Progress to Clarify How NOTCH3 Mutations Lead to CADASIL, a Hereditary Cerebral Small Vessel Disease
Abstract
1. Introduction
2. Overview of CADASIL
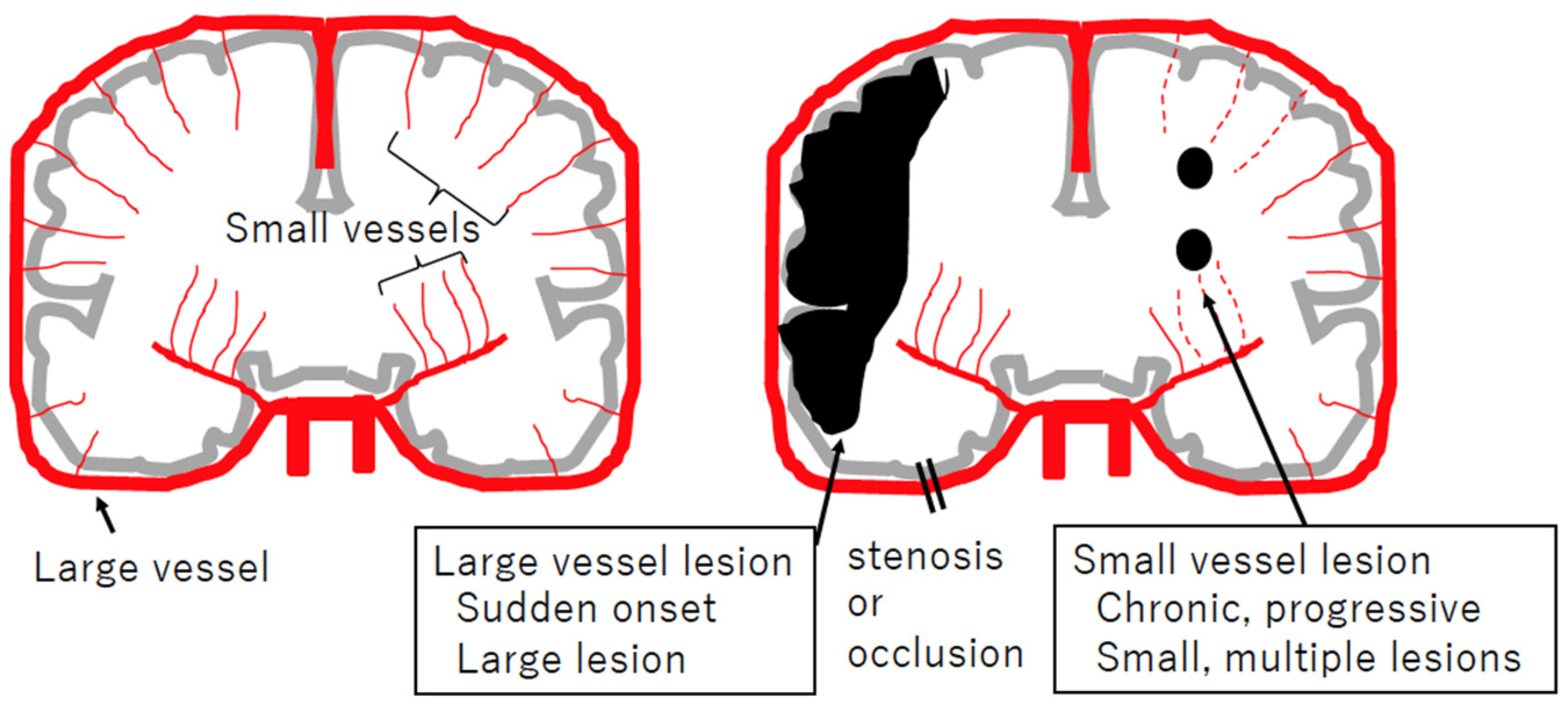
2.1. Cerebral Small Vessel Diseases
2.2. History of Disease Concept of CADASIL

2.3. Clinical Characteristics of CADASIL
2.4. Pathology of CADASIL
2.5. Genetics of CADASIL
2.5.1. Autosomal Dominant Inheritance
2.5.2. Typical Cysteine-Altering Mutations in EGFr of NOTCH3
2.5.3. Atypical Mutations in NOTCH3
2.5.4. EGFr Location–Phenotype Correlations
3. NOTCH3 and Notch Signaling
3.1. Protein Structure of NOTCH3 Receptor
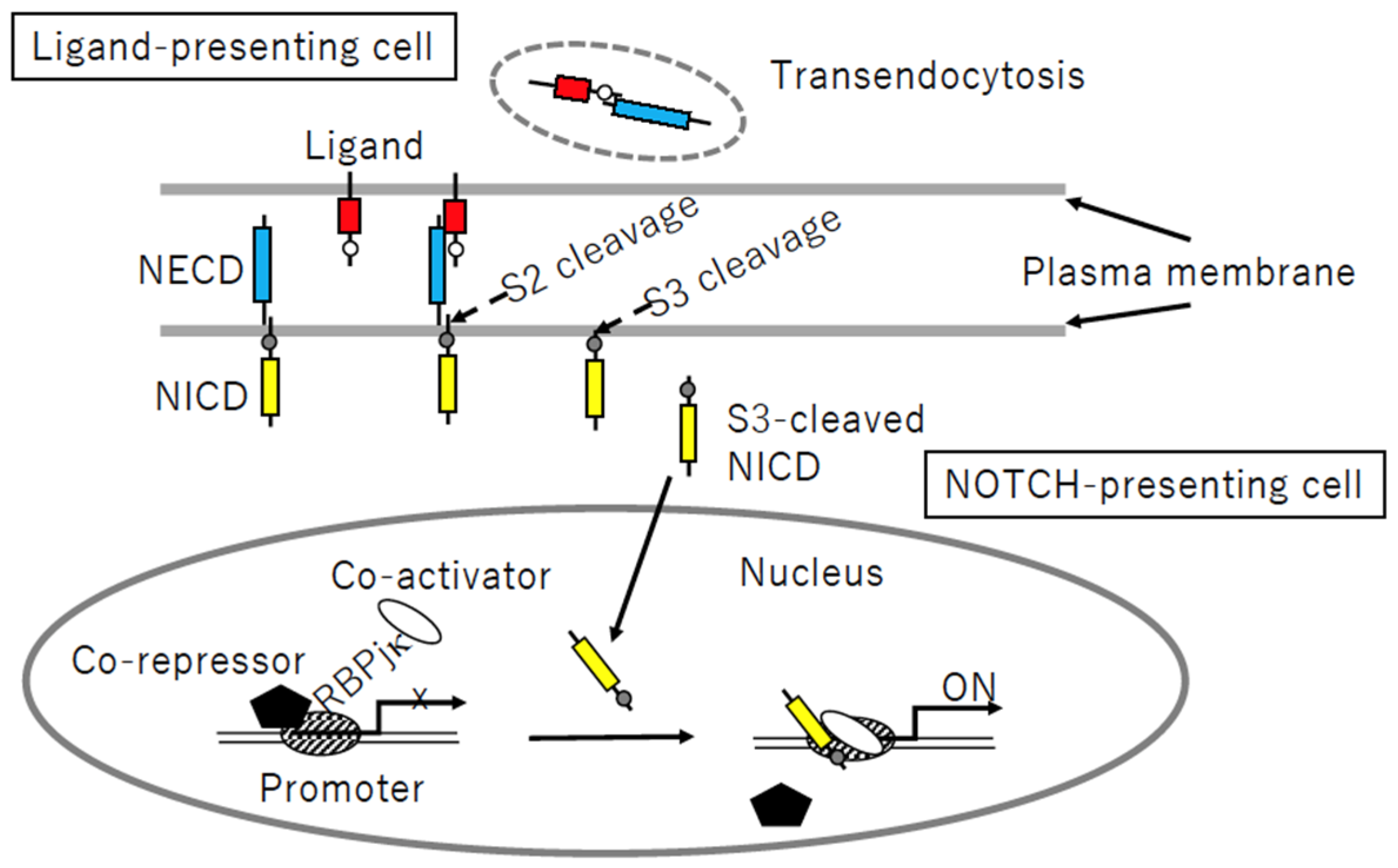
3.2. Notch Signaling Process
3.3. Localization of NOTCH Family Members and Their Ligands in Vessels
3.4. Evaluation of Notch Signaling
4. Strategy to Clarify Pathological Mechanism of CADASIL
4.1. Mouse Models of CADASIL
4.1.1. Transgenic Mice
| Mouse (Line) | Mutation (EGFr) | Transgene Expression 1 | Brain Pathology | Vascular Physiology |
|---|---|---|---|---|
| Knock-in | ||||
| Notch3R170C/R170C [78,81] | p.Arg169Cys (EGFr4) | not applicable | N3ECD accumulation at 4 months [78]; GOM deposition at 20 months [81] | Decreased passive diameter of isolated posterior cerebral arteries at 4 months [78] |
| Human NOTCH3 cDNA driven by murine SM22a promoter | ||||
| mN3+/+; TghN3 (WT) (line 46) [82] | Wild-type | 73% | No N3ECD accumulation; no GOM deposition | ND 2 |
| mN3+/+; TghN3(R90C) (line Ma) [71,80,82] | p.Arg90Cys (EGFr 2) | 86% | N3ECD accumulation and GOM deposition at 12 months [82] | Decreased flow-induced dilatation and increased pressure-induced myogenic tone in tail caudal arteries at 10–11 months [80] |
| mN3+/+; TghN3 (C428S) (line 10) [74] | p.Cys428Ser (EGFr 10) | 150% | N3ECD accumulation and GOM deposition at 8 months [74] | ND |
| PAC 3 containing genomic locus of rat Notch3 [72,78] | ||||
| TgNotch3R169C (line 88) | p.Arg169Cys (EGFr 4) | 400% | N3ECD accumulation at 1–2 months; GOM deposition at 5 months; white matter lesion (numerous vacuoles and loss of compact myelin with disorganized fibers) at 18–20 months [72] | Reduction of resting CBF 4 in gray matter (at 11–12 months) and white matter (at 18–20 months) [72]. Impaired cerebrovascular autoregulation at 5 months and attenuated functional hyperemia at 5–6 months [72]. Decreased increment of distensibility and decreased passive diameter in isolated posterior cerebral arteries at 6 and 2 months, respectively [78] |
| TgNotch3R169C (line 92) | 200% | N3ECD accumulation and GOM deposition [25] | ND | |
| TgNotch3WT (line 129) | Wild-type | 400% | No N3ECD accumulation: no GOM deposition up to 20 months [72] | No impairment of resting CBF, cerebrovascular autoregulation, or functional hyperemia [72]. |
| BAC 5 containing genomic locus of human NOTCH3 [73,79] | ||||
| tgN3MUT (line 350) | p.Arg182Cys (EGFr 4) | 350% | N3ECD accumulation detected at 6 weeks: GOM deposition detected at 5–6 months [73]; no white matter lesion detected [73] | No functional deficit in CBF [79]. |
| tgN3MUT (line 200) | 200% | N3ECD accumulation detected at 3 months [73] | ND | |
| tgN3MUT (line 150) | 150% | N3ECD accumulation detected at 5 months [73] | ND | |
| tgN3MUT (line 100) | 100% | N3ECD accumulation detected at 12 months [73] | No functional deficit in CBF [79]. | |
| tgN3WT | Wild-type | 100% | No N3ECD accumulation; no GOM deposition up to 20 months [73] | No functional deficit in CBF [79]. |
| Human NOTCH3 cDNA, SM22-Cre mediated conditional knock-in into ROSA26 locus [75] | ||||
| NOTCH3C455R | p.Cys455Arg (EGFr 11) | ND | GOM deposition at 6 months [75] | ND |
| NOTCH3R1031C | p.Arg1031Cys (EGFr 26) | ND | GOM deposition at 12 months [75] | ND |
4.1.2. Knock-In Mice
4.2. Cellular Model of CADASIL
4.2.1. Human VSMC Cells
4.2.2. iPSC-Derived Mural Cells
4.3. In Vitro Aggregation Assay
4.4. Drosophila Melanogaster and CADASIL
4.4.1. Drosophila Melanogaster and Human Neurological Diseases
4.4.2. Drosophila Notch Alleles Mimicking CADASIL-Causing Mutations
| N Allele | Fly Mutation (EGFr) | Wing Phenotype | Residue Corresponding to Human NOTCH3 (EGFr) | CADASIL Mutations Reported at the Residue 1 |
|---|---|---|---|---|
| FlyBase [105] | ||||
| Nnd−3 | p.Cys105Phe (EGFr 2) | Notching | p.Cys87 (EGFr 2) | p.Cys87Arg [44]/Tyr [44]/Phe [107] |
| NMcd5 | p.Cys739Tyr (EGFr 18) | Wild-type | p.Cys681 (EGFr 17) | not reported |
| NAx−59b | p.Cys972Gly (EGFr 24) | Abruptex | p.Cys875 (EGFr 22) | not reported |
| NAx-M1 | p.Cys999Tyr (EGFr 25) | Abruptex | p.Cys901 (EGFr 23) | not reported |
| Yamamoto et al., 2012 [106] | ||||
| egf8-C2S | p.Cys343Ser (EGFr 8) | Notching | p.Cys285 (EGFr 7) | p.Cys285Arg [108] |
| egf8-C2Y | p.Cys343Tyr (EGFr 8) | Notching | p.Cys285 (EGFr 7) | p.Cys285Arg [108] |
| egf8-C6S | p.Cys369Ser (EGFr 8) | Notching | p.Cys311 (EGFr 7) | p.Cys311Ser [109,110]/Gly [111] |
| egf9-C5Y | p.Cys398Tyr (EGFr 9) | Notching | p.Cys340 (EGFr 8) | p.Cys340Phe [8]/Trp [112] |
| egf9-C6S | p.Cys407Ser (EGFr 9) | Notching | p.Cys349 (EGFr 8) | not reported |
| egf10-C2S | p.Cys413Ser (EGFr 10) | Notching | p.Cys355 (EGFr 9) | p.Cys355Ser 2 [113] |
| egf11-C1S | p.Cys453Ser (EGFr 11) | Notching | p.Cys395 (EGFr 10) | p.Cys395Arg [44] |
| egf13-C2S | p.Cys535Ser (EGFr 13) | Wild-type | p.Cys478 (EGFr 12) | p.Cys478Tyr [114] |
| egf25-C2S | p.Cys993Ser (EGFr 25) | Notching | p.Cys896 (EGFr 23) | not reported |
| egf29-C2S | p.Cys1155Ser (EGFr 29) | Abruptex | p.Cys1055 (EGFr 27) | not reported |
| egf34-C1Y | p.Cys1341Tyr (EGFr 34) | Wild-type | p.Cys1250 (EGFr 32) | p.Cys1250Trp [115]/Gly 2 [113] |
| N Allele | Mutation (EGFr) | Bristle Formation 1 | Lateral Inhibition 2 | Inductive Signaling 3 | Intracellular Trafficking (Localization) |
|---|---|---|---|---|---|
| NX | p.Cys343Ser (EGFr 8) | Absent | Neurogenic | Depletion | Abnormal (loss of expression) |
| NOmicron | p.Cys343Tyr (EGFr 8) | Absent | Neurogenic | Depletion | Abnormal (ER 4) |
| NGamma | p.Cys398Tyr (EGFr 9) | Absent | Neurogenic | Depletion | Abnormal (ER) |
| NS | p.Cys407Ser (EGFr 9) | Absent | Neurogenic | Depletion | Abnormal (ER) |
| NIota | p.Cys413Ser (EGFr 10) | Absent | Neurogenic | Depletion | Abnormal (ER) |
| NG | p.Cys535Ser (EGFr 13) | Absent | Brain deformation | Depletion | Normal |
| NZeta | p.Cys993Ser (EGFr 25) | Absent | Neurogenic | Depletion | Abnormal (ER) |
| NH | p.Cys1155Ser (EGFr 29) | Absent | Normal | Normal | Abnormal (Early endosomes) |
| NJ | p.Cys1341Tyr (EGFr 34) | Absent | Normal | Normal | Normal |
5. Notch Signaling in CADASIL
5.1. Biological Role of NOTCH3 Signaling in Vessels
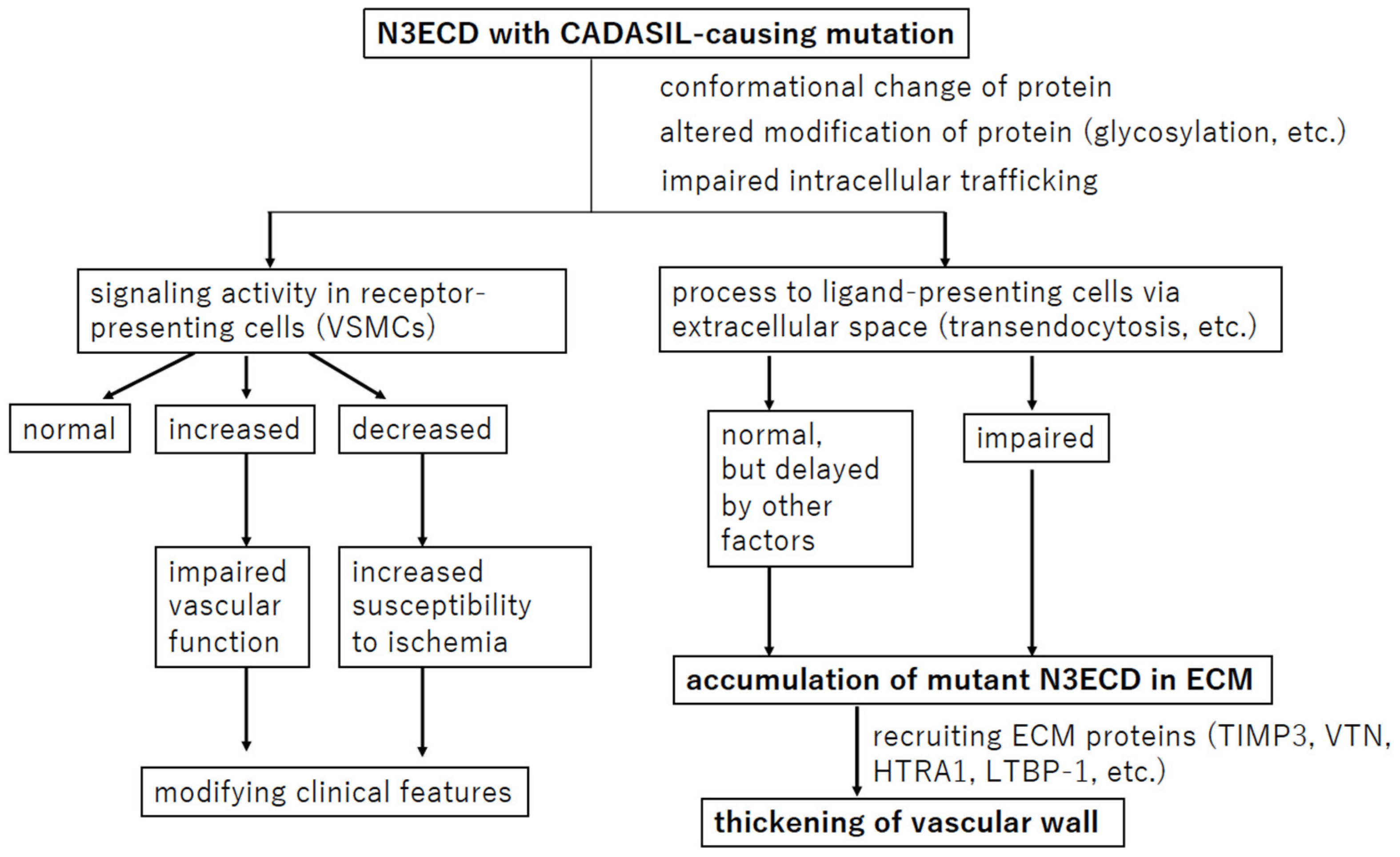
5.2. NOTCH3 Signaling Process in CADASIL Pathophysiology
5.2.1. NOTCH3 Signaling Activity in CADASIL
5.2.2. Transendocytosis of N3ECD
5.2.3. Cis-Interaction
5.2.4. Glycosylation of N3ECD
6. Protein Accumulation or Aggregation in CADASIL
6.1. TIMP3 and VTN
6.2. LTBP-1 and HTRA1
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chabriat, H.; Joutel, A.; Tournier-Lasserve, E.; Bousser, M.G. CADASIL: Yesterday, today, tomorrow. Eur. J. Neurol. 2020, 27, 1588–1595. [Google Scholar] [CrossRef]
- Chabriat, H.; Joutel, A.; Dichgans, M.; Tournier-Lasserve, E.; Bousser, M.G. Cadasil. Lancet Neurol. 2009, 8, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Joutel, A. The NOTCH3ECDcascade hypothesis of cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy disease. Neurol. Clin. Neurosci. 2015, 3, 1–6. [Google Scholar] [CrossRef]
- Caplan, L.R. Stroke: Etiology, classification, and epidemiology. In UpToDate; Kasner, S.E., Dashe, J.F., Eds.; Wolters Kluwer: Alphen aan den Rijn, The Netherlands, 2022. [Google Scholar]
- Rost, N.S. Definition, etiology, and clinical manifestations of transient ischemic attack. In UpToDate; Kasner, S.E., Dashe, J.F., Eds.; Wolters Kluwer: Alphen aan den Rijn, The Netherlands, 2022. [Google Scholar]
- Pantoni, L. Cerebral small vessel disease: From pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol. 2010, 9, 689–701. [Google Scholar] [CrossRef]
- Dichgans, M.; Pulit, S.L.; Rosand, J. Stroke genetics: Discovery, biology, and clinical applications. Lancet Neurol. 2019, 18, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Rutten, J.W.; Dauwerse, H.G.; Gravesteijn, G.; van Belzen, M.J.; van der Grond, J.; Polke, J.M.; Bernal-Quiros, M.; Lesnik Oberstein, S.A. Archetypal NOTCH3 mutations frequent in public exome: Implications for CADASIL. Ann. Clin. Transl. Neurol. 2016, 3, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Tournier-Lasserve, E.; Joutel, A.; Melki, J.; Weissenbach, J.; Lathrop, G.M.; Chabriat, H.; Mas, J.L.; Cabanis, E.A.; Baudrimont, M.; Maciazek, J.; et al. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy maps to chromosome 19q12. Nat. Genet. 1993, 3, 256–259. [Google Scholar] [CrossRef]
- Baudrimont, M.; Dubas, F.; Joutel, A.; Tournier-Lasserve, E.; Bousser, M.G. Autosomal dominant leukoencephalopathy and subcortical ischemic stroke. A clinicopathological study. Stroke 1993, 24, 122–125. [Google Scholar] [CrossRef]
- Gutiérrez-Molina, M.; Caminero Rodríguez, A.; Martínez García, C.; Arpa Gutiérrez, J.; Morales Bastos, C.; Amer, G. Small arterial granular degeneration in familial Binswanger’s syndrome. Acta Neuropathol. 1994, 87, 98–105. [Google Scholar] [CrossRef]
- Ruchoux, M.M.; Guerouaou, D.; Vandenhaute, B.; Pruvo, J.P.; Vermersch, P.; Leys, D. Systemic vascular smooth muscle cell impairment in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Acta Neuropathol. 1995, 89, 500–512. [Google Scholar]
- Tikka, S.; Baumann, M.; Siitonen, M.; Pasanen, P.; Poyhonen, M.; Myllykangas, L.; Viitanen, M.; Fukutake, T.; Cognat, E.; Joutel, A.; et al. CADASIL and CARASIL. Brain Pathol. 2014, 24, 525–544. [Google Scholar] [CrossRef] [PubMed]
- Wielaard, R.; Bornebroek, M.; Ophoff, R.A.; Winter-Warnars, H.A.; Scheltens, P.; Frants, R.R.; Ferrari, M.D.; Haan, J. A four-generation Dutch family with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL), linked to chromosome 19p13. Clin. Neurol. Neurosurg. 1995, 97, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Joutel, A.; Corpechot, C.; Ducros, A.; Vahedi, K.; Chabriat, H.; Mouton, P.; Alamowitch, S.; Domenga, V.; Cecillion, M.; Marechal, E.; et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 1996, 383, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, M.; Arnold, M.; Bersano, A.; Burlina, A.; Chabriat, H.; Debette, S.; Enzinger, C.; Federico, A.; Filla, A.; Finsterer, J.; et al. Monogenic cerebral small-vessel diseases: Diagnosis and therapy. Consensus recommendations of the European Academy of Neurology. Eur. J. Neurol. 2020, 27, 909–927. [Google Scholar] [CrossRef]
- Mizuno, T. Diagnosis, pathomechanism and treatment of CADASIL. Rinsho Shinkeigaku 2012, 52, 303–313. [Google Scholar] [CrossRef]
- Mizuno, T.; Muranishi, M.; Torugun, T.; Tango, H.; Nagakane, Y.; Kudeken, T.; Kawase, Y.; Kawabe, K.; Oshima, F.; Yaoi, T.; et al. Two Japanese CADASIL families exhibiting Notch3 mutation R75P not involving cysteine residue. Intern. Med. 2008, 47, 2067–2072. [Google Scholar]
- Di Donato, I.; Bianchi, S.; De Stefano, N.; Dichgans, M.; Dotti, M.T.; Duering, M.; Jouvent, E.; Korczyn, A.D.; Lesnik-Oberstein, S.A.; Malandrini, A.; et al. Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy (CADASIL) as a model of small vessel disease: Update on clinical, diagnostic, and management aspects. BMC Med. 2017, 15, 41. [Google Scholar] [CrossRef]
- Katan, M.; Luft, A. Global Burden of Stroke. Semin. Neurol. 2018, 38, 208–211. [Google Scholar] [CrossRef]
- Buffon, F.; Porcher, R.; Hernandez, K.; Kurtz, A.; Pointeau, S.; Vahedi, K.; Bousser, M.G.; Chabriat, H. Cognitive profile in CADASIL. J. Neurol. Neurosurg. Psychiatry 2006, 77, 175–180. [Google Scholar] [CrossRef]
- Lesnik Oberstein, S.A.; van den Boom, R.; Middelkoop, H.A.; Ferrari, M.D.; Knaap, Y.M.; van Houwelingen, H.C.; Breuning, M.H.; van Buchem, M.A.; Haan, J. Incipient CADASIL. Arch. Neurol. 2003, 60, 707–712. [Google Scholar] [CrossRef]
- Tikka, S.; Mykkanen, K.; Ruchoux, M.M.; Bergholm, R.; Junna, M.; Poyhonen, M.; Yki-Jarvinen, H.; Joutel, A.; Viitanen, M.; Baumann, M.; et al. Congruence between NOTCH3 mutations and GOM in 131 CADASIL patients. Brain 2009, 132, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Dziewulska, D.; Lewandowska, E. Pericytes as a new target for pathological processes in CADASIL. Neuropathology 2012, 32, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Joutel, A. Pathogenesis of CADASIL: Transgenic and knock-out mice to probe function and dysfunction of the mutated gene, Notch3, in the cerebrovasculature. Bioessays 2011, 33, 73–80. [Google Scholar] [CrossRef]
- Joutel, A.; Andreux, F.; Gaulis, S.; Domenga, V.; Cecillon, M.; Battail, N.; Piga, N.; Chapon, F.; Godfrain, C.; Tournier-Lasserve, E. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J. Clin. Investig. 2000, 105, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, T.; Ragno, M.; Paolinelli, F.; Castellucci, C.; Scarpelli, M.; Morroni, M. CADASIL: Ultrastructural insights into the morphology of granular osmiophilic material. Brain Behav. 2017, 7, e00624. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Craggs, L.J.; Watanabe, A.; Booth, T.; Attems, J.; Low, R.W.; Oakley, A.E.; Kalaria, R.N. Brain microvascular accumulation and distribution of the NOTCH3 ectodomain and granular osmiophilic material in CADASIL. J. Neuropathol. Exp. Neurol. 2013, 72, 416–431. [Google Scholar] [CrossRef]
- Ishiko, A.; Shimizu, A.; Nagata, E.; Takahashi, K.; Tabira, T.; Suzuki, N. Notch3 ectodomain is a major component of granular osmiophilic material (GOM) in CADASIL. Acta Neuropathol. 2006, 112, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Hack, R.; Rutten, J.; Lesnik Oberstein, S. CADASIL. In GeneReviews®; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2000. [Google Scholar]
- Rutten, J.W.; Haan, J.; Terwindt, G.M.; van Duinen, S.G.; Boon, E.M.; Lesnik Oberstein, S.A. Interpretation of NOTCH3 mutations in the diagnosis of CADASIL. Expert Rev. Mol. Diagn. 2014, 14, 593–603. [Google Scholar] [CrossRef]
- Mizuno, T.; Mizuta, I.; Watanabe-Hosomi, A.; Mukai, M.; Koizumi, T. Clinical and Genetic Aspects of CADASIL. Front. Aging Neurosci. 2020, 12, 91. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Liao, Y.C.; Lee, Y.C.; Ihara, M.; Choi, J.C. Update on the Epidemiology, Pathogenesis, and Biomarkers of Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy. J. Clin. Neurol. 2023, 19, 12–27. [Google Scholar] [CrossRef]
- Duering, M.; Karpinska, A.; Rosner, S.; Hopfner, F.; Zechmeister, M.; Peters, N.; Kremmer, E.; Haffner, C.; Giese, A.; Dichgans, M.; et al. Co-aggregate formation of CADASIL-mutant NOTCH3: A single-particle analysis. Hum. Mol. Genet. 2011, 20, 3256–3265. [Google Scholar] [CrossRef] [PubMed]
- Dichgans, M.; Ludwig, H.; Muller-Hocker, J.; Messerschmidt, A.; Gasser, T. Small in-frame deletions and missense mutations in CADASIL: 3D models predict misfolding of Notch3v EGF-like repeat domains. Eur. J. Hum. Genet. 2000, 8, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Muino, E.; Gallego-Fabrega, C.; Cullell, N.; Carrera, C.; Torres, N.; Krupinski, J.; Roquer, J.; Montaner, J.; Fernandez-Cadenas, I. Systematic Review of Cysteine-Sparing NOTCH3 Missense Mutations in Patients with Clinical Suspicion of CADASIL. Int. J. Mol. Sci. 2017, 18, 1964. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Yoon, C.W.; Seo, S.W.; Ki, C.S.; Kim, Y.B.; Kim, J.W.; Bang, O.Y.; Lee, K.H.; Kim, G.M.; Chung, C.S.; et al. Spectrum of NOTCH3 mutations in Korean patients with clinically suspicious cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Neurobiol. Aging. 2014, 35, 726.e1–726.e6. [Google Scholar] [CrossRef] [PubMed]
- Ueda, A.; Ueda, M.; Nagatoshi, A.; Hirano, T.; Ito, T.; Arai, N.; Uyama, E.; Mori, K.; Nakamura, M.; Shinriki, S.; et al. Genotypic and phenotypic spectrum of CADASIL in Japan: The experience at a referral center in Kumamoto University from 1997 to 2014. J. Neurol. 2015, 262, 1828–1836. [Google Scholar] [CrossRef] [PubMed]
- Mukai, M.; Mizuta, I.; Watanabe-Hosomi, A.; Koizumi, T.; Matsuura, J.; Hamano, A.; Tomimoto, H.; Mizuno, T. Genotype-phenotype correlations and effect of mutation location in Japanese CADASIL patients. J. Hum. Genet. 2020, 65, 637–646. [Google Scholar] [CrossRef]
- Kim, Y.; Choi, E.J.; Choi, C.G.; Kim, G.; Choi, J.H.; Yoo, H.W.; Kim, J.S. Characteristics of CADASIL in Korea: A novel cysteine-sparing Notch3 mutation. Neurology 2006, 66, 1511–1516. [Google Scholar] [CrossRef]
- Mukai, M.; Mizuta, I.; Ueda, A.; Nakashima, D.; Kushimura, Y.; Noto, Y.I.; Ohara, T.; Itoh, K.; Ando, Y.; Mizuno, T. A Japanese CADASIL patient with homozygous NOTCH3 p.Arg544Cys mutation confirmed pathologically. J. Neurol. Sci. 2018, 394, 38–40. [Google Scholar] [CrossRef]
- Choi, J.C.; Song, S.K.; Lee, J.S.; Kang, S.Y.; Kang, J.H. Diversity of stroke presentation in CADASIL: Study from patients harboring the predominant NOTCH3 mutation R544C. J. Stroke Cerebrovasc. Dis. 2013, 22, 126–131. [Google Scholar] [CrossRef]
- Liao, Y.C.; Hsiao, C.T.; Fuh, J.L.; Chern, C.M.; Lee, W.J.; Guo, Y.C.; Wang, S.J.; Lee, I.H.; Liu, Y.T.; Wang, Y.F.; et al. Characterization of CADASIL among the Han Chinese in Taiwan: Distinct Genotypic and Phenotypic Profiles. PLoS ONE 2015, 10, e0136501. [Google Scholar] [CrossRef]
- Opherk, C.; Peters, N.; Herzog, J.; Luedtke, R.; Dichgans, M. Long-term prognosis and causes of death in CADASIL: A retrospective study in 411 patients. Brain 2004, 127, 2533–2539. [Google Scholar] [CrossRef]
- Rutten, J.W.; Van Eijsden, B.J.; Duering, M.; Jouvent, E.; Opherk, C.; Pantoni, L.; Federico, A.; Dichgans, M.; Markus, H.S.; Chabriat, H.; et al. The effect of NOTCH3 pathogenic variant position on CADASIL disease severity: NOTCH3 EGFr 1–6 pathogenic variant are associated with a more severe phenotype and lower survival compared with EGFr 7-34 pathogenic variant. Genet. Med. 2019, 21, 676–682. [Google Scholar] [CrossRef]
- Hack, R.J.; Gravesteijn, G.; Cerfontaine, M.N.; Santcroos, M.A.; Gatti, L.; Kopczak, A.; Bersano, A.; Duering, M.; Rutten, J.W.; Lesnik Oberstein, S.A.J. Three-tiered EGFr domain risk stratification for individualized NOTCH3-small vessel disease prediction. Brain 2023, 146, 2913–2927. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S. Making sense out of missense mutations: Mechanistic dissection of Notch receptors through structure-function studies in Drosophila. Dev. Growth Differ. 2020, 62, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Low, W.C.; Santa, Y.; Takahashi, K.; Tabira, T.; Kalaria, R.N. CADASIL-causing mutations do not alter Notch3 receptor processing and activation. Neuroreport 2006, 17, 945–949. [Google Scholar] [CrossRef]
- Wang, T.; Baron, M.; Trump, D. An overview of Notch3 function in vascular smooth muscle cells. Prog. Biophys. Mol. Biol. 2008, 96, 499–509. [Google Scholar] [CrossRef]
- Rebay, I.; Fleming, R.J.; Fehon, R.G.; Cherbas, L.; Cherbas, P.; Artavanis-Tsakonas, S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: Implications for Notch as a multifunctional receptor. Cell 1991, 67, 687–699. [Google Scholar] [CrossRef]
- Arboleda-Velasquez, J.F.; Lopera, F.; Lopez, E.; Frosch, M.P.; Sepulveda-Falla, D.; Gutierrez, J.E.; Vargas, S.; Medina, M.; Martinez De Arrieta, C.; Lebo, R.V.; et al. C455R notch3 mutation in a Colombian CADASIL kindred with early onset of stroke. Neurology 2002, 59, 277–279. [Google Scholar] [CrossRef]
- Joutel, A.; Monet, M.; Domenga, V.; Riant, F.; Tournier-Lasserve, E. Pathogenic mutations associated with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy differently affect Jagged1 binding and Notch3 activity via the RBP/JK signaling Pathway. Am. J. Hum. Genet. 2004, 74, 338–347. [Google Scholar] [CrossRef]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Chapman, M.; Evans, K.; Azevedo, L.; Hayden, M.; Heywood, S.; Millar, D.S.; Phillips, A.D.; et al. The Human Gene Mutation Database (HGMD((R))): Optimizing its use in a clinical diagnostic or research setting. Hum. Genet. 2020, 139, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Pippucci, T.; Maresca, A.; Magini, P.; Cenacchi, G.; Donadio, V.; Palombo, F.; Papa, V.; Incensi, A.; Gasparre, G.; Valentino, M.L.; et al. Homozygous NOTCH3 null mutation and impaired NOTCH3 signaling in recessive early-onset arteriopathy and cavitating leukoencephalopathy. EMBO Mol. Med. 2015, 7, 848–858. [Google Scholar] [CrossRef] [PubMed]
- Greisenegger, E.K.; Llufriu, S.; Chamorro, A.; Cervera, A.; Jimenez-Escrig, A.; Rappersberger, K.; Marik, W.; Greisenegger, S.; Stogmann, E.; Kopp, T.; et al. A NOTCH3 homozygous nonsense mutation in familial Sneddon syndrome with pediatric stroke. J. Neurol. 2021, 268, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Stellingwerff, M.D.; Nulton, C.; Helman, G.; Roosendaal, S.D.; Benko, W.S.; Pizzino, A.; Bugiani, M.; Vanderver, A.; Simons, C.; van der Knaap, M.S. Early-Onset Vascular Leukoencephalopathy Caused by Bi-Allelic NOTCH3 Variants. Neuropediatrics 2022, 53, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.T.; Miyamoto, A.; Olsen, S.L.; D’Souza, B.; Yao, C.; Weinmaster, G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 2007, 176, 445–458. [Google Scholar] [CrossRef]
- van Tetering, G.; Vooijs, M. Proteolytic cleavage of Notch: “HIT and RUN”. Curr. Mol. Med. 2011, 11, 255–269. [Google Scholar] [CrossRef]
- Prakash, N.; Hansson, E.; Betsholtz, C.; Mitsiadis, T.; Lendahl, U. Mouse Notch 3 expression in the pre- and postnatal brain: Relationship to the stroke and dementia syndrome CADASIL. Exp. Cell Res. 2002, 278, 31–44. [Google Scholar] [CrossRef][Green Version]
- Hofmann, J.J.; Luisa Iruela-Arispe, M. Notch expression patterns in the retina: An eye on receptor–ligand distribution during angiogenesis. Gene Expr. Patterns 2007, 7, 461–470. [Google Scholar] [CrossRef]
- Hofmann, J.J.; Iruela-Arispe, M.L. Notch signaling in blood vessels: Who is talking to whom about what? Circ. Res. 2007, 100, 1556–1568. [Google Scholar] [CrossRef]
- O’Hare, M.; Arboleda-Velasquez, J.F. Notch Signaling in Vascular Endothelial and Mural Cell Communications. Cold Spring Harb. Perspect. Med. 2022, 12, a041159. [Google Scholar] [CrossRef]
- High, F.A.; Lu, M.M.; Pear, W.S.; Loomes, K.M.; Kaestner, K.H.; Epstein, J.A. Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development. Proc. Natl. Acad. Sci. USA 2008, 105, 1955–1959. [Google Scholar] [CrossRef] [PubMed]
- Breikaa, R.M.; Denman, K.; Ueyama, Y.; McCallinhart, P.E.; Khan, A.Q.; Agarwal, G.; Trask, A.J.; Garg, V.; Lilly, B. Loss of Jagged1 in mature endothelial cells causes vascular dysfunction with alterations in smooth muscle phenotypes. Vasc. Pharmacol. 2022, 145, 107087. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Krebs, L.T.; Gridley, T. Patent ductus arteriosus in mice with smooth muscle-specific Jag1 deletion. Development 2010, 137, 4191–4199. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Barbur, I.; Calderon, A.; Banerjee, S.; Proweller, A. Notch signaling regulates arterial vasoreactivity through opposing functions of Jagged1 and Dll4 in the vessel wall. Am. J. Physiol. Heart Circ. Physiol. 2018, 315, H1835–H1850. [Google Scholar] [CrossRef] [PubMed]
- Haritunians, T.; Chow, T.; De Lange, R.P.; Nichols, J.T.; Ghavimi, D.; Dorrani, N.; St Clair, D.M.; Weinmaster, G.; Schanen, C. Functional analysis of a recurrent missense mutation in Notch3 in CADASIL. J. Neurol. Neurosurg. Psychiatry 2005, 76, 1242–1248. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Hansson, E.M.; Tikka, S.; Lanner, F.; Sahlgren, C.; Farnebo, F.; Baumann, M.; Kalimo, H.; Lendahl, U. Notch signaling regulates platelet-derived growth factor receptor-beta expression in vascular smooth muscle cells. Circ. Res. 2008, 102, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Stockhausen, M.T.; Sjolund, J.; Axelson, H. Regulation of the Notch target gene Hes-1 by TGFalpha induced Ras/MAPK signaling in human neuroblastoma cells. Exp. Cell Res. 2005, 310, 218–228. [Google Scholar] [CrossRef]
- Ruchoux, M.M.; Domenga, V.; Brulin, P.; Maciazek, J.; Limol, S.; Tournier-Lasserve, E.; Joutel, A. Transgenic Mice Expressing Mutant Notch3 Develop Vascular Alterations Characteristic of Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy. Am. J. Pathol. 2003, 162, 329–342. [Google Scholar] [CrossRef]
- Joutel, A.; Monet-Lepretre, M.; Gosele, C.; Baron-Menguy, C.; Hammes, A.; Schmidt, S.; Lemaire-Carrette, B.; Domenga, V.; Schedl, A.; Lacombe, P.; et al. Cerebrovascular dysfunction and microcirculation rarefaction precede white matter lesions in a mouse genetic model of cerebral ischemic small vessel disease. J. Clin. Investig. 2010, 120, 433–445. [Google Scholar] [CrossRef]
- Rutten, J.W.; Klever, R.R.; Hegeman, I.M.; Poole, D.S.; Dauwerse, H.G.; Broos, L.A.; Breukel, C.; Aartsma-Rus, A.M.; Verbeek, J.S.; van der Weerd, L.; et al. The NOTCH3 score: A pre-clinical CADASIL biomarker in a novel human genomic NOTCH3 transgenic mouse model with early progressive vascular NOTCH3 accumulation. Acta Neuropathol. Commun. 2015, 3, 89. [Google Scholar] [CrossRef]
- Monet-Lepretre, M.; Bardot, B.; Lemaire, B.; Domenga, V.; Godin, O.; Dichgans, M.; Tournier-Lasserve, E.; Cohen-Tannoudji, M.; Chabriat, H.; Joutel, A. Distinct phenotypic and functional features of CADASIL mutations in the Notch3 ligand binding domain. Brain 2009, 132, 1601–1612. [Google Scholar] [CrossRef]
- Arboleda-Velasquez, J.F.; Manent, J.; Lee, J.H.; Tikka, S.; Ospina, C.; Vanderburg, C.R.; Frosch, M.P.; Rodriguez-Falcon, M.; Villen, J.; Gygi, S.; et al. Hypomorphic Notch 3 alleles link Notch signaling to ischemic cerebral small-vessel disease. Proc. Natl. Acad. Sci. USA 2011, 108, E128–E135. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Bacskai, B.J.; Ayata, C. Genetic animal models of cerebral vasculopathies. Prog. Mol. Biol. Transl. Sci. 2012, 105, 25–55. [Google Scholar] [CrossRef]
- Ayata, C. CADASIL: Experimental insights from animal models. Stroke 2010, 41, S129–S134. [Google Scholar] [CrossRef]
- Baron-Menguy, C.; Domenga-Denier, V.; Ghezali, L.; Faraci, F.M.; Joutel, A. Increased Notch3 Activity Mediates Pathological Changes in Structure of Cerebral Arteries. Hypertension 2017, 69, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Gravesteijn, G.; Munting, L.P.; Overzier, M.; Mulder, A.A.; Hegeman, I.; Derieppe, M.; Koster, A.J.; van Duinen, S.G.; Meijer, O.C.; Aartsma-Rus, A.; et al. Progression and Classification of Granular Osmiophilic Material (GOM) Deposits in Functionally Characterized Human NOTCH3 Transgenic Mice. Transl. Stroke Res. 2020, 11, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Dubroca, C.; Lacombe, P.; Domenga, V.; Maciazek, J.; Levy, B.; Tournier-Lasserve, E.; Joutel, A.; Henrion, D. Impaired vascular mechanotransduction in a transgenic mouse model of CADASIL arteriopathy. Stroke 2005, 36, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Wallays, G.; Nuyens, D.; Silasi-Mansat, R.; Souffreau, J.; Callaerts-Vegh, Z.; Van Nuffelen, A.; Moons, L.; D’Hooge, R.; Lupu, F.; Carmeliet, P.; et al. Notch3 Arg170Cys knock-in mice display pathologic and clinical features of the neurovascular disorder cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2881–2888. [Google Scholar] [CrossRef]
- Monet, M.; Domenga, V.; Lemaire, B.; Souilhol, C.; Langa, F.; Babinet, C.; Gridley, T.; Tournier-Lasserve, E.; Cohen-Tannoudji, M.; Joutel, A. The archetypal R90C CADASIL-NOTCH3 mutation retains NOTCH3 function in vivo. Hum. Mol. Genet. 2007, 16, 982–992. [Google Scholar] [CrossRef]
- Lundkvist, J.; Zhu, S.; Hansson, E.M.; Schweinhardt, P.; Miao, Q.; Beatus, P.; Dannaeus, K.; Karlstrom, H.; Johansson, C.B.; Viitanen, M.; et al. Mice carrying a R142C Notch 3 knock-in mutation do not develop a CADASIL-like phenotype. Genesis 2005, 41, 13–22. [Google Scholar] [CrossRef]
- Viitanen, M.; Sundstrom, E.; Baumann, M.; Poyhonen, M.; Tikka, S.; Behbahani, H. Experimental studies of mitochondrial function in CADASIL vascular smooth muscle cells. Exp. Cell Res. 2013, 319, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Panahi, M.; Yousefi Mesri, N.; Samuelsson, E.B.; Coupland, K.G.; Forsell, C.; Graff, C.; Tikka, S.; Winblad, B.; Viitanen, M.; Karlstrom, H.; et al. Differences in proliferation rate between CADASIL and control vascular smooth muscle cells are related to increased TGFbeta expression. J. Cell. Mol. Med. 2018, 22, 3016–3024. [Google Scholar] [CrossRef] [PubMed]
- Neves, K.B.; Harvey, A.P.; Moreton, F.; Montezano, A.C.; Rios, F.J.; Alves-Lopes, R.; Nguyen Dinh Cat, A.; Rocchicciolli, P.; Delles, C.; Joutel, A.; et al. ER stress and Rho kinase activation underlie the vasculopathy of CADASIL. JCI Insight 2019, 4, e131344. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kojima, K.; Taura, D.; Sone, M.; Washida, K.; Egawa, N.; Kondo, T.; Minakawa, E.N.; Tsukita, K.; Enami, T.; et al. Human iPS cell-derived mural cells as an in vitro model of hereditary cerebral small vessel disease. Mol. Brain 2020, 13, 38. [Google Scholar] [CrossRef] [PubMed]
- Kast, J.; Hanecker, P.; Beaufort, N.; Giese, A.; Joutel, A.; Dichgans, M.; Opherk, C.; Haffner, C. Sequestration of latent TGF-beta binding protein 1 into CADASIL-related Notch3-ECD deposits. Acta Neuropathol. Commun. 2014, 2, 96. [Google Scholar] [CrossRef] [PubMed]
- Zellner, A.; Scharrer, E.; Arzberger, T.; Oka, C.; Domenga-Denier, V.; Joutel, A.; Lichtenthaler, S.F.; Muller, S.A.; Dichgans, M.; Haffner, C. CADASIL brain vessels show a HTRA1 loss-of-function profile. Acta Neuropathol. 2018, 136, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Craggs, L.J.; Fenwick, R.; Oakley, A.E.; Ihara, M.; Kalaria, R.N. Immunolocalization of platelet-derived growth factor receptor-beta (PDGFR-beta) and pericytes in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL). Neuropathol. Appl. Neurobiol. 2015, 41, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Tikka, S.; Ng, Y.P.; Di Maio, G.; Mykkanen, K.; Siitonen, M.; Lepikhova, T.; Poyhonen, M.; Viitanen, M.; Virtanen, I.; Kalimo, H.; et al. CADASIL mutations and shRNA silencing of NOTCH3 affect actin organization in cultured vascular smooth muscle cells. J. Cereb. Blood Flow Metab. 2012, 32, 2171–2180. [Google Scholar] [CrossRef]
- Wollenweber, F.A.; Hanecker, P.; Bayer-Karpinska, A.; Malik, R.; Bazner, H.; Moreton, F.; Muir, K.W.; Muller, S.; Giese, A.; Opherk, C.; et al. Cysteine-sparing CADASIL mutations in NOTCH3 show proaggregatory properties in vitro. Stroke 2015, 46, 786–792. [Google Scholar] [CrossRef]
- Lee, S.J.; Zhang, X.; Wu, E.; Sukpraphrute, R.; Sukpraphrute, C.; Ye, A.; Wang, M.M. Structural changes in NOTCH3 induced by CADASIL mutations: Role of cysteine and non-cysteine alterations. J. Biol. Chem. 2023, 299, 104838. [Google Scholar] [CrossRef]
- McGurk, L.; Berson, A.; Bonini, N.M. Drosophila as an In Vivo Model for Human Neurodegenerative Disease. Genetics 2015, 201, 377–402. [Google Scholar] [CrossRef]
- Yamaguchi, M. (Ed.) Drosophila Models for Human Diseases; Springer: Singapore, 2018; Volume 1076. [Google Scholar]
- Bolus, H.; Crocker, K.; Boekhoff-Falk, G.; Chtarbanova, S. Modeling Neurodegenerative Disorders in Drosophila melanogaster. Int. J. Mol. Sci. 2020, 21, 3055. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, R.; Faber, A.; Sibon, O. Modelling in miniature: Using Drosophila melanogaster to study human neurodegeneration. Drug Discov. Today Dis. Models 2017, 25–26, 3–10. [Google Scholar] [CrossRef]
- Miller, A. The internal anatomy and histology of the imago of Drosophila melanpgaster. In Biology of Drosophila; Demerec, M., Ed.; John Wiley & Sons: New York, NY, USA, 1950. [Google Scholar]
- Wolf, M.J.; Amrein, H.; Izatt, J.A.; Choma, M.A.; Reedy, M.C.; Rockman, H.A. Drosophila as a model for the identification of genes causing adult human heart disease. Proc. Natl. Acad. Sci. USA 2006, 103, 1394–1399. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006, 7, 678–689. [Google Scholar] [CrossRef]
- de Celis, J.F.; García-Bellido, A. Roles of the Notch gene in Drosophila wing morphogenesis. Mech. Dev. 1994, 46, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Fryxell, K.J.; Soderlund, M.; Jordan, T.V. An animal model for the molecular genetics of CADASIL. (Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy). Stroke 2001, 32, 6–11. [Google Scholar] [CrossRef][Green Version]
- de Celis, J.F.; Garcia-Bellido, A. Modifications of the notch function by Abruptex mutations in Drosophila melanogaster. Genetics 1994, 136, 183–194. [Google Scholar] [CrossRef]
- Perez, L.; Milan, M.; Bray, S.; Cohen, S.M. Ligand-binding and signaling properties of the Ax[M1] form of Notch. Mech. Dev. 2005, 122, 479–486. [Google Scholar] [CrossRef]
- FlyBase. Available online: http://flybase.org (accessed on 11 June 2020).
- Yamamoto, S.; Charng, W.L.; Rana, N.A.; Kakuda, S.; Jaiswal, M.; Bayat, V.; Xiong, B.; Zhang, K.; Sandoval, H.; David, G.; et al. A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science 2012, 338, 1229–1232. [Google Scholar] [CrossRef]
- Mizuta, I.; Watanabe-Hosomi, A.; Koizumi, T.; Mukai, M.; Hamano, A.; Tomii, Y.; Kondo, M.; Nakagawa, M.; Tomimoto, H.; Hirano, T.; et al. New diagnostic criteria for cerebral autosomal dominant arteriopathy with subcortical infarcts and leukocencephalopathy in Japan. J. Neurol. Sci. 2017, 381, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Okubo, M.; Doi, H.; Fukai, R.; Fujita, A.; Mitsuhashi, S.; Hashiguchi, S.; Kishida, H.; Ueda, N.; Morihara, K.; Ogasawara, A.; et al. GGC Repeat Expansion of NOTCH2NLC in Adult Patients with Leukoencephalopathy. Ann. Neurol. 2019, 86, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Moreton, F.C.; Razvi, S.S.; Davidson, R.; Muir, K.W. Changing clinical patterns and increasing prevalence in CADASIL. Acta Neurol. Scand. 2014, 130, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Gurung, A.; Bhattacharjee, S.; Nor, A.M. Headache in a middle-aged man due to a rare mutation in the NOTCH 3 gene. Neurol. India 2019, 67, 879–881. [Google Scholar] [PubMed]
- Qi, Y.; Li, H.; Yu, L. Case report: Mild leukoencephalopathy caused by a new mutation of NOTCH3 gene. Medicine 2023, 102, e33289. [Google Scholar] [CrossRef]
- Min, J.Y.; Park, S.J.; Kang, E.J.; Hwang, S.Y.; Han, S.H. Mutation spectrum and genotype-phenotype correlations in 157 Korean CADASIL patients: A multicenter study. Neurogenetics 2022, 23, 45–58. [Google Scholar] [CrossRef]
- Cho, B.P.H.; Jolly, A.A.; Nannoni, S.; Tozer, D.; Bell, S.; Markus, H.S. Association of NOTCH3 Variant Position with Stroke Onset and Other Clinical Features among Patients with CADASIL. Neurology 2022, 99, e430–e439. [Google Scholar] [CrossRef]
- Ozaki, K.; Irioka, T.; Ishikawa, K.; Mizusawa, H. CADASIL with a novel NOTCH3 mutation (Cys478Tyr). J. Stroke Cerebrovasc. Dis. 2015, 24, e61–e62. [Google Scholar] [CrossRef]
- del Río-Espínola, A.; Fernández-Cadenas, I.; Mendióroz, M.; Gutiérrez-Agulló, M.; Fernández, M.T.; Fernández-Morales, J.; Maisterra, O.; Domingues-Montanari, S.; García-Patos, V.; Solé, E.; et al. Novel human pathological mutations. Gene symbol: NOTCH3. Disease: CADASIL. Hum. Genet. 2010, 127, 474. [Google Scholar]
- Nurmahdi, H.; Hasegawa, M.; Mujizah, E.Y.; Sasamura, T.; Inaki, M.; Yamamoto, S.; Yamakawa, T.; Matsuno, K. Notch Missense Mutations in Drosophila Reveal Functions of Specific EGF-like Repeats in Notch Folding, Trafficking, and Signaling. Biomolecules 2022, 12, 1752. [Google Scholar] [CrossRef]
- Winkler, E.A.; Bell, R.D.; Zlokovic, B.V. Central nervous system pericytes in health and disease. Nat. Neurosci. 2011, 14, 1398–1405. [Google Scholar] [CrossRef] [PubMed]
- Krebs, L.T.; Xue, Y.; Norton, C.R.; Sundberg, J.P.; Beatus, P.; Lendahl, U.; Joutel, A.; Gridley, T. Characterization of Notch3-deficient mice: Normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis 2003, 37, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, T.; Takahashi, K.; Takimoto, H.; Tomizuka, K.; Hayasaka, M.; Tabira, T.; Hanaoka, K. Functional redundancy of the Notch gene family during mouse embryogenesis: Analysis of Notch gene expression in Notch3-deficient mice. Biochem. Biophys. Res. Commun. 2005, 331, 1154–1162. [Google Scholar] [CrossRef]
- Manini, A.; Pantoni, L. CADASIL from Bench to Bedside: Disease Models and Novel Therapeutic Approaches. Mol. Neurobiol. 2021, 58, 2558–2573. [Google Scholar] [CrossRef] [PubMed]
- Domenga, V.; Fardoux, P.; Lacombe, P.; Monet, M.; Maciazek, J.; Krebs, L.T.; Klonjkowski, B.; Berrou, E.; Mericskay, M.; Li, Z.; et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004, 18, 2730–2735. [Google Scholar] [CrossRef] [PubMed]
- Belin de Chantemele, E.J.; Retailleau, K.; Pinaud, F.; Vessieres, E.; Bocquet, A.; Guihot, A.L.; Lemaire, B.; Domenga, V.; Baufreton, C.; Loufrani, L.; et al. Notch3 is a major regulator of vascular tone in cerebral and tail resistance arteries. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 2216–2224. [Google Scholar] [CrossRef] [PubMed]
- Arboleda-Velasquez, J.F.; Zhou, Z.; Shin, H.K.; Louvi, A.; Kim, H.H.; Savitz, S.I.; Liao, J.K.; Salomone, S.; Ayata, C.; Moskowitz, M.A.; et al. Linking Notch signaling to ischemic stroke. Proc. Natl. Acad. Sci. USA 2008, 105, 4856–4861. [Google Scholar] [CrossRef]
- Fouillade, C.; Baron-Menguy, C.; Domenga-Denier, V.; Thibault, C.; Takamiya, K.; Huganir, R.; Joutel, A. Transcriptome analysis for Notch3 target genes identifies Grip2 as a novel regulator of myogenic response in the cerebrovasculature. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 76–86. [Google Scholar] [CrossRef]
- Henshall, T.L.; Keller, A.; He, L.; Johansson, B.R.; Wallgard, E.; Raschperger, E.; Mae, M.A.; Jin, S.; Betsholtz, C.; Lendahl, U. Notch3 is necessary for blood vessel integrity in the central nervous system. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 409–420. [Google Scholar] [CrossRef]
- Peters, N.; Opherk, C.; Zacherle, S.; Capell, A.; Gempel, P.; Dichgans, M. CADASIL-associated Notch3 mutations have differential effects both on ligand binding and ligand-induced Notch3 receptor signaling through RBP-Jk. Exp. Cell Res. 2004, 299, 454–464. [Google Scholar] [CrossRef]
- Kofler, N.M.; Cuervo, H.; Uh, M.K.; Murtomaki, A.; Kitajewski, J. Combined deficiency of Notch1 and Notch3 causes pericyte dysfunction, models CADASIL, and results in arteriovenous malformations. Sci. Rep. 2015, 5, 16449. [Google Scholar] [CrossRef] [PubMed]
- Watanabe-Hosomi, A.; Watanabe, Y.; Tanaka, M.; Nakagawa, M.; Mizuno, T. Transendocytosis is impaired in CADASIL-mutant NOTCH3. Exp. Neurol. 2012, 233, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Hiura, S.; Mashiko, T.; Matsumoto, T.; Itoh, M. Lunatic fringe promotes the aggregation of CADASIL NOTCH3 mutant proteins. Biochem. Biophys. Res. Commun. 2021, 557, 302–308. [Google Scholar] [CrossRef] [PubMed]
- del Alamo, D.; Rouault, H.; Schweisguth, F. Mechanism and significance of cis-inhibition in Notch signalling. Curr. Biol. 2011, 21, R40–R47. [Google Scholar] [CrossRef] [PubMed]
- Nandagopal, N.; Santat, L.A.; Elowitz, M.B. Cis-activation in the Notch signaling pathway. Elife 2019, 8, e37880. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.L.; Quail, E.; Cruickshank, M.N.; Ulgiati, D. To Be, or Notch to Be: Mediating Cell Fate from Embryogenesis to Lymphopoiesis. Biomolecules 2021, 11, 849. [Google Scholar] [CrossRef] [PubMed]
- Arboleda-Velasquez, J.F.; Rampal, R.; Fung, E.; Darland, D.C.; Liu, M.; Martinez, M.C.; Donahue, C.P.; Navarro-Gonzalez, M.F.; Libby, P.; D’Amore, P.A.; et al. CADASIL mutations impair Notch3 glycosylation by Fringe. Hum. Mol. Genet. 2005, 14, 1631–1639. [Google Scholar] [CrossRef]
- Monet-Lepretre, M.; Haddad, I.; Baron-Menguy, C.; Fouillot-Panchal, M.; Riani, M.; Domenga-Denier, V.; Dussaule, C.; Cognat, E.; Vinh, J.; Joutel, A. Abnormal recruitment of extracellular matrix proteins by excess Notch3ECD: A new pathomechanism in CADASIL. Brain 2013, 136, 1830–1845. [Google Scholar] [CrossRef]
- Capone, C.; Cognat, E.; Ghezali, L.; Baron-Menguy, C.; Aubin, D.; Mesnard, L.; Stohr, H.; Domenga-Denier, V.; Nelson, M.T.; Joutel, A. Reducing Timp3 or vitronectin ameliorates disease manifestations in CADASIL mice. Ann. Neurol. 2016, 79, 387–403. [Google Scholar] [CrossRef]
- Nozaki, H.; Nishizawa, M.; Onodera, O. Features of cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy. Stroke 2014, 45, 3447–3453. [Google Scholar] [CrossRef]
- ten Dijke, P.; Arthur, H.M. Extracellular control of TGFbeta signalling in vascular development and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Shiga, A.; Nozaki, H.; Yokoseki, A.; Nihonmatsu, M.; Kawata, H.; Kato, T.; Koyama, A.; Arima, K.; Ikeda, M.; Katada, S.; et al. Cerebral small-vessel disease protein HTRA1 controls the amount of TGF-beta1 via cleavage of proTGF-beta1. Hum. Mol. Genet. 2011, 20, 1800–1810. [Google Scholar] [CrossRef] [PubMed]
- Uemura, M.; Nozaki, H.; Kato, T.; Koyama, A.; Sakai, N.; Ando, S.; Kanazawa, M.; Hishikawa, N.; Nishimoto, Y.; Polavarapu, K.; et al. HTRA1-Related Cerebral Small Vessel Disease: A Review of the Literature. Front. Neurol. 2020, 11, 545. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.; Prinz, A.; Tetzlaff, F.; Weis, E.M.; Moll, I.; Rodriguez-Vita, J.; Oka, C.; Korff, T.; Fischer, A. Loss of the serine protease HTRA1 impairs smooth muscle cells maturation. Sci. Rep. 2019, 9, 18224. [Google Scholar] [CrossRef]
- Klose, R.; Adam, M.G.; Weis, E.M.; Moll, I.; Wustehube-Lausch, J.; Tetzlaff, F.; Oka, C.; Ehrmann, M.; Fischer, A. Inactivation of the serine protease HTRA1 inhibits tumor growth by deregulating angiogenesis. Oncogene 2018, 37, 4260–4272. [Google Scholar] [CrossRef]

| Human materials: autopsied brain tissue or vessels, skin biopsy specimens | |
| Histopathology, immunohistochemistry | |
| Biochemistry, proteomics | |
| Gene expression, transcriptome | |
| Animal models: transgenic mice | |
| Genetic approach to clarify pathophysiology | |
| Temporal analysis of disease process | |
| Histopathology, immunohistochemistry | |
| Biochemistry, proteomics | |
| Gene expression, transcriptome | |
| Cell cultures: VSMCs 1, iPS cell-derived mural cells, cell lines (HEK293, NIH 3T3, etc.) | |
| Analysis of Notch signaling activity | |
| Recreation of pathology of CADASIL | |
| Vaiability and proliferation | |
| In vitro | |
| Aggregation assay of N3ECDpeptides | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mizuta, I.; Nakao-Azuma, Y.; Yoshida, H.; Yamaguchi, M.; Mizuno, T. Progress to Clarify How NOTCH3 Mutations Lead to CADASIL, a Hereditary Cerebral Small Vessel Disease. Biomolecules 2024, 14, 127. https://doi.org/10.3390/biom14010127
Mizuta I, Nakao-Azuma Y, Yoshida H, Yamaguchi M, Mizuno T. Progress to Clarify How NOTCH3 Mutations Lead to CADASIL, a Hereditary Cerebral Small Vessel Disease. Biomolecules. 2024; 14(1):127. https://doi.org/10.3390/biom14010127
Chicago/Turabian StyleMizuta, Ikuko, Yumiko Nakao-Azuma, Hideki Yoshida, Masamitsu Yamaguchi, and Toshiki Mizuno. 2024. "Progress to Clarify How NOTCH3 Mutations Lead to CADASIL, a Hereditary Cerebral Small Vessel Disease" Biomolecules 14, no. 1: 127. https://doi.org/10.3390/biom14010127
APA StyleMizuta, I., Nakao-Azuma, Y., Yoshida, H., Yamaguchi, M., & Mizuno, T. (2024). Progress to Clarify How NOTCH3 Mutations Lead to CADASIL, a Hereditary Cerebral Small Vessel Disease. Biomolecules, 14(1), 127. https://doi.org/10.3390/biom14010127






