Peroxisomal NAD(H) Homeostasis in the Yeast Debaryomyces hansenii Depends on Two Redox Shuttles and the NAD+ Carrier, Pmp47
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Growth Conditions and Media
2.2. Molecular Biology Techniques
2.3. D. hansenii and S. cerevisiae Strain Construction
2.4. Tagging in the Genome
2.5. Fluorescence Microscopy and Image Processing
2.6. Western Blot Analysis
2.7. β-Oxidation Measurement
2.8. Mass Spectrometric Metabolite Analyses
3. Results
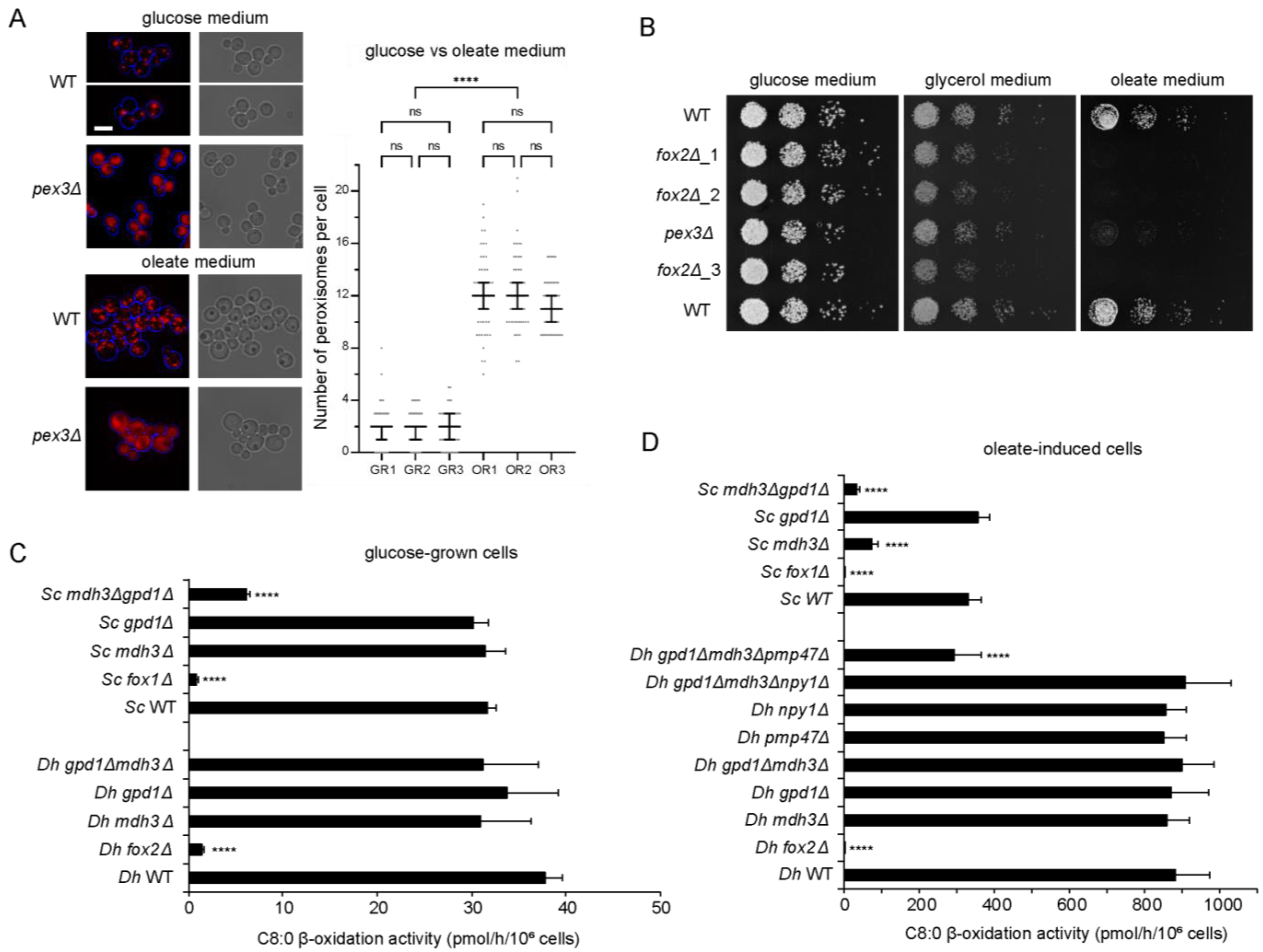
3.1. Fatty Acid Degradation Occurs Exclusively via Peroxisomal β-Oxidation in D. hansenii
3.2. Mdh3 and Gpd1 Are Localized to Peroxisomes but Not Essential for β-Oxidation Activity in D. hansenii
3.3. Pmp47, Together with Mdh3 and Gpd1, Is Essential for Normal β-Oxidation
3.4. Pmp47 Is a Peroxisomal NAD+ Transporter
3.5. Pmp47 Expression Restores Lysine Bradytrophy in S. cerevisiae mdh3Δgpd1Δ cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Navarrete, C.; Estrada, M.; Martínez, J.L. Debaryomyces Hansenii: An Old Acquaintance for a Fresh Start in the Era of the Green Biotechnology. World J. Microbiol. Biotechnol. 2022, 38, 99. [Google Scholar] [CrossRef] [PubMed]
- Yaguchi, A.; Rives, D.; Blenner, M. New Kids on the Block: Emerging Oleaginous Yeast of Biotechnological Importance. AIMS Microbiol. 2017, 3, 227–247. [Google Scholar] [CrossRef] [PubMed]
- Breuer, U.; Harms, H. Debaryomyces Hansenii—An Extremophilic Yeast with Biotechnological Potential. Yeast 2006, 23, 415–437. [Google Scholar] [CrossRef] [PubMed]
- de Figueiredo, P.; Li, L.; Nikolov, Z.; Shaw, B.D.; Dickman, M.B.; Louzada, E.S.; Sturino, J.M.; Chang, Y.Y. Transformation of Glycerol and Cellulosic Materials into High Energy Fuels. U.S. Patents 9,340,768, 17 May 2016. [Google Scholar]
- Alhajouj, S.; Turkolmez, S.; Abalkhail, T.; Hadi, Z.; Alwan, O.; Gilmour, D.J.; Mitchell, P.J.; Hettema, E.H. Efficient PCR-Based Gene Targeting in Isolates of the Non-Conventional Yeast Debaryomyces Hansenii. bioRxiv 2023. [Google Scholar] [CrossRef]
- Hiltunen, J.K.; Mursula, A.M.; Rottensteiner, H.; Wierenga, R.K.; Kastaniotis, A.J.; Gurvitz, A. The Biochemistry of Peroxisomal β-Oxidation in the Yeast Saccharomyces Cerevisiae. FEMS Microbiol. Rev. 2003, 27, 35–64. [Google Scholar] [CrossRef] [PubMed]
- Al-Saryi, N.A.; Al-Hejjaj, M.Y.; van Roermund, C.W.T.; Hulmes, G.E.; Ekal, L.; Payton, C.; Wanders, R.J.A.; Hettema, E.H. Two NAD-Linked Redox Shuttles Maintain the Peroxisomal Redox Balance in Saccharomyces Cerevisiae. Sci. Rep. 2017, 7, 11868. [Google Scholar] [CrossRef]
- van Roermund, C.W.T.; Elgersma, Y.; Singh, N.; Wanders, R.J.A.; Tabak, H.F. The Membrane of Peroxisomes in Saccharomyces Cerevisiae Is Impermeable to NAD(H) and Acetyl-CoA under in Vivo Conditions. EMBO J. 1995, 14, 3480–3486. [Google Scholar] [CrossRef]
- Chornyi, S.; IJlst, L.; van Roermund, C.W.T.; Wanders, R.J.A.; Waterham, H.R. Peroxisomal Metabolite and Cofactor Transport in Humans. Front. Cell Dev. Biol. 2021, 8, 1–24. [Google Scholar] [CrossRef]
- Bernhardt, K.; Wilkinson, S.; Weber, A.P.M.; Linka, N. A Peroxisomal Carrier Delivers NAD + and Contributes to Optimal Fatty Acid Degradation during Storage Oil Mobilization. Plant J. 2012, 69, 1–13. [Google Scholar] [CrossRef]
- van Roermund, C.W.T.; Schroers, M.G.; Wiese, J.; Facchinelli, F.; Kurz, S.; Wilkinson, S.; Charton, L.; Wanders, R.J.A.; Waterham, H.R.; Weber, A.P.M.; et al. The Peroxisomal NAD Carrier from Arabidopsis Imports NAD in Exchange with AMP. Plant Physiol. 2016, 171, 2127–2139. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, C.S.; Winston, F. A Ten-Minute DNA Preparation from Yeast Efficiently Releases Autonomous Plasmids for Transformaion of Escherichia Coli. Gene 1987, 57, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Akio, S. New Yeast-Escherichia Coli Shuttle Vectors Constructed with in Vitro Mutagenized Yeast Genes Lacking Six-Base Pair Restriction Sites. Gene 1988, 74, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Falcón, A.A.; Aris, J.P. Plasmid Accumulation Reduces Life Span in Saccharomyces Cerevisiae. J. Biol. Chem. 2003, 278, 41607–41617. [Google Scholar] [CrossRef]
- van Roermund, C.W.T.; IJlst, L.; Majczak, W.; Waterham, H.R.; Folkerts, H.; Wanders, R.J.A.; Hellingwerf, K.J. Peroxisomal Fatty Acid Uptake Mechanism in Saccharomyces Cerevisiae. J. Biol. Chem. 2012, 287, 20144–20153. [Google Scholar] [CrossRef]
- Schaub, Y.; Dünkler, A.; Walther, A.; Wendland, J. New PFA-Cassettes for PCR-Based Gene Manipulation in Candida Albicans. J. Basic. Microbiol. 2006, 46, 416–429. [Google Scholar] [CrossRef]
- Defosse, T.A.; Courdavault, V.; Coste, A.T.; Clastre, M.; de Bernonville, T.D.; Godon, C.; Vandeputte, P.; Lanoue, A.; Touzé, A.; Linder, T.; et al. A Standardized Toolkit for Genetic Engineering of CTG Clade Yeasts. J. Microbiol. Methods 2018, 144, 152–156. [Google Scholar] [CrossRef]
- Minhas, A.; Biswas, D.; Mondal, A.K. Development of Host and Vector for High-Efficiency Transformation and Gene Disruption in Debaryomyces Hansenii. FEMS Yeast Res. 2009, 9, 95–102. [Google Scholar] [CrossRef]
- Ekal, L.; Alqahtani, A.M.; Hettema, E.H. The Dynamin-Related Protein Vps1 and the Peroxisomal Membrane Protein Pex27 Function Together during Peroxisome Fission. J. Cell Sci. 2023, 136, jcs246348. [Google Scholar] [CrossRef]
- van Roermund, C.; Hettema, E. Characterization of Yeast Peroxisomes: Enrichment of Peroxisomal Fractions and Analysis of β-Oxidation Activity. In Peroxisomes: Methods and Protocols; Springer: New York, NY, USA, 2023; pp. 321–331. ISBN 9781071630471. [Google Scholar]
- Herzog, K.; Pras-Raves, M.L.; Vervaart, M.A.T.; Luyf, A.C.M.; van Kampen, A.H.C.; Wanders, R.J.A.; Waterham, H.R.; Vaz, F.M. Lipidomic Analysis of Fibroblasts from Zellweger Spectrum Disorder Patients Identifies Disease-Specific Phospholipid Ratios. J. Lipid Res. 2016, 57, 1447. [Google Scholar] [CrossRef]
- Veenhuis, M.; Mateblowski, M.; Kunau, W.H.; Harder, W. Proliferation of Microbodies in Saccharomyces Cerevisiae. Yeast 1987, 3, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Reuß, O.; Vik, Å.; Kolter, R.; Morschhäuser, J. The SAT1 Flipper, an Optimized Tool for Gene Disruption in Candida Albicans. Gene 2004, 341, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Neuberger, G.; Maurer-Stroh, S.; Eisenhaber, B.; Hartig, A.; Eisenhaber, F. Motif Refinement of the Peroxisomal Targeting Signal 1 and Evaluation of Taxon-Specific Differences. J. Mol. Biol. 2003, 328, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Petriv, O.I.; Tang, L.; Titorenko, V.I.; Rachubinski, R.A. A New Definition for the Consensus Sequence of the Peroxisome Targeting Signal Type 2. J. Mol. Biol. 2004, 341, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.A.; Cerutti, L.; Hulo, N.; Gattiker, A.; Falquet, L.; Pagni, M.; Bairoch, A.; Bucher, P. PROSITE: A Documented Database Using Patterns and Profiles as Motif Descriptors. Brief Bioinform. 2002, 3, 265–274. [Google Scholar] [CrossRef]
- Sigrist, C.J.A.; De Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and Continuing Developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef] [PubMed]
- McCammon, M.T.; Dowds, C.A.; Orth, K.; Moomaw, C.R.; Slaughter, C.A.; Goodman, J.M. Sorting of Peroxisomal Membrane Protein PMP47 from Candida Boidinii into Peroxisomal Membranes of Saccharomyces Cerevisiae. J. Biol. Chem. 1990, 265, 20098–20105. [Google Scholar] [CrossRef]
- Todisco, S.; Agrimi, G.; Castegna, A.; Palmieri, F. Identification of the Mitochondrial NAD+ Transporter in Saccharomyces Cerevisiae. J. Biol. Chem. 2006, 281, 1524–1531. [Google Scholar] [CrossRef]
- Einerhand, A.W.; Kos, W.T.; Distel, B.; Tabak, H.F. Characterization of a Transcriptional Control Element Involved in Proliferation of Peroxisomes in Yeast in Response to Oleate. Eur. J. Biochem. 1993, 214, 323–331. [Google Scholar] [CrossRef]
- Xu, W.; Dunn, C.A.; Bessman, M.J. Cloning and Characterization of the NADH Pyrophosphatases from Caenorhabditis Elegans and Saccharomyces Cerevisiae, Members of a Nudix Hydrolase Subfamily. Biochem. Biophys. Res. Commun. 2000, 273, 753–758. [Google Scholar] [CrossRef]
- Plett, A.; Charton, L.; Linka, N. Peroxisomal Cofactor Transport. Biomolecules 2020, 10, 1174. [Google Scholar] [CrossRef] [PubMed]
- Chornyi, S.; Costa, C.F.; IJlst, L.; Fransen, M.; Wanders, R.J.A.; van Roermund, C.W.T.; Waterham, H.R. Human Peroxisomal NAD+/NADH Homeostasis Is Regulated by Two Independent NAD(H) Shuttle Systems. Free Radic. Biol. Med. 2023, 206, 22–32. [Google Scholar] [CrossRef] [PubMed]
- McCammon, M.; McNew, J.; Willy, P.; Goodman, J. An Internal Region of the Peroxisomal Membrane Protein PMP47 Is Essential for Sorting to Peroxisomes. J. Cell Biol. 1994, 124, 915–925. [Google Scholar] [CrossRef]
- Sulter, G.J.; Waterham, H.R.; Vrieling, E.G.; Goodman, J.M.; Harder, W.; Veenhius, M. Expression and Targeting of a 47 KDa Integral Peroxisomal Membrane Protein of Candida Boidinii in Wild Type and a Peroxisome-Deficient Mutant of Hansenula Polymorpha. FEBS Lett. 1993, 315, 211–216. [Google Scholar] [CrossRef]
- Fukao, Y.; Hayashi, Y.; Mano, S.; Hayashi, M.; Nishimura, M. Developmental Analysis of a Putative ATP/ADP Carrier Protein Localized on Glyoxysomal Membranes During the Peroxisome Transition in Pumpkin Cotyledons. Plant Cell Physiol. 2001, 42, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, M.; Metzl-Raz, E.; Bürgi, J.; Yifrach, E.; Drwesh, L.; Fadel, A.; Peleg, Y.; Rapaport, D.; Wilmanns, M.; Barkai, N.; et al. Uncovering Targeting Priority to Yeast Peroxisomes Using an In-Cell Competition Assay. Proc. Natl. Acad. Sci. USA 2020, 117, 21432–21440. [Google Scholar] [CrossRef]
- Münsterkötter, M.; Steinberg, G. The Fungus Ustilago Maydis and Humans Share Disease-Related Proteins That Are Not Found in Saccharomyces Cerevisiae. BMC Genom. 2007, 8, 473. [Google Scholar] [CrossRef]
- Camões, F.; Islinger, M.; Guimarães, S.C.; Kilaru, S.; Schuster, M.; Godinho, L.F.; Steinberg, G.; Schrader, M. New Insights into the Peroxisomal Protein Inventory: Acyl-CoA Oxidases and -Dehydrogenases Are an Ancient Feature of Peroxisomes. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 111–125. [Google Scholar] [CrossRef]
- Jones, E.W. Proteinase Mutants of Saccharomyces Cerevisiae. Genetics 1977, 85, 23–33. [Google Scholar] [CrossRef]
- Elgersma, Y.; van den Berg, M.; Tabak, H.F.; Distel, B. An Efficient Positive Selection Procedure for the Isolation of Peroxisomal Import and Peroxisome Assembly Mutants of Saccharomyces Cerevisiae. Genetics 1993, 135, 731–740. [Google Scholar] [CrossRef]
- IJlst, L.; van Roermund, C.W.T.; Iacobazzi, V.; Oostheim, W.; Ruiter, J.P.N.; Williams, J.C.; Palmieri, F.; Wanders, R.J.A. Functional Analysis of Mutant Human Carnitine Acylcarnitine Translocases in Yeast. Biochem. Biophys. Res. Commun. 2001, 280, 700–706. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turkolmez, S.; Chornyi, S.; Alhajouj, S.; IJlst, L.; Waterham, H.R.; Mitchell, P.J.; Hettema, E.H.; van Roermund, C.W.T. Peroxisomal NAD(H) Homeostasis in the Yeast Debaryomyces hansenii Depends on Two Redox Shuttles and the NAD+ Carrier, Pmp47. Biomolecules 2023, 13, 1294. https://doi.org/10.3390/biom13091294
Turkolmez S, Chornyi S, Alhajouj S, IJlst L, Waterham HR, Mitchell PJ, Hettema EH, van Roermund CWT. Peroxisomal NAD(H) Homeostasis in the Yeast Debaryomyces hansenii Depends on Two Redox Shuttles and the NAD+ Carrier, Pmp47. Biomolecules. 2023; 13(9):1294. https://doi.org/10.3390/biom13091294
Chicago/Turabian StyleTurkolmez, Selva, Serhii Chornyi, Sondos Alhajouj, Lodewijk IJlst, Hans R. Waterham, Phil J. Mitchell, Ewald H. Hettema, and Carlo W. T. van Roermund. 2023. "Peroxisomal NAD(H) Homeostasis in the Yeast Debaryomyces hansenii Depends on Two Redox Shuttles and the NAD+ Carrier, Pmp47" Biomolecules 13, no. 9: 1294. https://doi.org/10.3390/biom13091294
APA StyleTurkolmez, S., Chornyi, S., Alhajouj, S., IJlst, L., Waterham, H. R., Mitchell, P. J., Hettema, E. H., & van Roermund, C. W. T. (2023). Peroxisomal NAD(H) Homeostasis in the Yeast Debaryomyces hansenii Depends on Two Redox Shuttles and the NAD+ Carrier, Pmp47. Biomolecules, 13(9), 1294. https://doi.org/10.3390/biom13091294





