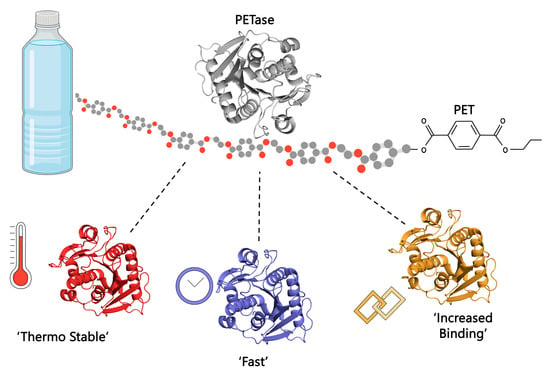
Engineering Plastic Eating Enzymes Using Structural Biology
Abstract
:1. Introduction
2. Molecular Features of Plastic-Degrading Enzymes
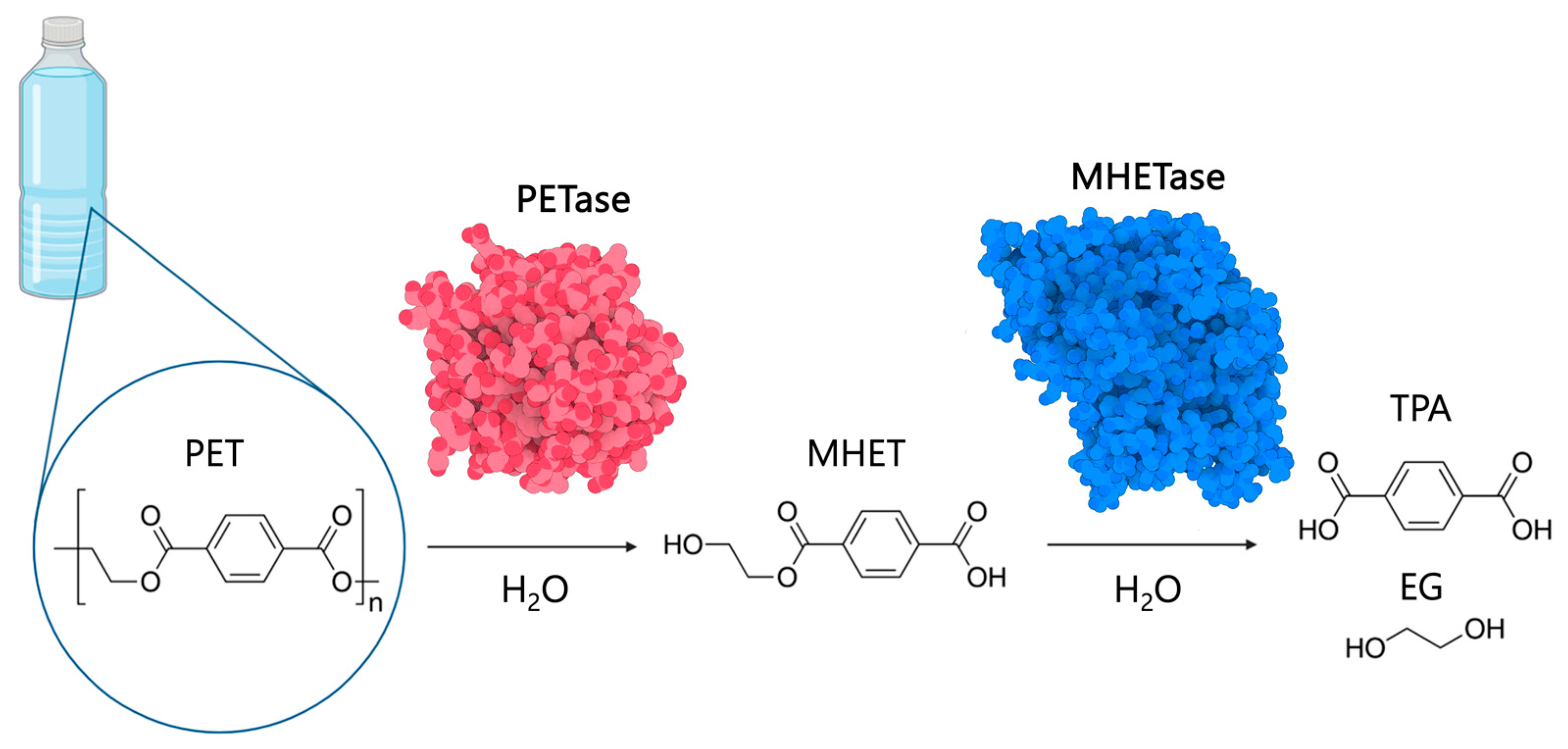
2.1. The PETase Catalytic Mechanism
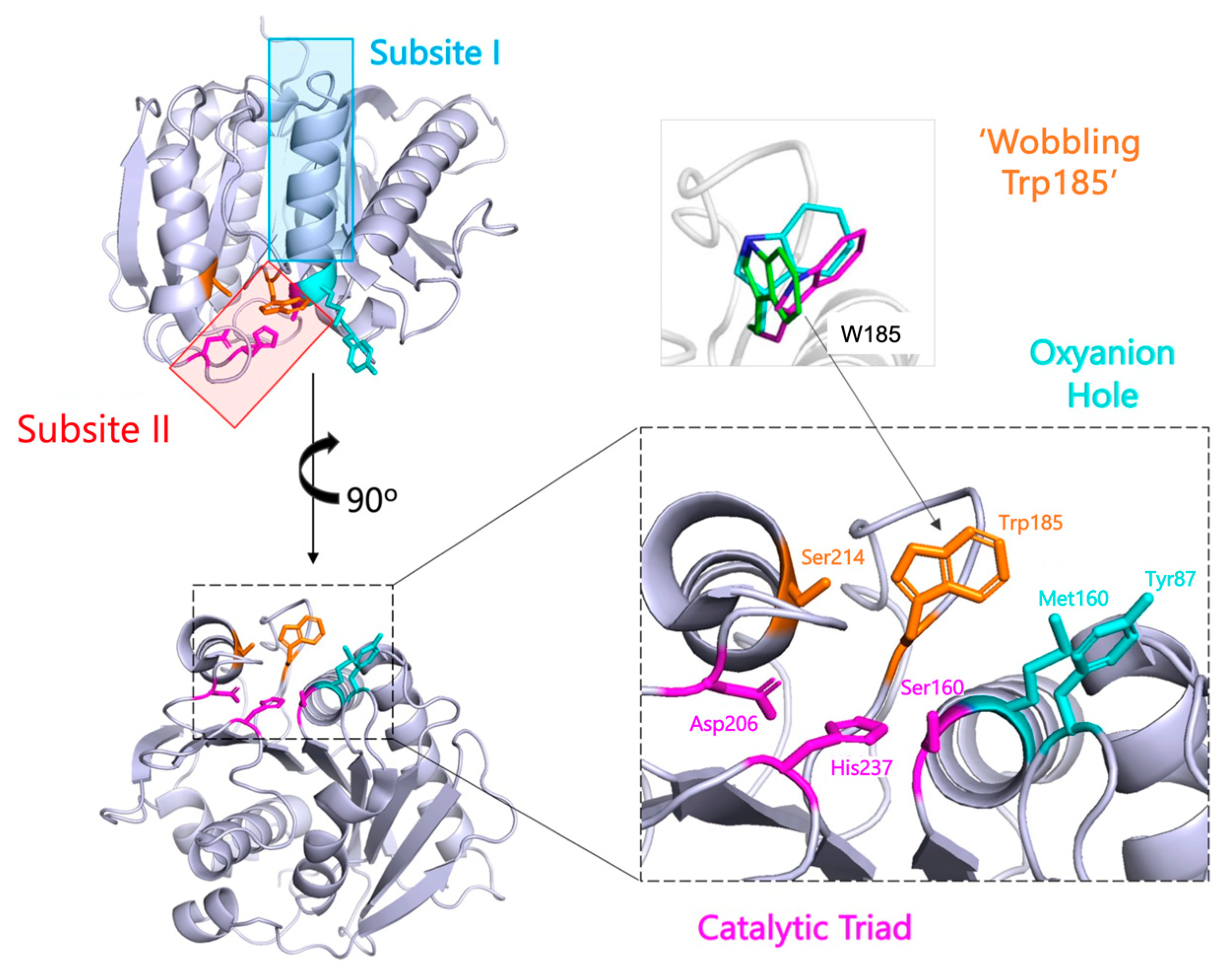
2.1.1. Comparison of PETase with the Homologous Cutinases
2.1.2. Efforts to Understand the Enzyme-Substrate Complex
2.1.3. Identification of Unique Enzyme Feature—Wobbling Trp
2.2. Engineering the Enzyme to Enhance Efficiency
2.2.1. Developing a Thermostable Enzyme to Enhance Efficiency
2.2.2. Industrial Applications and Challenges
2.2.3. Increase Efficiency under Environmental Conditions
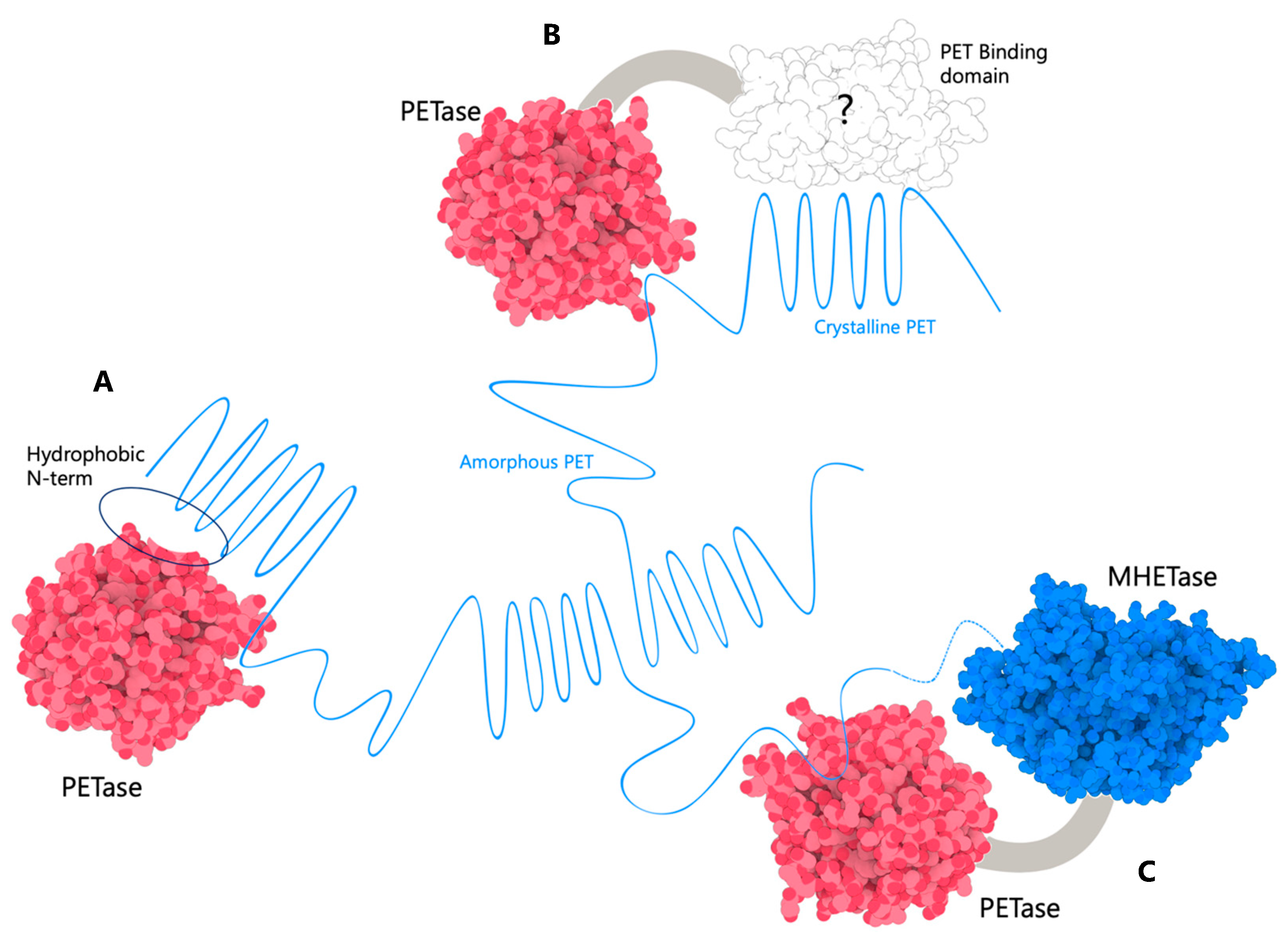
2.2.4. Overcoming the Hydrophobicity of Plastics and PET to Increase Efficiency
2.2.5. A More Collaborative Global Approach for the Future Plastic Management
3. Future Directions and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Danopoulos, E.; Twiddy, M.; West, R.; Rotchell, J.M. A rapid review and meta-regression analyses of the toxicological impacts of microplastic exposure in human cells. J. Hazard. Mater. 2022, 427, 127861. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Zimmermann, W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb. Biotechnol. 2017, 10, 1308–1322. [Google Scholar] [CrossRef] [PubMed]
- Tokiwa, Y.; Suzuki, T. Hydrolysis of polyesters by lipases. Nature 1977, 270, 76–78. [Google Scholar] [CrossRef]
- Martínez, A.; Maicas, S. Cutinases: Characteristics and insights in industrial production. Catalysts 2021, 11, 1194. [Google Scholar] [CrossRef]
- Gomes, D.; Matamá, T.; Cavaco-Paulo, A.; Takaki, G.; Salgueiro, A. Production of heterologous cutinases by E. coli and improved enzyme formulation for application on plastic degradation. Electron. J. Biotechnol. 2013, 16, 3. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Tomasello, G.; Armenia, I.; Molla, G. The Protein Imager: A full-featured online molecular viewer interface with server-side HQ-rendering capabilities. Bioinformatics 2020, 36, 2909–2911. [Google Scholar] [CrossRef] [PubMed]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveira, R.L.; Pollard, B.C.; Dominick, G.; Duman, R.; El Omari, K.; et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, E4350–E4357. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.; Kim, W.; Shroff, R.; Acosta, D.J.; Alexander, B.R.; Cole, H.O.; Zhang, Y.; et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 2022, 604, 662–667. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S.; et al. An engineered pet depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Palm, G.J.; Reisky, L.; Böttcher, D.; Müller, H.; Michels, E.A.; Walczak, M.C.; Berndt, L.; Weiss, M.S.; Bornscheuer, U.T.; Weber, G. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat. Commun. 2019, 10, 1717. [Google Scholar] [CrossRef]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.-Y.; Shin, T.J.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 2018, 9, 382. [Google Scholar] [CrossRef]
- Han, X.; Liu, W.; Huang, J.-W.; Ma, J.; Zheng, Y.; Ko, T.-P.; Xu, L.; Cheng, Y.-S.; Chen, C.-C.; Guo, R.-T. Structural insight into catalytic mechanism of pet hydrolase. Nat. Commun. 2017, 8, 2106. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with alphafold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Fecker, T.; Galaz-Davison, P.; Engelberger, F.; Narui, Y.; Sotomayor, M.; Parra, L.P.; Ramírez-Sarmiento, C.A. Active site flexibility as a hallmark for efficient pet degradation by I. sakaiensis petase. Biophys. J. 2018, 114, 1302–1312. [Google Scholar] [CrossRef]
- Zeng, W.; Li, X.; Yang, Y.; Min, J.; Huang, J.-W.; Liu, W.; Niu, D.; Yang, X.; Han, X.; Zhang, L.; et al. Substrate-binding mode of a thermophilic pet hydrolase and engineering the enzyme to enhance the hydrolytic efficacy. ACS Catal. 2022, 12, 3033–3040. [Google Scholar] [CrossRef]
- Visual Feature: Beat Plastic Pollution. Available online: https://www.unep.org/interactives/beat-plastic-pollution/ (accessed on 25 August 2023).
- Maurya, A.; Bhattacharya, A.; Khare, S.K. Enzymatic remediation of polyethylene terephthalate (PET)–based polymers for effective management of plastic wastes: An overview. Front. Bioeng. Biotechnol. 2020, 8, 602325. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, C.; Zhu, S.; Wei, R.; Yin, C.-C. Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis. Biochem. Biophys. Res. Commun. 2019, 508, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Barth, M.; Oeser, T.; Wei, R.; Then, J.; Schmidt, J.; Zimmermann, W. Effect of hydrolysis products on the enzymatic degradation of polyethylene terephthalate nanoparticles by a polyester hydrolase from Thermobifida fusca. Biochem. Eng. J. 2015, 93, 222–228. [Google Scholar] [CrossRef]
- Bell, E.; Smithson, R.; Kilbride, S.; Foster, J.; Hardy, F.; Ramachandran, S.; Tedstone, A.; Haigh, S.; Garforth, A.; Day, P.; et al. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 2022, 5, 673–681. [Google Scholar]
- Erickson, E.; Gado, J.E.; Avilán, L.; Bratti, F.; Brizendine, R.K.; Cox, P.A.; Gill, R.; Graham, R.; Kim, D.-J.; König, G.; et al. Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity. Nat. Commun. 2022, 13, 7850. [Google Scholar] [CrossRef] [PubMed]
- Gambarini, V.; Pantos, O.; Kingsbury, J.M.; Weaver, L.; Handley, K.M.; Lear, G. PlasticDB: A database of microorganisms and proteins linked to plastic biodegradation. Database 2022, 2022, baac008. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D. Three ways to solve the plastics pollution crisis. Nature 2023, 616, 234–237. [Google Scholar] [CrossRef]
- Peng, B.-Y.; Su, Y.; Chen, Z.; Chen, J.; Zhou, X.; Benbow, M.E.; Criddle, C.S.; Wu, W.-M.; Zhang, Y. Biodegradation of polystyrene by dark (tenebrio obscurus) and yellow (tenebrio molitor) mealworms (Coleoptera: Tenebrionidae). Environ. Sci. Technol. 2019, 53, 5256–5265. [Google Scholar] [CrossRef]
- Zhang, Z.; Peng, H.; Yang, D.; Zhang, G.; Zhang, J.; Ju, F. Polyvinyl chloride degradation by a bacterium isolated from the gut of insect larvae. Nat. Commun. 2022, 13, 5360. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Huo, Y.; Yang, Y. Microbial degradation and valorization of plastic wastes. Front. Microbiol. 2020, 11, 442. [Google Scholar] [CrossRef]
- Meyer-Cifuentes, I.E.; Öztürk, B. MLE046 is a marine mesophilic mhetase-like enzyme. Front. Microbiol. 2021, 12, 693985. [Google Scholar] [CrossRef]
- Biundo, A.; Ribitsch, D.; Steinkellner, G.; Gruber, K.; Guebitz, G.M. Polyester hydrolysis is enhanced by a truncated esterase: Less is more. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef]
- Payne, C.M.; Knott, B.C.; Mayes, H.B.; Hansson, H.; Himmel, M.E.; Sandgren, M.; Ståhlberg, J.; Beckham, G.T. Fungal cellulases. Chem. Rev. 2015, 115, 1308–1448. [Google Scholar] [CrossRef]
- Pan, X.; Kortemme, T. Recent advances in de Novo Protein Design: Principles, methods, and applications. J. Biol. Chem. 2021, 296, 100558. [Google Scholar] [CrossRef]
- Knott, B.C.; Erickson, E.; Allen, M.D.; Gado, J.E.; Graham, R.; Kearns, F.L.; Pardo, I.; Topuzlu, E.; Anderson, J.J.; Austin, H.P.; et al. Characterization and engineering of a two-enzyme system for plastics depolymerization. Proc. Natl. Acad. Sci. USA 2020, 117, 25476–25485. [Google Scholar] [CrossRef] [PubMed]
- Artzi, L.; Bayer, E.A.; Moraïs, S. Cellulosomes: Bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 2016, 15, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Revolution Plastics. Available online: https://www.port.ac.uk/research/themes/sustainability-and-the-environment/revolution-plastics (accessed on 23 August 2023).
- Earth Negotiations Bulletin. Summary Report 29 May–2 June 2023. Available online: https://enb.iisd.org/plastic-pollution-marine-environment-negotiating-committee-inc2-summary (accessed on 25 August 2023).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barclay, A.; Acharya, K.R. Engineering Plastic Eating Enzymes Using Structural Biology. Biomolecules 2023, 13, 1407. https://doi.org/10.3390/biom13091407
Barclay A, Acharya KR. Engineering Plastic Eating Enzymes Using Structural Biology. Biomolecules. 2023; 13(9):1407. https://doi.org/10.3390/biom13091407
Chicago/Turabian StyleBarclay, Amelia, and K. Ravi Acharya. 2023. "Engineering Plastic Eating Enzymes Using Structural Biology" Biomolecules 13, no. 9: 1407. https://doi.org/10.3390/biom13091407
APA StyleBarclay, A., & Acharya, K. R. (2023). Engineering Plastic Eating Enzymes Using Structural Biology. Biomolecules, 13(9), 1407. https://doi.org/10.3390/biom13091407