli, F.B.; Laurent, S. Protein-Nanoparticle Interactions: Opportunities and Challenges. Chem. Rev. 2011, 111, 5610–5637. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Kalhor, H.R.; Laurent, S.; Lynch, I. Protein Fibrillation and Nanoparticle Interactions: Opportunities and Challenges. Nanoscale 2013, 5, 2570. [Google Scholar] [CrossRef]
- Shen, L.; Adachi, T.; Vanden Bout, D.; Zhu, X.Y. A Mobile Precursor Determines Amyloid-β Peptide Fibril Formation at Interfaces. J. Am. Chem. Soc. 2012, 134, 14172–14178. [Google Scholar] [CrossRef]
- Hajiraissi, R.; Hanke, M.; Yang, Y.; Duderija, B.; Gonzalez Orive, A.; Grundmeier, G.; Keller, A. Adsorption and Fibrillization of Islet Amyloid Polypeptide at Self-Assembled Monolayers Studied by QCM-D, AFM, and PM-IRRAS. Langmuir 2018, 34, 3517–3524. [Google Scholar] [CrossRef] [PubMed]
- Cheung, D.L. Effect of Surface Chemistry on Islet Amyloid Polypeptide Conformation. Biointerphases 2020, 15, 051001. [Google Scholar] [CrossRef] [PubMed]
- Vácha, R.; Linse, S.; Lund, M. Surface Effects on Aggregation Kinetics of Amyloidogenic Peptides. J. Am. Chem. Soc. 2014, 136, 11776–11782. [Google Scholar] [CrossRef]
- Radic, S.; Davis, T.P.; Ke, P.C.; Ding, F. Contrasting Effects of Nanoparticle–Protein Attraction on Amyloid Aggregation. RSC Adv. 2015, 5, 105489–105498. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Xu, L.; Thompson, D. Revisiting the Earliest Signatures of Amyloidogenesis: Roadmaps Emerging from Computational Modeling and Experiment. WIREs Comput. Mol. Sci. 2018, 8, e1359. [Google Scholar] [CrossRef]
- Close, W.; Neumann, M.; Schmidt, A.; Hora, M.; Annamalai, K.; Schmidt, M.; Reif, B.; Schmidt, V.; Grigorieff, N.; Fändrich, M. Physical Basis of Amyloid Fibril Polymorphism. Nat. Commun. 2018, 9, 699. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.M.; Selkoe, D.J. Aβ Oligomers—A Decade of Discovery. J. Neurochem. 2007, 101, 1172–1184. [Google Scholar] [CrossRef]
- Taneja, V.; Verma, M.; Vats, A. Toxic Species in Amyloid Disorders: Oligomers or Mature Fibrils. Ann. Indian Acad. Neurol. 2015, 18, 138. [Google Scholar] [CrossRef]
- Hervás, R.; Oroz, J.; Galera-Prat, A.; Goñi, O.; Valbuena, A.; Vera, A.M.; Gómez-Sicilia, À.; Losada-Urzáiz, F.; Uversky, V.N.; Menéndez, M.; et al. Common Features at the Start of the Neurodegeneration Cascade. PLoS Biol. 2012, 10, e1001335. [Google Scholar] [CrossRef]
- Nagel-Steger, L.; Owen, M.C.; Strodel, B. An Account of Amyloid Oligomers: Facts and Figures Obtained from Experiments and Simulations. ChemBioChem 2016, 17, 657–676. [Google Scholar] [CrossRef]
- Ozboyaci, M.; Kokh, D.B.; Corni, S.; Wade, R.C. Modeling and Simulation of Protein-Surface Interactions: Achievements and Challenges. Q. Rev. Biophys. 2016, 49, 1–45. [Google Scholar] [CrossRef]
- Walsh, T.R. Pathways to Structure-Property Relationships of Peptide-Materials Interfaces: Challenges in Predicting Molecular Structures. Accounts Chem. Res. 2017, 50, 1617–1624. [Google Scholar] [CrossRef]
- Brancolini, G.; Corazza, A.; Vuano, M.; Fogolari, F.; Mimmi, M.C.; Bellotti, V.; Stoppini, M.; Corni, S.; Esposito, G. Probing the Influence of Citrate-Capped Gold Nanoparticles on an Amyloidogenic Protein. ACS Nano 2015, 9, 2600–2613. [Google Scholar] [CrossRef]
- Bellucci, L.; Ardèvol, A.; Parrinello, M.; Lutz, H.; Lu, H.; Weidner, T.; Corni, S. The Interaction with Gold Suppresses Fiber-like Conformations of the Amyloid β (16–22) Peptide. Nanoscale 2016, 8, 8737–8748. [Google Scholar] [CrossRef] [PubMed]
- Gladytz, A.; Abel, B.; Risselada, H.J. Gold-Induced Fibril Growth: The Mechanism of Surface-Facilitated Amyloid Aggregation. Angew. Chem. Int. Ed. 2016, 55, 11242–11246. [Google Scholar] [CrossRef] [PubMed]
- Bellucci, L.; Bussi, G.; Di Felice, R.; Corni, S. Fibrillation-Prone Conformations of the Amyloid-β-42 Peptide at the Gold/Water Interface. Nanoscale 2017, 9, 2279–2290. [Google Scholar] [CrossRef] [PubMed]
- Soltani, N.; Gholami, M.R. Increase in the β-Sheet Character of an Amyloidogenic Peptide upon Adsorption onto Gold and Silver Surfaces. ChemPhysChem 2017, 18, 526–536. [Google Scholar] [CrossRef]
- Wang, S.T.; Lin, Y.; Todorova, N.; Xu, Y.; Mazo, M.; Rana, S.; Leonardo, V.; Amdursky, N.; Spicer, C.D.; Alexander, B.D.; et al. Facet-Dependent Interactions of Islet Amyloid Polypeptide with Gold Nanoparticles: Implications for Fibril Formation and Peptide-Induced Lipid Membrane Disruption. Chem. Mater. 2017, 29, 1550–1560. [Google Scholar] [CrossRef]
- Tavanti, F.; Pedone, A.; Menziani, M.C. Disclosing the Interaction of Gold Nanoparticles with Aβ(1–40) Monomers through Replica Exchange Molecular Dynamics Simulations. Int. J. Mol. Sci. 2020, 22, 26. [Google Scholar] [CrossRef]
- Somers, K.P.; Cheung, D.L. The Amyloidogenic Peptide Amyloid Beta(16–22) Displays Facet Dependent Conformation on Metal Surfaces. Biophysica 2022, 2, 135–153. [Google Scholar] [CrossRef]
- Earl, D.J.; Deem, M.W. Parallel Tempering: Theory, Applications, and New Perspectives. Phys. Chem. Chem. Phys. 2005, 7, 3910–3916. [Google Scholar] [CrossRef] [PubMed]
- Laio, A.; Gervasio, F.L. Metadynamics: A Method to Simulate Rare Events and Reconstruct the Free Energy in Biophysics, Chemistry and Material Science. Rep. Prog. Phys. 2008, 71, 126601. [Google Scholar] [CrossRef]
- Ilie, I.M.; Caflisch, A. Simulation Studies of Amyloidogenic Polypeptides and Their Aggregates. Chem. Rev. 2019, 119, 6956–6993. [Google Scholar] [CrossRef] [PubMed]
- Strodel, B. Amyloid Aggregation Simulations: Challenges, Advances and Perspectives. Curr. Opin. Struct. Biol. 2021, 67, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Matthes, D.; Gapsys, V.; Daebel, V.; De Groot, B.L. Mapping the Conformational Dynamics and Pathways of Spontaneous Steric Zipper Peptide Oligomerization. PLoS ONE 2011, 6, e19129. [Google Scholar] [CrossRef]
- Schor, M.; Mey, A.S.J.S.; Noé, F.; Macphee, C.E. Shedding Light on the Dock-Lock Mechanism in Amyloid Fibril Growth Using Markov State Models. J. Phys. Chem. Lett. 2015, 6, 1076–1081. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Balaji, P.V. Molecular Events during the Early Stages of Aggregation of GNNQQNY: An All Atom MD Simulation Study of Randomly Dispersed Peptides. J. Struct. Biol. 2015, 192, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhao, C.; Zhao, J.; Wang, J.; Yang, J.C.; Yu, X.; Zheng, J. Comparative Molecular Dynamics Study of Aβ Adsorption on the Self-Assembled Monolayers. Langmuir 2010, 26, 3308–3316. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, Q.; Liang, G.; Zheng, J. Molecular Dynamics Simulations of Low-Ordered Alzheimer β-Amyloid Oligomers from Dimer to Hexamer on Self-Assembled Monolayers. Langmuir 2011, 27, 14876–14887. [Google Scholar] [CrossRef]
- Kang, S.g.; Li, H.; Huynh, T.; Zhang, F.; Xia, Z.; Zhang, Y.; Zhou, R. Molecular Mechanism of Surface-Assisted Epitaxial Self-Assembly of Amyloid-like Peptides. ACS Nano 2012, 6, 9276–9282. [Google Scholar] [CrossRef]
- John, T.; Adler, J.; Elsner, C.; Petzold, J.; Krueger, M.; Martin, L.L.; Huster, D.; Risselada, H.J.; Abel, B. Mechanistic Insights into the Size-Dependent Effects of Nanoparticles on Inhibiting and Accelerating Amyloid Fibril Formation. J. Colloid Interface Sci. 2022, 622, 804–818. [Google Scholar] [CrossRef]
- Gladytz, A.; Wagner, M.; Häupl, T.; Elsner, C.; Abel, B. Structure-Making Effects of Metal Nanoparticles in Amyloid Peptide Fibrillation. Part. Part. Syst. Charact. 2015, 32, 573–582. [Google Scholar] [CrossRef]
- Hamley, I.W. The Amyloid Beta Peptide: A Chemist’s Perspective. Role in Alzheimer’s and Fibrillization. Chem. Rev. 2012, 112, 5147–5192. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Cheng, Y.; Sun, M. Functionalized Gold Nanoparticles: Synthesis, Properties and Biomedical Applications. Chem. Rec. 2020, 20, 1474–1504. [Google Scholar] [CrossRef]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.B.; Rodger, P.M.; Corni, S.; Walsh, T.R. GolP-CHARMM: First-principles Based Force Fields for the Interaction of Proteins with Au(111) and Au(100). J. Chem. Theory Comput. 2013, 9, 1616–1630. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D.; Bashford, D.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; Joseph-McCarthy, D.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins †. J. Phys. Chem. 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Bjelkmar, P.; Larsson, P.; Cuendet, M.A.; Hess, B.; Lindahl, E. Implementation of the {CHARMM} Force Field in {GROMACS}: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models. J. Chem. Theory Comput. 2010, 6, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Piana, S.; Lindorff-Larsen, K.; Shaw, D.E. How Robust Are Protein Folding Simulations with Respect to Force Field Parameterization? Biophys. J. 2011, 100, L47–L49. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Iori, F.; Corni, S. Including Image Charge Effects in the Molecular Dynamics Simulations of Molecules on Metal Surfaces. J. Comput. Chem. 2008, 29, 1656–1666. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A Toolkit for the Analysis of MolecularDynamics Simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Colletier, J.P.; Laganowsky, A.; Landau, M.; Zhao, M.; Soriaga, A.B.; Goldschmidt, L.; Flot, D.; Cascio, D.; Sawaya, M.R.; Eisenberg, D. Molecular Basis for Amyloid-β Polymorphism. Proc. Natl. Acad. Sci. USA 2011, 108, 16938–16943. [Google Scholar] [CrossRef]
- Meisl, G.; Kirkegaard, J.B.; Arosio, P.; Michaels, T.C.T.; Vendruscolo, M.; Dobson, C.M.; Linse, S.; Knowles, T.P.J. Molecular Mechanisms of Protein Aggregation from Global Fitting of Kinetic Models. Nat. Protoc. 2016, 11, 252–272. [Google Scholar] [CrossRef] [PubMed]
- Palafox-Hernandez, J.P.; Tang, Z.; Hughes, Z.E.; Li, Y.; Swihart, M.T.; Prasad, P.N.; Walsh, T.R.; Knecht, M.R. Comparative Study of Materials-Binding Peptide Interactions with Gold and Silver Surfaces and Nanostructures: A Thermodynamic Basis for Biological Selectivity of Inorganic Materials. Chem. Mater. 2014, 26, 4960–4969. [Google Scholar] [CrossRef]
- Futera, Z.; Blumberger, J. Adsorption of Amino Acids on Gold: Assessing the Accuracy of the GolP-CHARMM Force Field and Parametrization of Au–S Bonds. J. Chem. Theory Comput. 2019, 15, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Balbach, J.J.; Ishii, Y.; Antzutkin, O.N.; Leapman, R.D.; Rizzo, N.W.; Dyda, F.; Reed, J.; Tycko, R. Amyloid Fibril Formation by Aβ 16-22, a Seven-Residue Fragment of the Alzheimer’s β-Amyloid Peptide, and Structural Characterization by Solid State NMR. Biochemistry 2000, 39, 13748–13759. [Google Scholar] [CrossRef]
- Li, H.; Luo, Y.; Derreumaux, P.; Wei, G. Carbon Nanotube Inhibits the Formation of β-Sheet-Rich Oligomers of the Alzheimer’s Amyloid-β(16-22) Peptide. Biophys. J. 2011, 101, 2267–2276. [Google Scholar] [CrossRef] [PubMed]
- Tjernberg, L.O.; Näslund, J.; Lindqvist, F.; Johansson, J.; Karlström, A.R.; Thyberg, J.; Terenius, L.; Nordstedt, C. Arrest of -Amyloid Fibril Formation by a Pentapeptide Ligand. J. Biol. Chem. 1996, 271, 8545–8548. [Google Scholar] [CrossRef] [PubMed]
- Hajiraissi, R.; Hanke, M.; Gonzalez Orive, A.; Duderija, B.; Hofmann, U.; Zhang, Y.; Grundmeier, G.; Keller, A. Effect of Terminal Modifications on the Adsorption and Assembly of hIAPP(20–29). ACS Omega 2019, 4, 2649–2660. [Google Scholar] [CrossRef] [PubMed]
- Morriss-Andrews, A.; Shea, J.E. Simulations of Protein Aggregation: Insights from Atomistic and Coarse-Grained Models. J. Phys. Chem. Lett. 2014, 5, 1899–1908. [Google Scholar] [CrossRef]
- Illig, A.M.; Strodel, B. Performance of Markov State Models and Transition Networks on Characterizing Amyloid Aggregation Pathways from MD Data. J. Chem. Theory Comput. 2020, 16, 7825–7839. [Google Scholar] [CrossRef]
- Peter, E.K.; Shea, J.E. An Adaptive Bias – Hybrid MD/kMC Algorithm for Protein Folding and Aggregation. Phys. Chem. Chem. Phys. 2017, 19, 17373–17382. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheung, D.L. Aggregation of an Amyloidogenic Peptide on Gold Surfaces. Biomolecules 2023, 13, 1261. https://doi.org/10.3390/biom13081261
Cheung DL. Aggregation of an Amyloidogenic Peptide on Gold Surfaces. Biomolecules. 2023; 13(8):1261. https://doi.org/10.3390/biom13081261
Chicago/Turabian StyleCheung, David L. 2023. "Aggregation of an Amyloidogenic Peptide on Gold Surfaces" Biomolecules 13, no. 8: 1261. https://doi.org/10.3390/biom13081261
APA StyleCheung, D. L. (2023). Aggregation of an Amyloidogenic Peptide on Gold Surfaces. Biomolecules, 13(8), 1261. https://doi.org/10.3390/biom13081261








